Abstract
Telomeres are structures composed of repetitive DNA and proteins that protect the chromosomal ends in eukaryotic cells from fusion or degradation, thus contributing to genomic stability. Although telomere length varies between species, in all organisms studied telomere length appears to be controlled by a dynamic equilibrium between elongating mechanisms (mainly addition of repeats by the enzyme telomerase) and nucleases that shorten the telomeric sequences. Two previous studies have analyzed a collection of yeast deletion strains (deleted for nonessential genes) and found over 270 genes that affect telomere length (Telomere Length Maintenance or TLM genes). Here we complete the list of TLM by analyzing a collection of strains carrying hypomorphic alleles of most essential genes (DAmP collection). We identify 87 essential genes that affect telomere length in yeast. These genes interact with the nonessential TLM genes in a significant manner, and provide new insights on the mechanisms involved in telomere length maintenance. The newly identified genes span a variety of cellular processes, including protein degradation, pre-mRNA splicing and DNA replication.
INTRODUCTION
Telomeres are the specialized DNA–protein structures at the ends of eukaryotic chromosomes. Telomeric DNA is composed of highly repetitive sequences [such as (TTAGGG)n in humans and (C1–3A/TG1–3)n in yeast (1)]. The telomeric structure performs a capping function by which it defines the end of the chromosome as a native edge, rather than the aberrant structure of a DNA double-strand break (DSB). This telomeric capping allows cells with linear chromosomes to function properly, while maintaining efficient mechanisms for DSB repair. When telomeric capping is abolished, activation of a DSB repair mechanism may cause telomere–telomere fusions resulting in chromosomal aberrations. Thereby, by performing their capping function telomeres carry out an essential role in maintaining chromosomal stability and integrity (2,3).
A linear DNA molecule cannot be completely replicated by the cellular DNA polymerase due to the ‘end replication problem’ (4). All DNA polymerases need a primer to initiate DNA replication. This primer is later removed from the DNA by the DNA polymerases arriving from the upstream direction. At telomeres no upstream fragment is present, resulting in a gap of unreplicated DNA (5). As a consequence, in most somatic cells, chromosomes lose information from their ends during each DNA replication cycle. This eventually leads to senescence and cell death (6). In germ cells, and some proliferating somatic cells, telomeres are maintained by telomerase, a cellular reverse transcriptase that copies a short template sequence within its own RNA into the telomeric sequence (6,7). When telomerase is inactivated, telomeres shorten from one division to the other leading eventually to cell death (8–10). Therefore telomerase performs an essential function, allowing proper replication of the chromosomes.
Telomerase elongates the telomere, adding one repeat at a time to the telomeric single-stranded substrate. This results in elongation of the single stranded DNA (ssDNA) at the end of the chromosome (7). Following telomerase action, the DNA polymerases can replicate the complementary strand, creating a double-stranded DNA (dsDNA) molecule (11,12). Addition of new sequences by telomerase is typically tightly regulated, resulting in the telomeres of many organisms being kept within particular size ranges. For example, while repetitive sequences in yeast telomeres span an average of 350 bp, human telomeres exceed several kilobases (kb) (1).
In all organisms studied, telomere length seems to be controlled by a dynamic equilibrium between elongating mechanisms (such as telomerase) and nucleases that shorten the telomeric sequences (13). Nevertheless, telomere length in all organisms tested tends to be remarkably stable, indicating that telomere length is under strict genetic control. Two recent genome wide studies in yeast have identified genes that affect telomere length (TLM, or telomere length maintenance genes). A collection of 4750 yeast strains, each of them deleted for a particular gene, was screened by Southern blot analysis for genes that, when mutated, caused telomere lengthening or shortening (14,15). Together, these studies identified 272 TLM genes (representing ∼5% of all nonessential genes); these include not only genes affecting expected categories, such as DNA and RNA metabolism, chromatin remodeling, etc., but also many genes affecting seemingly unrelated processes [e.g. vesicular transport (16)]. The large number of genes that affect telomere length homeostasis underscores the importance of this process. Many cellular functions seem to contribute to telomere length maintenance, and an initial unified model that integrates these activities has been recently proposed (17).
While the previous studies have focused on investigating the role of nonessential genes, two new methodologies have been developed lately to create a systematic collection of mutants defective in all essential genes in the yeast. The first collection consists of temperature-sensitive mutants (18): haploid cells can be grown at the permissive temperature, and only show their defective phenotype at the restrictive temperature. Here, we take advantage of a second collection, the Decreased Abundance by mRNA Perturbation library (DAmP) (19,20), which consists of hypomorphic alleles of most of the yeast essential genes. Screening this collection of 739 mutants, we have identified 86 genes that affect telomere length: 37 of them show long telomeres, whereas 49 show short telomeres (strikingly, a larger percentage of genes involved in the TLM system than that found in the nonessential gene set). Our results complete the list of telomere-affecting genes and add new insights on the mechanisms involved in telomere length maintenance.
MATERIALS AND METHODS
Yeast strain collection
The hypomorphic library developed by Weismann and co-workers (19,20) was used. Mutations were confirmed by PCR, and the relationship between the tlm phenotype and the KanMX insertion was confirmed by tetrad analysis of 20 individual mutants, after crosses to the isogenic BY4741/2 yeast strains. BY4741: (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0); BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). In all 20 cases, analysis of two tetrads showed a direct correspondence between the tlm phenotype and resistance to G418.
Telomere length measurement
Telomeric Southern blots were carried out as in (14). PCR fragments containing telomeric sequences and a genomic region that hybridizes to two size marker bands (2044 and 779 bp) were used as probes. The telomere length was measured with the GelQuant software using the size marker bands as reference. Telomere length was ≈1250 bp in wt cells [composed of the sub-telomeric region (≈900 bp) and the telomere repeats (≈350 bp)]. At least three independent colonies of each strain were analyzed by Southern blot to ensure reproducibility. Whenever discrepancies were observed, the strains were re-created and three additional independent colonies were tested.
PCR
The identity of a sample of 20 mutants was determined by PCR analysis, as described in (14).
Comparison with previous screens
We assembled protein–protein interaction (PPI) data from public databases and recent publications to construct a comprehensive PPI network of yeast (21–23). The PPIs were assigned confidence scores based on the experimental evidence available for each interaction using a logistic regression model adapted from (24).
To test the set of TLM genes identified by the DAmP screen, we examined their interactions with a set S of nonessential TLM genes discovered on previous screens using the hypergeometric P-value:
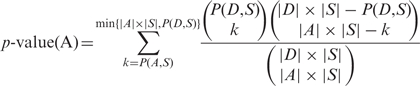 |
where D denotes the entire set of genes in the DAmP collection, and A denotes the genes on the DAmP collection that exhibited defects in telomere length. For two sets of proteins X1 and X2, we denote the number of interactions between them by P(X1, X2). We use two definitions for P: (i) considering the entire set of interactions regardless of their confidence, and (ii) considering only interactions with confidence level above 0.5. Notably, the set of genes that were discovered on previous screens and have PPI information available contains a total of 250 genes [167 from Askree et al. (14), and 147 from Gatbonton et al. (15)].
RESULTS AND DISCUSSION
Telomere length maintenance is a complex trait, affected by a large number of genes. Two previous systematic genome-wide studies in the yeast Saccharomyces cerevisiae have uncovered 272 nonessential genes that, when mutated, cause either telomere lengthening or shortening (14,15). This large collection of mutants, encompassing ∼5% of the yeast genome, is composed of haploid cells in which individual genes were deleted, one at a time. In order to complete the screening of all yeast genes, including those that are inviable when deleted, we took advantage of a collection of hypomorphic mutants, encompassing 842 out of the 1033 essential yeast genes (20). Briefly, in this strain collection a KanMX cassette (conferring resistance to the drug G418) was inserted within the 3′ UTR of all essential genes. In some cases, this insertion resulted in lethality (only 842 haploid strains could be created); however, a large proportion of the viable strains thus created exhibit scorable phenotypes, suggesting that the KanMX insertion destabilized the expression of the disrupted gene (20).
Due to technical reasons (many strains failed to grow or to yield appropriate DNA preparations) we analyzed a total of 739 DAmP mutants for telomere length. At least three individual colonies of each strain in the DAmP collection were grown in rich medium, and their DNA was subjected to a telomeric Southern blot analysis. Each Southern blot also included triplicates of the wild-type isogenic strain. Marker PCR fragments, containing a genomic region that hybridizes to two bands (2044- and 779-bp long), were included in the labeled probes along with the telomeric probe. Telomere length in all strains was extremely reproducible, with a standard variation of <10% among repeats of the same strain. After an initial screen, all strains suspected as having a significant telomere length phenotype were re-tested, starting from fresh duplicate cultures. PCR analysis and tetrad analysis were used to confirm the identity of each individual mutant, and the co-segregation between telomere length and resistance to G418 (conferred by the KanMX allele at the DAmP allele). Figure 1 shows several examples of various tetrads thus analyzed. In total, our screen uncovered 86 essential genes whose DAmP allele leads to altered telomere length. Tables 1 and 2 show the list of genes that, when mutated, exhibit short and long phenotypes, respectively, divided by category. Figure 2A shows representative Southern blot analysis of various tlm DAmP alleles. Figure 2B shows all the tlm DAmP alleles sorted by relative length compared to the wild type. As in our previous screen (14) a slight bias is seen towards mutants exhibiting short telomeres (49 versus 37). It is not clear whether this represents a biological outcome (i.e. it is easier to cause telomere shortening by mutation than to cause telomere lengthening) or a technical bias (e.g. the probe used may underestimate long telomeres). For a discussion on these possibilities, see ref. 1). The frequency of tlm mutants in the DAmP collection (86/739 = 11.6%) is much higher than the one obtained among the non-essential mutants (250/4770 = 5.2%). This reflects the centrality of the telomeric pathways: TLM genes are enriched for essential functions. Notably, the frequency calculated for the essential collection is likely to be an under-estimate, as we have no efficient way of evaluating how well the DAmP methodology fares in creating hypomorphic alleles (19). See below for a discussion of this point.
Figure 1.
Tetrad analysis of DAmP tlm mutants. To confirm that the tlm phenotype observed was caused by the KanMX insertion, tetrad analysis was carried out after crossing to a wild-type yeast strain. In 20/20 cases, resistance to G418 (provided by the KanMX cassette) co-segregated with telomere length. Tetrad results of five representative mutants are shown. The white horizontal line represents the average wild-type telomere length.
Table 1.
List of S. cerevisiae essential-DAmP genes that exhibit ‘short’ telomere phenotype
| Gene | Function |
|---|---|
| Telomere-related | |
| TBF1 | Binds to TTAGGG repeats within subtelomeric anti-silencing regions. |
| Proteasome/SCF | |
| PRE2 | Beta 5 subunit of the 20S proteasome. |
| PRE5 | Alpha 6 subunit of the 20S proteasome. |
| RPN5 | Non-ATPase regulatory subunit of the 26S proteasome lid. |
| RPN6 | Non-ATPase regulatory subunit of the 26S proteasome lid. |
| RPN7 | Non-ATPase regulatory subunit of the 26S proteasome. |
| RPN12 | Subunit of the 19S regulatory particle of the 26S proteasome lid. |
| RPT3 | One of six ATPases of the 19S regulatory particle of the 26S proteasome. |
| RPT5 | One of six ATPases of the 19S regulatory particle of the 26S proteasome. |
| HRT1 | RING finger containing subunit of Skp1-Cullin-F-box ubiquitin protein ligases (SCF). |
| CDC34 | Ubiquitin-conjugating enzyme (E2) and catalytic subunit of SCF complex. |
| Splicing | |
| PRP4 | Splicing factor, component of the U4/U6-U5 snRNP complex. |
| PRP22 | RNA helicase of the DEAH-box family/ATP-dependent RNA helicase. |
| PRP31 | Splicing factor, component of the U4/U6-U5 snRNP complex. |
| PRP38 | Splicing factor, component of the U4/U6-U5 snRNP complex. |
| PRP43 | RNA helicase of the DEAH-box family |
| NTR2 | Involved in spliceosome disassembly, forms a complex with Prp43. |
| Arp2/3 complex | |
| ARP2 | Essential component of the Arp2/3 complex. |
| ARP3 | Essential component of the Arp2/3 complex. |
| ARC35 | Essential component of the Arp2/3 complex. |
| ARC15 | Essential component of the Arp2/3 complex. |
| Secretion and Golgi traffic | |
| TRS23 | One of 10 subunits of the TRAPP complex (ER to Golgi traffic). |
| COG3 | Component of the conserved oligomeric Golgi complex (vesicle to Golgi traffic). |
| YPP1 | Cargo-transport protein involved in endocytosis. |
| RNA processing and Transcription | |
| FCP1 | Essential for dephosphorylation of RNA polymerase II large subunit (Rpo21p). |
| RNA14 | Cleavage and polyadenylation factor I (CF I) component. |
| RNA15 | Cleavage and polyadenylation factor I (CF I) component |
| RPB7 | RNA polymerase II subunit B16. |
| RGR1 | Subunit of the RNA polymerase II mediator complex |
| YDR396W | overlaps NCB2:Subunit of the NC2 transcription regulator complex (with Bur6p). |
| rRNA processing/nucleolus | |
| FAL1 | Nucleolar protein required for maturation of 18S rRNA. |
| POP7 | Subunit of both RNase MRP (pre-rRNA processing), and RNase P (tRNA processing). |
| TSR1 | Protein required for processing of 20S pre-rRNA. |
| NET1 | Core subunit of the RENT complex (nucleolar). |
| MDN1 | Midasin, remodeling and export of 60S ribosomal subunits. |
| NOC4 | Nucleolar protein, forms a complex with Nop14p. |
| MTR3 | 3′5′ exoribonuclease, exosome subunit; nucleolar protein involved in export of mRNA. |
| tRNA processing | |
| SEN54 | Subunit of the tRNA splicing endonuclease. |
| tRNA synthetases | |
| KRS1 | Lysyl-tRNA synthetase. |
| ALA1 | Cytoplasmic alanyl-tRNA synthetase. |
| APC/cyclosome | |
| APC4 | Subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C). |
| DNA Replication | |
| ORC5 | Subunit of the origin recognition complex. |
| PSF3 | Subunit of the GINS complex, a putative helicase with a role in DNA replication. |
| Kinetochore | |
| TID3 | Component of the evolutionarily conserved kinetochore-associated Ndc80 complex. |
| DAD2 | Essential subunit of the Dam1 complex (aka DASH complex). |
| Miscellaneous | |
| SAM35 | Mitochondrial outer membrane protein, member of the SAM complex. |
| SAH1 | S-adenosyl-l-homocysteine hydrolase. |
| TUB2 | Beta-tubulin; associates with alpha-tubulin (Tub1p and Tub3p) to form tubulin dimmer. |
| ILV5 | Acetohydroxyacid reductoisomerase, mitochondrial protein. |
| NAM9 | Mitochondrial ribosomal component of the small subunit |
Descriptions are from the Saccharomyces genome database (SGD: http://www.yeastgenome.org/).
Table 2.
List of S. cerevisiae essential-DAmP genes that exhibit ‘long’ telomere phenotype
| Gene | Function |
|---|---|
| Telomere-related | |
| STN1 | Telomere end-binding and capping protein. |
| CDC13 | Telomere single-stranded DNA-binding protein. |
| DNA replication | |
| YDL163W | Overlaps CDC9: DNA ligase. |
| YDL165w | Overlaps CDC9: DNA ligase. |
| YOR218C | Overlaps RFC1: clamp loader, loads PCNA to initiate replication. |
| PRI1 | Subunit of DNA primase. |
| MCM3 | Replicative helicase. |
| MCM6 | Replicative helicase. |
| APC/cyclosome | |
| CDC16 | Subunit of the anaphase-promoting complex/cyclosome. |
| Transcription | |
| RPB5 | RNA polymerase subunit ABC27. |
| SPN1 | Protein that interacts with Spt6p and co-purifies with Spt5p and RNA polymerase II |
| NHP2 | Nuclear protein related to mammalian high-mobility group (HMG) proteins. |
| SUMO/Ubiquitin | |
| SMT3 | SUMO (small ubiquitin-like modifier). |
| RPS31 | Ubiquitin. |
| Chromatin remodeling | |
| RSC4 | Component of the RSC chromatin-remodeling complex. |
| RSC8 | Component of the RSC chromatin-remodeling complex. |
| Secretion and Golgi traffic | |
| SEC20 | Membrane glycoprotein, v-SNARE (Golgi to ER). |
| SEC63 | Essential subunit of Sec63 complex (import to the ER). |
| TRS20 | Component of the TRAPP complex (cis-Golgi). |
| YJL032W | Overlaps BET4: ER–Golgi transport. |
| MCD4 | ER protein involved in GPI anchor synthesis. |
| GPI8 | Subunit of the ER GPI transamidase complex. |
| CMD1 | Calmodulin; Ca++-binding protein. Required for vacuolar fusion. |
| Splicing | |
| SAD1 | Conserved zinc-finger domain protein involved in pre-mRNA splicing, required for assembly of U4 snRNA into the U4/U6 particle. |
| AAR2 | Component of the U5 snRNP, required for splicing of U3 precursors. |
| rRNA/tRNA processing | |
| YLR317W | Overlaps TAD3: Subunit of tRNA-specific adenosine-34 deaminase. |
| LSM8 | RNA degradation, tRNA/rRNA modification and splicing. |
| CBF5 | Pseudouridine synthase, catalytic subunit of box H/ACA snoRNPs. |
| YLR198C | Overlaps NOP56: Nucleolar component of the box C/D snoRNP complexes that direct 2′-O-methylation of pre-rRNA during its maturation. |
| Miscellaneous | |
| YLR230W | Overlaps CDC42: small Rho-like GTPase essential for cell polarity. |
| NCP1 | NADP-cytochrome P450 reductase, ergosterol synthesis. |
| BBP1 | Protein required for the spindle pole body (SPB) duplication |
| CDC19 | Pyruvate kinase, last step in glycolysis. |
| HEM4 | Heme biosynthetic pathway. |
| YLR339C | Overlaps RPP0: conserved ribosomal protein P0. |
| GCD1 | Gamma subunit of the translation initiation factor eIF2B, the guanine-nucleotide exchange factor for eIF2. |
Descriptions are from the Saccharomyces genome database (SGD: http://www.yeastgenome.org/).
Figure 2.
(A) Representative Southern blots of DAmP strains. The white horizontal line represents the average wild-type telomere length. The white arrow points to pre2-DAmP mutants, which shows short telomeres, whereas black arrow points to rpb5-DAmP mutant, which exhibit long telomeres. The DAmP alleles of NOG2, USO1 and CCT4 do not show tlm phenotypes. (B) tlm DAmP alleles sorted by relative length. The telomere length of all the tlm DAmP alleles was measured and compared to the wild-type length (here defined as 1.0). The wild-type telomere length is about 350 bp.
The genes identified in our screen may be divided into various categories, according to their known functions (Tables 1 and 2). Below, we discuss the main categories individually:
Telomere maintenance and DNA replication
Our screen uncovered only a small number of the essential genes previously known to affect telomere length. This is most likely due to the fact that even hypomorphic mutations in these genes lead to continuous telomere shortening and to cell death. For example, the genes encoding the catalytic subunit of telomerase, Est2, and its two associated proteins Est1 and Est3 (25), were absent from our collection (due to their low cell viability), as did other essential genes encoding DNA polymerases. We did, however, find genes encoding some of the known components of telomeres: Cdc13, a telomeric ssDNA binding protein involved in telomere capping and protection (26) and its partner Stn1, which, together with Cdc13, helps in the recruiting of telomerase (27) (Table 2).
We also identified several genes involved in DNA replication: mutations in the PRI1 primase gene, or in the MCM3/MCM6 genes, encoding subunits of the replicative helicase, caused elongated telomeres. Similar results were observed for mutations in the ORFs YDL163w and YDL165w, which flank or overlap the gene CDC9, encoding the only essential DNA ligase of yeast. Thus, it appears that, as previously suggested (28), defects in some aspects of DNA replication may cause telomere elongation. This is consistent with the fact that the nonessential genes RAD27, ELG1, PIF1 and POL32, also involved in various aspects of DNA replication, cause elongated telomeres upon deletion (14).
Proteasome
Mutations in eight genes encoding proteasomal subunits (PRE2, PRE5, RPT3, RPT5, RPN5, RPN6, RPN7 and RPN12) lead to short telomeres [Table 1, Figures 1 and 2; results regarding RPN6, RPN12 and RPT3 have been reported in another publication (Yosef et al., in press)]. These mutations affect different substructures (e.g. base and lid), as well as functional portions (e.g. ATPases and non-ATPases) of the proteasome. In addition, mutations in components of the SCF machinery, required to add ubiquitin moieties to proteins in order to send them to degradation, cause the same phenotype (e.g. HRT1 and CDC34). Therefore, our results suggest that proteasomal function (and not individual subunits) plays an important role in telomere elongation. Similar conclusions were reached in a more elaborate network-based analysis of protein–protein interaction data (Yosef et al., in press). Lack of degradation of a protein with a role in telomere shortening (such as a nuclease or one of its regulators) may lead to higher abundance levels, causing increased telomere shortening. Alternatively, proteasome inactivation may lead to accumulation of a repressor of telomerase activity [such as Rif1, Rif2 (29) or Pif1 (30)], thus changing the equilibrium towards a shorter length.
Pre-mRNA splicing
Six genes with roles in pre-mRNA splicing lead to short telomeres when mutated: PRP4, PRP22, PRP32, PRP31, PRP38, PRP43 and NTR2 (Table 1).
Pre-mRNA splicing is a complex reaction involving dozens of proteins, and consists of two consecutive catalytic reactions. Interestingly, all the genes identified seem to play a role in the structural changes in the spliceosome associated with the second catalytical splicing reaction (Prp4, Prp31 and Prp38), or with the disassembly of the spliceosome after completing the reaction (Prp22, Prp43 and Ntr2). Notably, Prp22 and Prp43 (along with its auxiliary subunit Ntr2) are both DEAH-box RNA-dependent ATPase/ATP-dependent RNA helicases that mediate ATP-dependent mRNA release from the spliceosome, unwinding RNA duplexes (31,32). Thus, it appears that spliceosomal disassembly, but not assembly, or other stages of pre-mRNA splicing, affects telomere length. The significance of this observation is still unclear. Although pre-mRNA splicing is a global cellular mechanism, in yeast only about 250 genes carry introns, and specific effects of individual splicing components on the splicing of particular genes have been observed (33,34).
Transcription and RNA processing
Telomere length regulation requires a delicate balance between more then 270 nonessential genes (14,15) and close to 90 essential ones, as this paper suggests. Diminishing the RNA transcription or processing of Telomerase core components or their regulators may have an influence on telomere length. Several of these regulators may be present in limiting levels in the nucleus, and thus may be strongly affected by even slight variations in transcript levels. For example, previous work has shown that a subgroup of the tlm genes influences telomere length by affecting the abundance of TLC1, the telomerase RNA component (35). Two of the short tlm mutants identified in this study involve an RNA polymerase II subunit (Rpb7) or a regulator of RNA polymerase II activity (Fcp1) (36,37) (Table 1). Interestingly, mutations in a different RNA polymerase II subunit, Rpb5, lead to elongated telomeres (Table 2). In addition, we found that mutations in the RNA14 and RNA15 genes lead to a significant shortening of telomeres. RNA14 and RNA15 encode components of the cleavage and polyadenylation factor I (CF I) complex, required for mRNA end processing (38). It was previously shown that RNA15 is required for poly(A) addition to the telomerase RNA (39). This gene plays also a role in the processing of TERRA (telomeric repeat-containing RNA), telomeric-specific transcripts of unknown function created by RNA polymerase II (40). Further work is needed to determine the precise mechanism by which mRNA processing affects telomere length.
The Arp2/3 complex
This complex is a highly conserved actin nucleation center required for the motility and integrity of actin patches (41). It is involved in endocytosis and in membrane growth and polarity processes. The Arp2/3 complex is a seven-protein complex containing two actin-related proteins, Arp2p and Arp3p, and five nonactin-related proteins, Arc15p, Arc18p, Arc19p, Arc35p and Arc40p. We have identified four of the genes encoding these proteins: ARP2, ARP3, ARC15 and ARC35 as short tlm DAmP mutants (Table 1). ARC18 and ARC19 were not present in our collection, and the DAmP allele of ARC40 exhibited normal telomere length. Either this particular subunit is dispensable for the telomere-related function of the Arp2/3 complex, or, more likely, the DAmP allele of this gene is not tight enough to produce a visible phenotype. As in the case of the proteasome, it is likely that the whole complex is involved in a process that helps regulate telomere length. Among the many roles attributed to the Arp2/3 complex is the fusion of vacuoles during endocytosis, and internal cellular traffic (42). This may link the complex to the next category:
Vesicular traffic
One of the surprises that came out from the two genome-wide screens for nonessential tlm genes was that the largest category observed consisted of proteins involved in vesicular traffic (14,15). As expected, this category was also prominent among the essential genes, and included many proteins of the Golgi-ER traffic system. Surprisingly, however, whereas in the nonessential collection the tlm mutants in this category exhibited mainly short telomeres, among the DAmP collection most mutations (although not all) led to elongated telomeres (Tables 1 and 2). In this context, it is interesting to note that two members of the TRAPP complex, Trs20 and Trs23, involved in ER-Golgi traffic (43) exhibit opposite effects on telomere length (Tables 1 and 2). Several mechanisms have been proposed to explain how vesicular traffic may affect telomere length (16). For example, component(s) of the telomerase machinery may require vesicular traffic (in order to be modified, for example) to allow proper activity regulation (44). Alternatively, a yet-unknown structural relation may exist between vesicular traffic and telomere maintenance (e.g. through nuclear morphology). More experiments are required in order to solve this enigma.
Nucleolar proteins and rRNA processing components
A number of genes encoding proteins involved in nucleolar functions show a short telomere phenotype when mutated (Table 1). These include POP7, a member of the RNase MRP (45), and TSR1 and FAL1, both required for the processing of rRNA. Additional nucleolar proteins include NET1, a subunit of the RENT complex, NOC4, important for the maturation and nuclear export of 40S ribosomal subunits, and MTR3, encoding a nucleolar nuclease that also plays a role in export of ribosomal subunits. Finally, YDR396w is a dubious ORF, but neighbors UTP5, a subunit of the SSU processome complex, involved in production of 18S rRNA and assembly of small ribosomal subunits (46). Interestingly, not all the proteins involved in rRNA processing, when mutated, give raise to short telomeres. Mutations in the CBF5 and NOP56 genes, both subunits of snoRNPs complexes with roles in modifying nucleolar RNA, produce elongated telomeres. CBF5 encodes the catalytic subunit of a pseudouridine synthase enzyme, which modifies box H/ACA-containing small nucleolar ribonucleoproteins. NOP56 is a component of the box C/D-containing snoRNP complexes and directs 2′-O-methylation of pre-rRNA during its maturation (46).
Telomerase functions as a ribonucleoprotein, containing an RNA moiety (7). In higher eukaryotes, telomerase RNA includes sequences of the box H/ACA snoRNA family and associates with box H/ACA core snoRNP proteins (47). In yeast, telomerase RNA is not a box H/ACA snoRNA, but rather associates with Sm proteins (48). This suggests that either an Sm-binding site or a box H/ACA motif can supply the functions required for this RNA, such as stability, localization and association with various factors. Our identification of mutants in genes encoding the snoRNA modifying enzymes Cbf5 and Nop56 as having a ‘long Tlm’ phenotype supports this hypothesis. Further support is provided by a recent study, which found that the non-essential snoRNA methyltransferase Tgs1 plays a role in the hypermethylation of yeast telomerase RNA (49).
Ubiquitin and SUMO
Interestingly, our screen uncovered both the gene encoding ubiquitin (RPS31) and the one encoding SUMO (SMT3). Mutations in these two essential protein modifiers lead to elongated telomeres. A similar phenotype was seen when nonessential components of the SUMO machinery were analyzed for telomere length [e.g. NFI1/SIZ2 and SIZ1 (50)].
However, mutations in known nonessential members of the ubiquitin-modifying machinery [e.g. RAD6, BRE1 (15)], as well as mutations in the proteasome/SCF machinery required for ubiquitin-mediated protein degradation (Table 1), result in telomere shortening. This suggests that the RPS31 mutation may play a regulatory role on ubiquitin metabolism. Alternatively, it is possible that the phenotype observed is due to a role of the ribosomal protein that is co-translationally synthesized with the ubiquitin moiety (51). Most nonessential proteins of the small subunit of the ribosome cause telomere elongation when deleted (14,15).
RSC complex
Two essential components of the RSC complex, involved in chromatin remodeling, Rsc4 and Rsc8, were uncovered as long telomere tlm mutants by our screen. RSC is an abundant complex composed of 10 essential and seven nonessential proteins, with roles in transcription regulation, sister chromatid cohesion and genomic stability (52). Deletion of genes encoding other nonessential components of the RSC complex (HTL1, NPL6, LDB7) led to the same long tlm phenotype (14,15). Interestingly, a recent publication (53) that analyzed gene expression in the absence of RSC subunits found increased levels of transcription of genes involved in ER-Golgi traffic. Moreover, RSC mutants show resistance to brefeldin A, a compound that promotes Golgi disassembly (53). Thus, the RSC complex seems to act antagonistically to the vesicular traffic pathway, consistent with the opposing tlm phenotypes of most participants in these pathways.
Additional genes
Not all genes could be grouped by their known function. As with previous studies, we find a number of mutants with clear telomeric phenotype, whose alleged function is apparently unrelated to telomere biology. Further work is required to find out whether these represent proteins playing several independent roles, or whether connections may be found between telomere length maintenance and seemingly unrelated cellular pathways. For example, two members of the APC/cyclosome, the ubiquitin binding machinery in charge of controlling cell cycle progression, have been found: APC4 (Table 1) and CDC16 (Table 2). Interestingly, these two members of the same complex also exhibit opposite effects on telomere length.
Epistasis analysis
In order to start organizing the large number of TLM genes uncovered into functional pathways, we began an epistasis analysis of the DAmP mutants identified in the screen. We crossed several of the mutants to known telomere-affecting mutants, and created double mutants (at this stage of the same type, long or short). The telomere length of the double mutants was compared to that of each single mutant parent (obtained from the same cross), after an extensive number of generations, to ensure that telomeres can reach their final length. In principle, three types of interactions are expected (54,55): (i) epistasis: the phenotype of the double mutant is as severe as that of the most defective parent, the most likely explanation is that the two affected genes act in the same pathway of telomere length maintenance; (ii) additivity: the double mutant appears as the sum of the phenotypes of each parent – this result suggests that the two mutants affect independent pathways; and finally, (iii) synergism: the double mutant is markedly more defective than what would be expected from an additive relation. This category is harder to interpret, but it is usually assumed that the mutants affect competing pathways.
Figure 3 shows representative results of this analysis. Mutation of PRI1 causes a severe elongation of the telomeres, comparable to that seen in rif1, rif2 or pif1 mutants. The pri1 rif1 double mutant is no more defective than the single mutants, indicating epistatic relations, whereas pri1 is additive to rif2 and synergistic to pif1. Similar results were seen for other mutants affecting DNA replication, suggesting that Rif1 may play a role in the coordination between chromosomal replication and telomerase addition. Interestingly, mutations in SUMO (smt3) show a similar pattern of interactions (Figure 3), indicating a possible role of SUMO modification in this pathway.
Figure 3.
Representative Southern blots of telomere epistasis assay. The white horizontal line represents the average wild-type telomere length.
Figure 3 also shows an epistasis analysis of pre2, a component of the proteasome. It is possible to see that mutations in either TEL1, the yeast ATM ortholog (56), or in Xrs2 [a subunit of the MRX complex, (57)] are epistatic to proteasome defects, suggesting that the proteasome affects telomere length through the Tel1-MRX pathway (58). An attractive possibility is that a regulator in this pathway is degraded by the proteasome in a timely manner, to ensure proper telomere length maintenance. Alternatively, the effect may be indirect, as proteasome function may be needed for transcription of a regulator, or for signal transduction within the cell. Interestingly tbf1, a DNA-binding protein that plays roles in transcription and in silencing, but is also capable of telomere binding (59), affects telomere by the same pathway, as seen in Figure 3.Additional epistasis analysis of this kind should help us elucidate all the pathways affecting telomere length and their topology.
Comparison with previous screens
Examining the DAmP collection provides us with additional information on the regulation of telomere length, focusing on a set of genes, which were excluded on previous screens (14,15). Since the collection of genes analyzed in this paper and that of the combined previous studies are complementary and do not intersect, we used their relatedness in a protein-protein interaction network as a measuring rod to test whether the genes identified in the different studies are related. We discovered that proteins from the DAmP collection that exhibited a defect in telomere length show significantly more interactions with nonessential proteins discovered on the previous screens, compared with proteins from the DAmP collection that had no effect. Considering the entire available PPI data (see ‘Materials and Methods’ section), a protein from the library that exhibited a phenotype has on average 83% (P < 1e-5) more interactions with proteins discovered by Askree et al. (with an average of 0.9 interactions per protein with phenotype versus 0.49 interactions per protein without phenotype) and 87% (P = 1.1e-5) more interactions with proteins discovered by Gatbonton et al. (with an average of 0.76 interactions per protein with phenotype versus 0.4 interactions per protein without phenotype). Considering only high confidence interactions (P ≥ 0.5) we observe even more substantial differences: 89% more interactions (P < 1e-5) for the data set of Askree et al., and 97% more for Gatbonton et al. (P < 1e-5, see ‘Materials and Methods’ section).
Revisiting the DamP methodology
Clearly, the DAmP collection is a useful tool for screening phenotypes in a genome-wide fashion. For example, the ts collection (18) requires one to grow each individual mutant at its highest permissive temperature, whereas DAmP mutants can be grown in batches at a single temperature. However, it is also clear that it has some disadvantages: first, as the KanMX insertion is likely to affect differently each individual gene, the collection is somewhat heterogeneous, with some genes being more affected than others. The exact level of expression of each hypomorphic allele in the collection is not known. For the same reason, only 739 haploid strains [out of the 1033 essential genes (20)] could be tested. This is likely due to the fact that in many cases, the 3′ UTR insertion destabilizes the transcript at a level that is below that required for survival. For example, although we obtained the two ORFs that flank the CDC9 gene, encoding the yeast only replicative DNA ligase, the DAmP allele of that gene is apparently lethal.
Despite these disadvantages, the DAmP collection has allowed us to increase our knowledge on the cellular processes that affect telomere length regulation. A total of more than 350 genes, and possibly many more, affect the regulation of telomere length. Remarkably, many of these genes are additionally involved in a diverse array of other cellular functions (as evident from the large variety of processes involved), testifying to the extensively distributed nature of cellular processing in yeast. Somehow the cells are able to integrate this enormous amount of information into a final output that keeps length homeostasis constant. The mechanisms by which cells achieve such integration are the subject of intensive studies, and promise to unravel basic aspects of genetic expression and its regulation.
FUNDING
A Converging-Technologies grant from the Israel Science Foundation (to M.K., R.S. and E.R.); and grants from The US-Israel Bi-national Fund (BSF) and the Association for International Cancer Research (to M.K.). Funding for open access charge: Israel Science Foundation.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Zakian VA. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 2.Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 3.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 4.Watson JD. Origin of concatemeric T7 DNA. Nat. New. Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 5.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 6.Harrington L. Does the reservoir for self-renewal stem from the ends? Oncogene. 2004;23:7283–7289. doi: 10.1038/sj.onc.1207948. [DOI] [PubMed] [Google Scholar]
- 7.Harrington L. Biochemical aspects of telomerase function. Cancer Lett. 2003;194:139–154. doi: 10.1016/s0304-3835(02)00701-2. [DOI] [PubMed] [Google Scholar]
- 8.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 9.Niida H, Matsumoto T, Satoh H, Shiwa M, Tokutake Y, Furuichi Y, Shinkai Y. Severe growth defect in mouse cells lacking the telomerase RNA component. Nat. Genet. 1998;19:203–206. doi: 10.1038/580. [DOI] [PubMed] [Google Scholar]
- 10.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 12.Greider CW. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 13.Lydall D. Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways. J. Cell Sci. 2003;116:4057–4065. doi: 10.1242/jcs.00765. [DOI] [PubMed] [Google Scholar]
- 14.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rog O, Smolikov S, Krauskopf A, Kupiec M. The yeast VPS genes affect telomere length regulation. Curr. Genet. 2005;47:18–28. doi: 10.1007/s00294-004-0548-y. [DOI] [PubMed] [Google Scholar]
- 17.Shachar R, Ungar L, Kupiec M, Ruppin E, Sharan R. A systems-level approach to mapping the telomere length maintenance gene circuitry. Mol. Syst. Biol. 2008;4:172. doi: 10.1038/msb.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Aroya S, Coombes C, Kwok T, O'Donnell KA, Boeke JD, Hieter P. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell. 2008;30:248–258. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 20.Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 22.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 23.Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharan R, Ideker T, Kelley B, Shamir R, Karp RM. Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data. J. Comput. Biol. 2005;12:835–846. doi: 10.1089/cmb.2005.12.835. [DOI] [PubMed] [Google Scholar]
- 25.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 28.Smolikov S, Mazor Y, Krauskopf A. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc. Natl Acad. Sci. USA. 2004;101:1656–1661. doi: 10.1073/pnas.0307796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wotton D, Shore D. Novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 30.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 32.Schneider S, Hotz HR, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 33.Dahan O, Kupiec M. The Saccharomyces cerevisiae gene CDC40/PRP17 controls cell cycle progression through splicing of the ANC1 gene. Nucleic Acids Res. 2004;32:2529–2540. doi: 10.1093/nar/gkh574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jasiak AJ, Hartmann H, Karakasili E, Kalocsay M, Flatley A, Kremmer E, Strasser K, Martin DE, Soding J, Cramer P. Genome-associated RNA polymerase II includes the dissociable Rpb4/7 subcomplex. J. Biol. Chem. 2008;283:26423–26427. doi: 10.1074/jbc.M803237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh MH, Ye P, Zhang M, Hausmann S, Shuman S, Gnatt AL, Fu J. Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc. Natl Acad. Sci. USA. 2005;102:17314–17319. doi: 10.1073/pnas.0507987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross S, Moore CL. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapon C, Cech TR, Zaug AJ. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 40.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 41.Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr. Opin. Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 42.Jin M, Cai M. A novel function of Arp2p in mediating Prk1p-specific regulation of actin and endocytosis in yeast. Mol. Biol. Cell. 2008;19:297–307. doi: 10.1091/mbc.E07-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrowman J, Sacher M, Ferro-Novick S. TRAPP stably associates with the Golgi and is required for vesicle docking. EMBO J. 2000;19:862–869. doi: 10.1093/emboj/19.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teter SA, Klionsky DJ. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Semin. Cell Dev. Biol. 2000;11:173–179. doi: 10.1006/scdb.2000.0163. [DOI] [PubMed] [Google Scholar]
- 45.Houser-Scott F, Ziehler WA, Engelke DR. Saccharomyces cerevisiae nuclear ribonuclease P: structure and function. Methods Enzymol. 2001;342:101–117. doi: 10.1016/s0076-6879(01)42539-0. [DOI] [PubMed] [Google Scholar]
- 46.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 49.Franke J, Gehlen J, Ehrenhofer-Murray AE. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J. Cell Sci. 2008;121:3553–3560. doi: 10.1242/jcs.033308. [DOI] [PubMed] [Google Scholar]
- 50.Chen XL, Silver HR, Xiong L, Belichenko I, Adegite C, Johnson ES. Topoisomerase I-dependent viability loss in saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics. 2007;177:17–30. doi: 10.1534/genetics.107.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Vugt JJ, Ranes M, Campsteijn C, Logie C. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim. Biophys. Acta. 2007;1769:153–171. doi: 10.1016/j.bbaexp.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 53.Monahan BJ, Villen J, Marguerat S, Bahler J, Gygi SP, Winston F. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 2008;15:873–880. doi: 10.1038/nsmb.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 55.St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 57.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirano Y, Fukunaga K, Sugimoto K. Rif1 and Rif2 Inhibit Localization of Tel1 to DNA Ends. Mol. Cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]





