Abstract
Many genome-wide assays involve the generation of a subset (or representation) of the genome following restriction enzyme digestion. The use of enzymes sensitive to cytosine methylation allows high-throughput analysis of this epigenetic regulatory process. We show that the use of a dual-adapter approach allows us to generate genomic representations that includes fragments of <200 bp in size, previously not possible when using the standard approach of using a single adapter. By expanding the representation to smaller fragments using HpaII or MspI, we increase the representation by these isoschizomers to more than 1.32 million loci in the human genome, representing 98.5% of CpG islands and 91.1% of refSeq promoters. This advance allows the development of a new, high-resolution version of our HpaII-tiny fragment Enrichment by Ligation-mediated PCR (HELP) assay to study cytosine methylation. We also show that the MspI representation generates information about copy-number variation, that the assay can be used on as little as 10 ng of DNA and that massively parallel sequencing can be used as an alternative to microarrays to read the output of the assay, making this a powerful discovery platform for studies of genomic and epigenomic abnormalities.
INTRODUCTION
At present, there are several types of assays to test cytosine methylation on a genome-wide scale. Some depend on the relative enrichment of methylated or hypomethylated DNA, mapping back these fractions to the genome by microarray or, more recently, massively parallel sequencing techniques. Enrichment of a certain pattern of methylation can be performed using proteins binding to methylated DNA [antibodies (1,2) or methyl-binding domain proteins (3)], methylation-dependent restriction enzymes [mcrBC (4)] or methylation-sensitive restriction enzymes. The development of massively parallel sequencing promises to allow nucleotide-resolution, quantitative analysis of cytosine methylation throughout mammalian genomes, as demonstrated in Arabidopsis (5,6) although as we have discussed there remain significant practical problems to performing such assays at present (7). These problems include significant cost, due to the need to deeply resequence an entire genome but further complicated by the creation of noncomplementary forward and reverse strands, doubling the effective size of the reference genome, and the difficulty of mapping short, degenerate bisulphite-converted sequences to this reference genome, especially at unmethylated loci where the cytosines are all converted by bisulphite. The HpaII tiny fragment-Enrichment by Ligation-mediated PCR (HELP) assay (8) is a means of screening DNA methylation status at a large proportion of the ∼2.3 million CCGG sites throughout the genome (9). The HELP assay is based on the creation of a representation of the subset of unmethylated HpaII fragments in the genome by ligation-mediated PCR (LM-PCR), with the methylation-insensitive MspI representation serving as a control for experimental variability and genomic copy number heterogeneity. HELP is unusual among genome-wide techniques testing cytosine methylation for enriching hypomethylated loci specifically, whereas most other approaches specifically enrich methylated DNA, which constitutes the majority of the genome (10,11) and is disproportionately comprised of repetitive sequences (12). The HELP assay positively identifies hypomethylated loci by means of the HpaII representation, whereas techniques that enrich methylated DNA infer the presence of a hypomethylated locus by means of the absence of signal, for which there can be other, technical reasons.
Ligation-mediated (LM-) PCR is the fundamental technique used in the HELP assay. In our previous report, we used LM-PCR to amplify the fragments in a size range from 200 to 2000 bp, with little amplification of DNA of <200 bp (8). We show here that the use of a dual-adapter strategy combined with optimization of PCR conditions for high (C + G) content regions allows us to increase the HELP assay fragment representation to include smaller fragments. By expanding the size range of the genomic representation, we increase the number of loci that can be studied in the genome by ∼50%, especially in the most CG-dense regions of the genome, markedly increasing the resolution of this assay and demonstrating how other assays based on genomic representations could be improved substantially. We used the MspI representation to discover loci with altered copy number, and that we can use as little as 10 ng of starting material for the assay. In addition, we present data from the use of massively parallel sequencing of the HpaII and MspI representations, showing that these new technologies offer advantages over prior microarray approaches. The new high-resolution HELP assay is therefore a significant improvement on the prior protocol that is supported by an opensource informatic pipeline for data analysis (13).
METHODS
Cell preparation and DNA purification
GM06990 B lymphoblastoid cells were cultured in RPMI 1640 with 15% FBS, 1% glutamine and antibiotics. The cells were harvested and washed twice by PBS and stored in –70°C. The H1 human embryonic stem cells (hESCs, NIH code WA01 from Wicell Research Institute, Madison WI) were cultured on matrigel (BD Biosciences, San Diego). Amplified hESC pluripotency was assessed by flow cytometry with SSEA4, CD24 and Oct4 markers. Erythroid progenitors derived from hESCs were produced as previously described (14). The hESCs were differentiated by co-culture on immortalized human fetal hepatocytes (FH-B-hTERT) for 2 weeks. CD34-positive cells were then magnetically sorted and placed in a serum-free liquid culture for two weeks. The erythroid characteristics of the differentiated cells were assessed by flow cytometry with CD71 and CD235a (glycophorin A) markers. Primary acute lymphoblastic leukemia cells were obtained from Our Lady of Mercy Cancer Center, approved by Institutional Review Board of the Albert Einstein College of Medicine. One million fresh CD34 + bone marrow progenitors were purchased from Allcells (Emerville, CA). DNA was extracted from these cells and checked for purity and amount by spectrometry (Nanodrop, Wilmington, DE).
LM-PCR with a two adapter set
The primers used were JHpaII12 (5′-CGGCTGTTCATG-3′), JHpaII24 (5′-CGACGTCGACTATCCATGAACAGC-3′), NHpaII12 (5′-CGGCTTCCCTCG-3′) and NHpaII24 (5′-GCAACTGTGCTATCCGAGGGAAGC-3′). Our previous protocol (8) was modified with the additional adapter set (NHpaII12/NHpaII24). Five micrograms of genomic DNA were digested by either HpaII or MspI and purified by phenol/chloroform extraction and ethanol precipitation. Each 1 μg of digested genomic DNA was ligated by T4 DNA ligase using four oligos (JHpaII12 and NHpaII12, JHpaII24 and NHpaII24, each 40 mM) in a final volume of 33 μl. The ligated genomic fragments were diluted and used as the template for the LM-PCR. The optimization of PCR conditions for LM-PCR was performed using our prior PCR conditions (8) with/without betaine or dimethyl sulfoxide. PCR products were assessed by gel electrophoresis and purified using a PCR purification kit (Qiagen). The concentrations of PCR products were measured by spectrometry. The intensities of DNA from gel images were processed using ImageJ and Photoshop (Adobe).
To improve the PCR conditions, we tested higher concentrations of magnesium, and explored the use of betaine as a means of improving the amplification of (G + C)-rich templates (15). Dimethyl sulfoxide was also tested with and without betaine, but this failed to enhance short fragment amplification (data not shown).
Array design, hybridization and data analysis
We custom-designed two microarrays (Roche-NimbleGen, Inc.) for these experiments. One interrogated the 1% of the genome studied by the ENCODE consortium in their pilot phase using oligonucleotides for each of the 18 529 HpaII/MspI fragments of 50–2000 bp in these ENCODE regions (16). The second tested the >1.32 million loci throughout the human genome that have HpaII sites 50–2000 bp apart and have unique sequences between them that allow oligonucleotide design. The HELP samples were labeled for microarray analysis as described previously (17) using Cy3- or Cy5-conjugated random primers. The HpaII and MspI representations were co-hybridized to the microarray and scanned to quantify the 532- and 635-nm fluorescence at each oligonucleotide on the microarray. The quantile-normalized HpaII/MspI log2 ratios were calculated as previously described (13). We added a category of intermediate methylation to our analysis by defining all HpaII signals falling below the 99% centile of the distribution of random probes when scanning the HpaII channel, lower-intensity signals in which we could assign methylation status with less confidence. Those signals exceeding the 99% centile were assigned as hypomethylated, while methylated loci were defined as previously described (13).
Correlation of genomic annotations with HELP cytosine methylation data
The start and end positions of MspI fragments were computationally calculated from the hg17 assembly of the human genome sequence at the UCSC Genome Browser (genome.ucsc.edu). Using these DNA sequences, their base compositional characteristics [(C + G) mononucleotide percent, CG dinucleotide frequency per 1 kb] were computationally calculated. The genomic positions of the MspI fragments were used to quantify the overlap with annotations such as CpG islands (cpgislandExt table), CG clusters (18), retroelements (rmsk table) and the 2-kb region flanking refSeq transcription start sites.
Use of limited amounts of starting material
Ten nanograms of genomic DNA (representing ∼2000 mammalian diploid cell equivalents) from human melanoma samples were digested overnight by HpaII or MspI (New England Biolabs, Inc., Ipswich, MA) in separate 20-μl reactions. The digested DNA fragments were purified by phenol/chloroform/isoamyl alcohol (25:24:1 by volume, pH 5.2, Thermo Fisher Scientific Inc., Waltham, MA) and ethanol precipitated in the presence of glycogen. After washing with 70% ethanol, the DNA pellet was resuspended in 5 μl of Tris–HCl (pH 8.0). Overnight adapter ligation was set up in a 10-μl reaction, scaling the proportional amount of adapter down by a factor of 30 compared with 1 μg HELP, with T4 DNA ligase (Invitrogen Corp., Carlsbad, California). Adapter-ligated DNA fragments were amplified by two rounds of PCR [67 mM Tris–HCl, 4 mM MgCl2, 16 mM (NH4)2SO4, 0.35% β-mercaptoethanol, 0.5 μg/μl BSA, Taq DNA polymerase] using 72°C for 10 min, 15 cycles of 95°C for 30 s and 72°C for 3 min, with a final extension of 72°C for 10 min. For the first round of PCR, 2.5 μl (MspI) or 5 μl (HpaII) of the adapter ligation mixture was used in a 100-μl reaction, of which 1 μl of PCR product was used for a second 100-μl reaction.
Analysis of DNA copy-number variation using the MspI representation
The analysis of HELP data used the published analytical pipeline mentioned earlier (13). The MspI and HpaII signal intensities were summarized for each HpaII fragment as the average of its component probe-level signal intensities. Background noise thresholds were calculated using random probes as 2.5 median absolute deviations (MAD) above the median of MspI and HpaII signal intensities. Those fragments with HpaII above background but MspI below background were removed from consideration. A subset of fragments (those between 250 and 1000 bp) was then selected and centered to align the distribution ranges, with further removal of datapoints that exceeded 2 MADs from the center. Log ratios of MspI signal intensities were calculated for the test sample as a function of a normal cell type. The data were then divided by chromosome and analyzed for copy-number variation using DNAcopy (19), available through BioConductor for the R Statistical Package. Copy-number array (CNA) objects were smoothed and circular binary segmentation applied, with default parameters and change points <1 standard deviation removed.
Massively parallel sequencing of HELP representations
To eliminate adapters from LM-PCR products, we digested the purified LM-PCR product with MspI and concentrated the digested product using isopropanol precipitation. The digested product was loaded onto a 5% acrylamide gel, which was run to resolve DNA fragments in the <1000-bp size range. The DNA in the gel was visualized using visible spectrum transillumination following staining with SYBR Gold (Invitrogen), allowing us to excise the fragments of ∼100–600 bp in size, in doing so eliminating the LM-PCR adapters. The DNA was extracted from the gel pieces using Miniprep columns (Qiagen), following which the eluted DNA was precipitated by isopropanol and purified with the PCR purification kit (Qiagen). Illumina libraries were prepared using their standard protocol, and single end sequencing was performed with a single lane per library, 47 bp per read. The first 32 bp were aligned to the reference genome using ELAND (Illumina), and these alignments were analysed using a local copy of the UCSC genome browser database and a downloaded copy of the human genome sequence (hg18 release). All HpaII sites in the genome were identified using a custom set of PERL and BASH shell scripts and loaded into a MySQL database. The HpaII site locations were then compared to the genomic loci corresponding to the Illumina reads using custom PERL scripts and modules based on the DBI PERL MySQL interface. The number of reads per locus (excluding those not starting with CGG) was also calculated using MySQL queries after the locations of the Illumina reads were loaded into the MySQL database. The HpaII representation was normalised in terms of that produced by MspI using the sequencing equivalent of the microarray intensity-dependent ratio (20) of the components as follows: H × 2[(H–M)/(H+M)] + M × (2[(H–M)/(H+M)] – 1), where H = number of HpaII reads and M = number of MspI reads. The results of these HELP and HELP-seq assays were compared against bisulphite MassArray (Sequenom) data from the KCNQ1 promoter region using primers and conditions described in Supplementary Table 1.
RESULTS
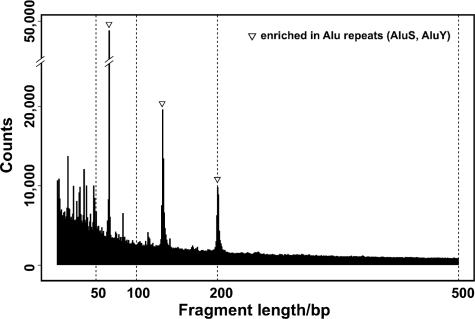
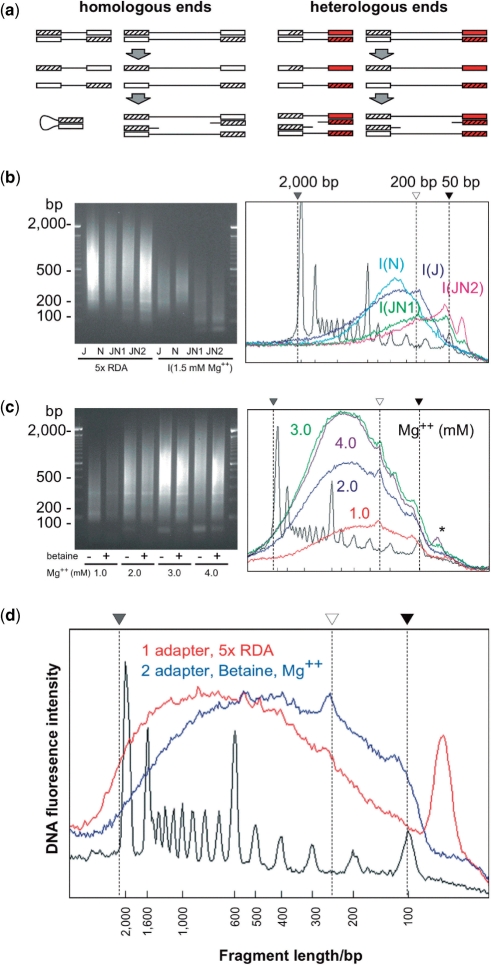
Our original assay failed to represent HpaII/MspI fragments <200 bp despite their relative abundance in the genome, as shown by our in silico digestion of the human genome at HpaII/MspI digestion sites (Figure 1). Of all of the HpaII/MspI fragments in human DNA, 44.5% of fragments in the 200–2000-bp range are represented in the original HELP assay, while the 22.5% in the 50–200-bp range were not generated using our original protocol (Table 1). We considered two possible reasons for this. First, the regions with HpaII/MspI CCGG sites at higher frequency are enriched in (C + G) mononucleotide content, making them relatively difficult to amplify by conventional PCR techniques. The (C + G) content of the HpaII/MspI fragments of 50–2000-bp averages 60.0%, in contrast to the 47.7% for 200–2000-bp fragments. Second, the presence of a ligated adapter sequence may cause intramolecular self-annealing for shorter fragments preferentially, preventing PCR amplification of these hairpin (panhandle) structures (21). To test the second hypothesis, we added a second adaptor for the ligation step. This is expected to provide the ligated product with heterologous ends at 50% frequency (Figure 2a), preventing intramolecular annealing in this subpopulation of molecules. We find that the addition of the second adapter shifts the size range of PCR products compared with the use of a single adapter (Figure 2b). We conclude that the use of a single adapter for genomic representations fails to amplify fragments <200 bp because of a strong panhandle effect, a problem alleviated by the use of heterologous adapters.
Figure 1.
In silico analysis of human HpaII/MspI fragments by length. The numbers of fragments computationally generated from the reference human genome sequence were plotted by length, demonstrating higher frequencies of shorter fragments. The three peaks observed (at 69, 135 and 204 bp) are due to the presence of Alu SINEs (mainly AluS and AluY), peaks that can also be observed in the ethidium bromide staining of the MspI reference representation in Figure 2 (b, c).
Table 1.
Annotation of human HpaII/MspI fragments in high-resolution HELP
| Original representation | Added representation | Combined representation | ||
|---|---|---|---|---|
| Total | Size range/bp | 200–2000 | 50–199 | 50–2000 |
| Number | 1 016 980 | 514 387 | 1 531 367 | |
| CpG islands | Number overlapped | 23 881 | 23 716 | 27 035 |
| Proportion represented | 87.00% | 86.40% | 98.50% | |
| Mean number of fragments/locus | 1.6 | 3.9 | 5.5 | |
| CG clusters | Number overlapped | 42 226 | 39 206 | 43 740 |
| Proportion overlapped | 95.10% | 88.30% | 98.60% | |
| Mean number of fragments/locus | 2.3 | 3.9 | 6.3 | |
| refSeq | Number overlapped | 16 848 | 14 923 | 17 223 |
| promoters* | Proportion overlapped | 89.10% | 78.90% | 91.10% |
| Mean number of fragments/locus | 2.6 | 4.2 | 6.8 |
*2 kb flanking transcription start site.
Figure 2.
Optimization of the HELP protocol for high-resolution representations. In (a) we show data representing the results of the two-adapter approach. Homologous ends using a single adapter can produce a hairpin structure in short fragments, an event that should be eliminated in 50% of fragments when using dual adapters to create heterologous ends. In (b) we demonstrate that the use of a single adapter (J, N) fails to create the smaller fragments seen when used together (JN, replicate experiments annotated as JN1, JN2). We also show that the type of buffer used, Mg2+ concentration and betaine combine to improve representation and yield (b, c). The original (red) and improved (blue) conditions for creating representations are compared in (d) as densitometric plots of gel electrophoreses, demonstrating the shift toward representation of PCR products of ≤100 bp. As the adapters contribute ∼50 bp to the size of the product, the representation of HpaII/MspI fragments in the genome includes those as small as 50 bp in this new protocol.
These improvements in size range representation came at the expense of yield, which may be due to the increased (C + G) mononucleotide content of the template. By increasing the concentration of magnesium and introducing betaine to the PCR reaction (Figure 2c), we could generate amounts of PCR product comparable to the original protocol. The amplification products shown in Figure 2d include the adapter sequences (∼50 bp), indicating that we are also amplifying products ≤50 bp in size while preserving our ability to amplify up to 2000 bp.
Our next concern was whether the shorter fragments generated could be labeled and hybridized to a microarray as successfully as larger fragments. We custom-designed a NimbleGen DNA methylation 385K microarray (Roche-NimbleGen Inc.) using oligonucleotides for each of the 18 529 HpaII/MspI fragments of 50–2000 bp in the ENCODE regions of the genome (16). We prepared HpaII and MspI representations from the GM06990 cell line using the optimized conditions described above (Figure 2d), labeling the representations by random priming and co-hybridizing the labeled representations to the microarray. This customized ENCODE array also included a set of 10 100 oligonucleotides representing random sequences. To assess the microarray data, we defined the background fluorescence level using a threshold of 2.5 median absolute deviations (2.5 MAD) above the median intensity of these control oligonucleotides. We have found this value to define consistently the distinctive population of loci in HpaII representations that does not amplify due to local methylation, and applied the same parameter to define signal intensities in the MspI channel, indicating loci that have failed to amplify adequately (Supplementary Figure 1). In the current experiment, we observed a failure rate of 5–9% in total, but only 2–4% were in the size range of 50–199 bp (Supplementary Figure 1). When the HpaII representation was studied, we observed a bimodal distribution, demonstrating that the two-adapter HELP method discriminates between methylated and unmethylated loci (Supplementary Figure 1).
In addition, the scatterplots and Pearson's correlation coefficients for log2 MspI intensities and HpaII/MspI intensity ratios illustrated that both technical and biological replicates show high correlation in MspI intensities (98–99%) and HpaII/MspI ratios (95–99%) (Supplementary Figure 1). We conclude that the labeling and hybridization of the additional representation of shorter fragments is as efficient and reproducible as the original method.
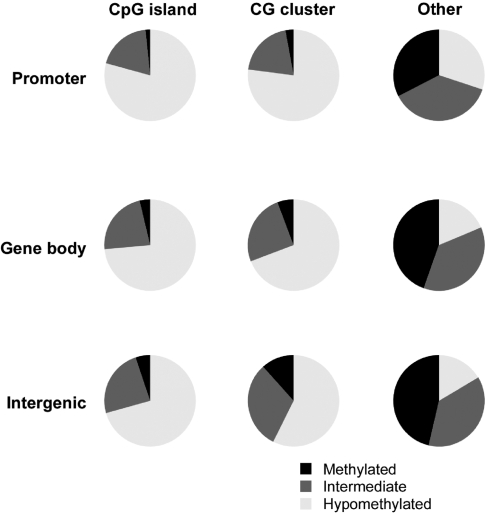
We then used a genome-wide microarray consisting of >1.32 million loci for the human genome to test the cytosine methylation pattern in human embryonic stem cells. We show the results of the methylation patterns observed in Figure 3. We see that CG-dense loci [CpG islands or CG clusters (18)] and promoters are less methylated than the CG-depleted sequences within gene bodies and intergenic sequences. These are results concordant with prior expectations (22) and demonstrate the assay to be able to report expected patterns of methylation in a variety of sequence contexts.
Figure 3.
The distribution of cytosine methylation by genomic compartment. The distribution of cytosine methylation using the new high-resolution genome-wide HELP microarray is categorized as hypomethylated (white), methylated (black) and intermediate (grey) based on thresholds categorising the bimodal distribution of HpaII/MspI ratios. The results show relative hypomethylation of CpG islands or CG clusters (18) whether at promoters, within gene bodies or intergenically compared with other sequences. This is consistent with expectations for the distribution of methylation and indicates that the assay is capable of defining methylation states in a range of genomic contexts.
Adaptation of protocol to limited starting amounts of DNA: nanoHELP
A limitation in many epigenomic assays is their requirement for large amounts of sample. As the HELP assay involves a PCR amplification step, we tested whether we could use substantially less starting amounts of DNA and still get reproducible results. We performed the assay using 1 µg of DNA as a reference sample and 10 ng of the same DNA for LM-PCR, using two rounds of amplification with 15 cycles per round to stay within the linear stage of amplification. We show in Figure 4 that we can amplify the DNA robustly from the 10-ng samples, and that microarray hybridizations generate correlations of HpaII/MspI ratios measured by R-values in excess of 0.96. We conclude that HELP can be used on amounts of DNA that represent approximately the amount in 2000 cells, and is likely to be amenable to further scaling to even more limited starting amounts.
Figure 4.
HELP with limited amounts of DNA template: nanoHELP. To test whether HELP could be used with more limited quantities of DNA than previously attempted, we generated representations using 1 µg and 10 ng of material from the same DNA sample, using the PCR conditions shown (a). It is apparent that the conditions used generate representations from the 10-ng samples similar in quantity to those from the 1 µg sample. These representations were used for HELP microarray hybridization to test whether the smaller sample amounts reproduced the methylation profile of the 1 µg sample. HpaII/MspI log ratios were correlated for the 1 µg control and the three experimental replicates of 10 ng each. We found the limited sample quantities to generate profiles highly concordant with the 1 µg control, with R-values exceeding 0.96 in all cases (b). We conclude that HELP representations can be generated with DNA amounts equivalent to the yield from thousands of diploid mammalian cells.
Simultaneous copy-number analysis: HELP-CNV
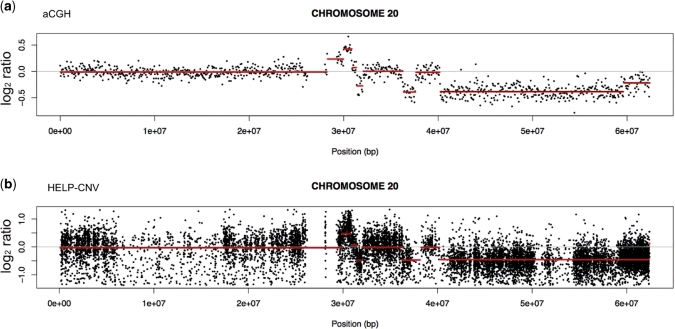
We investigated whether the HELP data could simultaneously be used to provide copy-number information, given that the MspI representation should be unaffected by cytosine methylation and instead dependent on the amount of DNA present. In a manner analogous to the ROMA technique (23), we compared the MspI (methylation-insensitive) representations of two different samples. One sample was a primary leukemia sample from a human subject that we have previously studied (24) and found to have copy-number changes by array comparative genomic hybridization (aCGH), the other was a normal CD34 + hematopoietic stem cell sample used as a control with presumed minimal copy-number variability. The copy-number variation analysis of HELP data (HELP-CNV) identified the same patterns of amplifications and deletions on chromosome 20 found using aCGH (Figure 5). The HELP-CNV approach allows the data generated as part of a HELP assay to harvest a second source of information about DNA copy number while testing cytosine methylation genome wide, a useful combination for diseases such as cancer.
Figure 5.
The MspI representation in a HELP assay can be used to detect copy-number variation: HELP-CNV. The MspI profile generated as part of the HELP assay is not affected by CG dinucleotide methylation and should instead be influenced by the amount of template available. We tested whether we could exploit this to detect copy-number variability (CNV) in a leukemia cell sample known to have amplifications and deletions (24) shown by microarray comparative genomic hybridization [aCGH, (a)]. In this representation of chromosome 20, we see regions with amplifications (increased log2 signal) and deletions (decreased signal). We performed a HELP assay on the same leukemia DNA sample and compared the MspI signal from each locus with the MspI signal from a HELP assay performed on a reference cell sample (normal CD34+ hematopoietic stem cells). These results are shown in (b). By performing the same segmentation analysis as for aCGH to define blocks of signal intensities, we found the MspI HELP data identify the same regions of amplification and deletion in these leukemic cells. HELP can thus give information about CNV simultaneously with its cytosine methylation analysis.
HELP studies using massively parallel sequencing: HELP-seq
The development of massively parallel sequencing has provided an alternative to microarrays for measuring the enrichment of genomic representations following chromatin immunoprecipitation (ChIP-seq). We sought to test whether the HELP representations could allow similar mapping of hypomethylated loci. Contaminating fragments in the assay can be recognized because they lack the HpaII/MspI site remnant (CGG) that starts each sequence read following the digestion and end-polishing when creating the libraries. In Table 2 we show a summary of the results for each library, including a proportion (∼3%) in each library that starts with a CGG but is not located at a canonical HpaII site (CCGG) in the reference genome sequence, indicating that we are identifying a substantial number of polymorphic HpaII sites in this ES cell DNA sample.
Table 2.
HELP-seq data
| Total reads | Low quality | Not aligned | Non-unique aligned reads | Unique aligned read | Number of unique aligned reads starting with CGG | Number of unique aligned reads starting with CGG at annotated HpaII sites | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MspI ES | 3 510 528 | 40 179 | 1.1% | 754 146 | 21.5% | 473 994 | 13.5% | 2 242 209 | 63.9% | 2 205 347 | 98.4% | 2 146 819 | 95.7% |
| HpaII ES | 5 872 425 | 65 788 | 1.1% | 1 358 445 | 23.1% | 88 096 | 1.5% | 4 360 096 | 74.2% | 4 322 845 | 99.1% | 4 259 942 | 97.7% |
| HpaII EP | 7 737 981 | 78 564 | 1.0% | 1 789 257 | 23.1% | 254 132 | 3.3% | 5 616 028 | 72.6% | 5 562 184 | 99.0% | 5 464 541 | 97.3% |
| Distinct loci defined by unique aligned reads | Number of distinct loci starting with CGG | Proportion of all distinct loci | Number of distinct loci starting with CGG at annotated Hpall sites | Proportion of distinct loci starting with CGG | Remaining distinct loci starting with CGG not at annotated HpaII sites | Proportion of distinct loci starting with CGG | |||||||
| MspI ES | 839 993 | 805 382 | 95.9% | 779 519 | 96.8% | 25 863 | 3.2% | ||||||
| HpaII ES | 656 320 | 624 143 | 95.1% | 607 028 | 97.3% | 17 115 | 2.7% | ||||||
| HpaII EP | 894 053 | 848 575 | 94.9% | 623 035 | 97.0% | 25 540 | 3.0% | ||||||
In the upper panel, we show the results of analysis of all of the reads obtained from the HELP-seq experiment. Most sequences mapped uniquely to the reference human genome, of which the vast majority had the CGG remnant of the original HpaII/MspI digestion. Multiple coincident reads defined the distinct loci described in the lower panel. Again, most loci had the CGG sequence and mapped to HpaII sites in the reference genome, but ∼ 3% of the distinct loci had evidence of an original HpaII site where none exists in the reference genome, defining polymorphic HpaII sites in our cell sample.
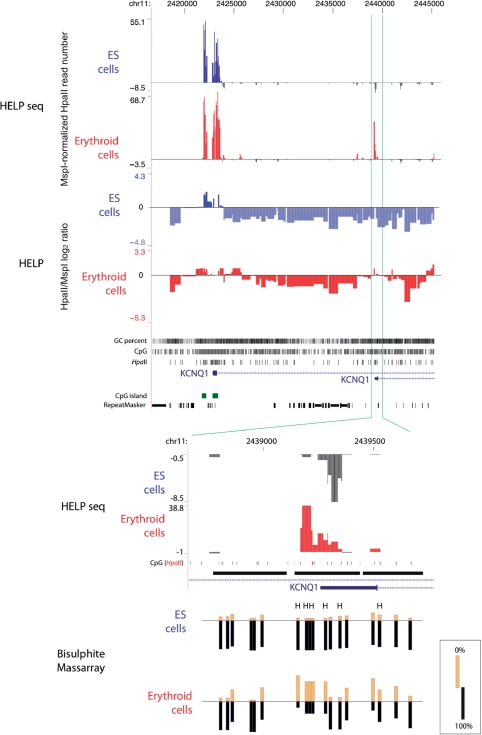
When we map these sites to the genome we observe ‘piling up’ of reads at HpaII sites, indicating their enrichment by the representation. We note that the control MspI representation is not enriched equally at all sites, a finding similar to those we have observed previously using microarray-based approaches (13). The HpaII representations are also heterogeneous but this is subject to the additional influence of cytosine methylation. We normalized the HpaII representation in terms of the MspI data and plotted wiggle tracks for the resulting data. In general, we found the microarray-based HELP and the HELP-seq data to be strongly concordant, as represented in Figure 6. We noted that the discordance of the assays appeared to be mostly in terms of HELP-seq identifying additional hypomethylated loci, such as the alternative, non-CpG island, non-CG cluster (18) promoter of KCNQ1, which appears to become hypomethylated in erythroid progenitor cells (Figure 6). We tested this locus using bisulphite MassArray and confirmed its hypomethylation (Figure 6), indicating that HELP-seq is more sensitive than microarray-based HELP in identifying hypomethylated loci. We note that HELP-seq allows mapping of sequences shorter than the 50-bp minimum that we can represent on a microarray, and can thus give more detailed information about CG-dense regions of the genome than when using microarrays.
Figure 6.
Adaptation of the HELP assay to use massively parallel sequencing: HELP-seq. Instead of using microarrays to map the representations to their genomic source, we sequenced the ends of the fragments using massively parallel sequencing (Illumina). We show the promoter region of the KCNQ1 gene as an example of the concordant data obtained, with hypomethylated sites represented as an upward/positive peak and methylated as downward/negative, in both the microarray-based (HELP) and Illumina-based (HELP-seq) assays. The HELP data are represented by quantile-normalized HpaII/MspI log2 ratios (13) while the HELP-seq data are represented by the number of HpaII reads per locus normalised to MspI reads for the same locus as described in the main text. Both embryonic stem (ES, red) and derived in vitro-differentiated erythroid progenitor (blue) cells are shown. The upstream (left) promoter of KCNQ1 is hypomethylated in both cell types, with a broader region of hypomethylation in erythroid compared with ES cells in both the HELP and HELP-seq studies. The downstream promoter appears to be methylated in ES cells in both assays, but microarray-based HELP weakly indicates hypomethylation in erythroid cells, whereas HELP-seq shows a strong hypomethylated signal in these cells. To determine which technique was more accurate, we performed validation using bisulphite MassArray (bottom), confirming the underlying hypomethylation of this locus in the erythroid and not ES cells, and demonstrating that HELP-seq offers improved capacity to identify sites of hypomethylated DNA in the genome.
DISCUSSION
Genomic representations by LM-PCR are used by a number of different applications, including representational oligonucleotide microarray analysis (ROMA) (23), high-density SNP microarrays (25) and other epigenomic assays testing cytosine methylation (26–28). The HELP assay uses representations from HpaII to distinguish methylated from unmethylated loci in the genome, with a concurrent MspI representation defining the full range of potential HpaII-amplifiable fragments. The more fragments that can be represented, the greater the level of detail we can achieve in ROMA, SNP or epigenomic analyses. By increasing the representation of shorter fragments using dual adapters and modified PCR conditions, we achieve greater coverage of CG-dense regions in particular. By computational analysis, we show that the proportional coverage for CpG islands or our updated definition of CG clusters (18) approaches the maximum possible (98.5% and 98.6%, respectively, Table 1). In addition, the number of fragments at each CG-dense locus and refSeq promoter is increased 2- to 3-fold compared with the previous representation, which enables a more detailed analysis of DNA methylation in these promoter regions. We demonstrate that these shorter fragments (50–200 bp) can be labeled with the random priming technique and hybridized to microarrays with signal ranges comparable to those of larger fragments. Our practical lower limit for microarray analysis is constrained by the size of the oligonucleotides we use (≥50 nt), making it difficult to represent fragments smaller than the oligonucleotides themselves. For massively parallel sequencing applications, the sequences generated are generally shorter (∼35 bp) and need only be long enough to allow accurate mapping to the reference genome, increasing the number of testable loci compared with microarrays.
This new representation allowed us to design high-resolution HELP microarrays for the human, mouse, rat and cow genomes, each representing over 1 million loci throughout these genomes. By using a high-density platform, we can represent these loci on a single microarray, allowing analysis of the entire genome in a single hybridization experiment. We performed a high-resolution HELP assay on human ES cells to test how the cytosine methylation patterns observed correlate with genomic annotations. We found relative hypomethylation of CpG islands and CG clusters compared with other sequences, and less methylation at promoters compared with gene bodies or intergenic regions, patterns consistent with prior observations about the distribution of cytosine methylation (22).
When moving epigenomic assays to the clinical setting, a critical issue to address is the potential to use the limited sample amounts that can be acquired from biopsies. As the generation of genomic representations in the HELP assay involves PCR, we tested whether the amplification step could allow us to generate adequate amounts of material from limited starting amounts of DNA. We used 10 ng of DNA in our studies as an amount representing approximately 2 × 103 cells, and found the profiles generated to be highly comparable with those generated using our usual microgram quantities of starting material. We conclude that the HELP assay can be used on relatively limited numbers of cells. Our ongoing studies are testing whether we can decrease the starting amounts of DNA still further.
We also found that the MspI representation on its own allows an accurate and detailed analysis of copy number. This approach is similar to the published ROMA technique (23) in that it uses genomic representations by methylation-insensitive restriction enzymes, but is substantially higher resolution with >1.32 million loci represented. With MspI/HpaII sites more frequent in (C + G) mononucleotide-rich regions which tend also to be more gene-rich (9), the MspI representation offers selectively higher resolution in gene-rich regions, which is potentially advantageous. The HELP-CNV application allows DNA to be tested for cytosine methylation and copy-number variation simultaneously, a valuable tool especially in cancer research, in which epigenomic alterations (29) and copy-number changes (30) are frequent.
A major advantage of HELP-seq compared with microarray-based HELP appears to be one of sensitivity of detection of hypomethylated loci. We believe that this is due to the lower background noise for sequencing compared with microarrays, which always generate a fluorescence intensity reading whether there is genuine signal present or not. There are other potential advantages of HELP-seq, including the capacity to detect allelic differences in methylation, information about repetitive sequences and the ability to detect events in HpaII fragments smaller than the 50-bp lower limit for microarray studies. We also note that we identified a substantial number of HpaII sites (∼3%) that are not present in the reference human genomic sequence, variability that would not be captured by or that would cause errors in microarray-based approaches.
The ideal assay to test cytosine methylation would test every CG dinucleotide individually and quantitatively throughout the genome, preserving information about cis-relationships of methylation states between CGs, and allowing high sample throughput. No such assay exists at present. While massively parallel sequencing-based approaches promise to make nucleotide-resolution studies possible (2,5,6,31), at present their sample quantity demands and costs remain daunting. The HELP assay falls into a category of assays that act to screen the genome at lower resolution, the ‘discovery’ step that defines loci for more detailed, nucleotide-resolution studies. Other assays in this category include methylation-dependent restriction enzyme assays (32) and affinity-based assays using antibodies (1) or other natural methyl-binding proteins (3). The comparative advantages of HELP include its capability of using a single array and the easy technical validation of results, as the focus is solely on methylation at the HpaII/MspI sites (CCGG) generating the representations. While HpaII/MspI sites constitute only ∼8% of the CG dinucleotides in the genome (9), the presence of methylation ‘states’ in cis that may extend over as much as 1 kb (33) allow discovery assays to flag interesting regions by testing a subset of CGs. When we measure the proportion of CGs in the human genome residing in proximity to the HpaII sites on the HELP microarray we used, we find that approximately two-thirds are within 1 kb of these sites (Supplementary Figure 2). While epigenomic discovery approaches directly test only a minority of CGs, they have the potential of ‘flagging’ the majority of CGs in the genome. We conclude that the high-resolution HELP assay is technically simple and robust and offers the capacity to test both the epigenome and copy-number variability, and has the potential for use with limited numbers of cells and adaptation to massively parallel sequencing platforms.
ADDENDUM
While this manuscript was in review, an assay fundamentally similar to HELP-seq, described as Methyl-seq, was published by Brunner et al. (34).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (NIH) [(NHGRI) R01 HG004401 to J.M.G, (NHLBI) R01 HL082946 to A.V., (NIA) P01 AG027734 and R01 AG18728 to R.F.T., and (NIGMS) T32 GM007288 to J.L.G.]; the Japan Society for the Promotion of Science (to M.O.). A Leukemia and Lymphoma Society Translational Research Award and funding from the Hershaft Foundation (to A.V.) and the Leukemia and Lymphoma Society (to A.M.). Funding for open access charge: NHGRI R01 HG004401.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the support of the Einstein Center for Epigenomics, the Bioinformatics Shared Resource and the Genomics Core Facility.
Footnotes
Present address: Mayumi Oda, Laboratory of Cellular Biochemistry, Department of Veterinary Medical Sciences/Animal Resource Sciences, University of Tokyo, Bunkyo-ku, Tokyo, Japan
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
REFERENCES
- 1.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 2.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 4.Holemon H, Korshunova Y, Ordway JM, Bedell JA, Citek RW, Lakey N, Leon J, Finney M, McPherson JD, Jeddeloh JA. MethylScreen: DNA methylation density monitoring using quantitative PCR. Biotechniques. 2007;43:683–693. doi: 10.2144/000112597. [DOI] [PubMed] [Google Scholar]
- 5.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeddeloh JA, Greally JM, Rando OJ. Reduced-representation methylation mapping. Genome Biol. 2008;9:231. doi: 10.1186/gb-2008-9-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 10.Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 11.Kunnath L, Locker J. Characterization of DNA methylation in the rat. Biochim. Biophys. Acta. 1982;699:264–271. doi: 10.1016/0167-4781(82)90116-6. [DOI] [PubMed] [Google Scholar]
- 12.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 13.Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, Zheng X, Kim K, Greally JM. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24:1161–1167. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivier EN, Qiu C, Velho M, Hirsch RE, Bouhassira EE. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp. Hematol. 2006;34:1635–1642. doi: 10.1016/j.exphem.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ENCODE consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 18.Glass JL, Thompson RF, Khulan B, Figueroa ME, Olivier EN, Oakley EJ, Van Zant G, Bouhassira EE, Melnick A, Golden A, et al. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 2007;35:6798–6807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 20.Dabney AR, Storey JD. Normalization of two-channel microarrays accounting for experimental design and intensity-dependent relationships. Genome Biol. 2007;8:R44. doi: 10.1186/gb-2007-8-3-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 23.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa ME, Reimers M, Thompson RF, Ye K, Li Y, Selzer RR, Fridriksson J, Paietta E, Wiernik P, Green RD, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS ONE. 2008;3:e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 27.Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sato M, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- 28.Yuan E, Haghighi F, White S, Costa R, McMinn J, Chun K, Minden M, Tycko B. A single nucleotide polymorphism chip-based method for combined genetic and epigenetic profiling: validation in decitabine therapy and tumor/normal comparisons. Cancer Res. 2006;66:3443–3451. doi: 10.1158/0008-5472.CAN-05-3739. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 30.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 31.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 33.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009 doi: 10.1101/gr.088773.108. (in press, available online) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.