Abstract
Fibroblast growth factor receptor 3 (FGFR3) plays an important role in cartilage development. Although upregulation of FGFR3 expression in response to bone morphogenetic protein-2 (BMP-2) has been reported, the molecular mechanisms remain unknown. In this study, we used in vivo approaches to characterize BMP-2-induced alterations in the chromatin organization of the FGFR3 core promoter. Chromatin immunoprecipitation analysis demonstrated that the binding of Brg1, a component of the SWI/SNF remodeling complex, may selectively remodel a chromatin region (encompassing nucleotide –90 to +35), uncovering the transcription start site and three Sp1-binding sites, as revealed by nuclease digestion hypersensitivity assays. We then showed an increase in the association of Sp1 with the proximal promoter, followed by the recruitment of p300, resulting in a change of the histone ‘code’, such as in phosphorylation and methylation. Collectively, our study results suggest a model for BMP-2-induced FGFR3 expression in which the core promoter architecture is specifically regulated.
INTRODUCTION
Growth factors that have been implicated in chondrogenesis include fibroblast growth factors (FGFs), platelet-derived growth factor, bone morphogenetic proteins (BMPs) and transforming growth factor. Fibroblast growth factor receptors (FGFRs) constitute a family of four members with tyrosine kinase activity; among these, FGFR3 plays an important role in cartilage development. Three inherited human dwarfism syndromes—hypochondroplasia, achondroplasia and thanatophoric dysplasia—are caused by missense mutations in the FGFR3 gene. These mutations lead to different levels of receptor activation, which correlate well with the severity of the human phenotypes (1).
FGFR3 has been identified in the cartilage primordia of developing mouse long bones. Several mouse models mimicking the human achondroplasia phenotype have been created by expressing mutated forms of FGFR3 in the developing cartilage anlagen (2–4). These mice display a severe shortening of the appendicular skeletal elements due to reduced regions of proliferating and hypertrophic chondrocytes. Additionally, mice carrying a targeted deletion of FGFR3 are characterized by increased regions of proliferating and hypertrophic chondrocytes (5,6). These studies have led to the conclusion that FGF signaling is a negative regulator of chondrocyte proliferation and differentiation.
Interestingly, evidence from other reports indicates a role for FGFR3 signaling in promoting chondrocyte differentiation. For example, FGF18 has been identified as a selective ligand for FGFR3 in mouse limb bud mesenchymal cells, which suppressed proliferation and promoted their differentiation and production of the cartilage matrix (7). These observations support those from in vivo studies of the growth plates of human thanatophoric dwarfs (8) and the results of in vitro studies of human articular chondrocytes (9), as well as those of transgenic mice expressing constitutively active FGFR3 in the articular joints (10) and examinations of mouse limb explants (11). The apparent discrepancies may arise from a combination of the different experimental models employed and the use of different surrogate markers for proliferation and cartilage synthesis, and also from the lack of discrimination between the stages of differentiation under investigation.
BMPs are present in the mesenchyme and are necessary for aggregation of mesenchymal cells and maturation of chondrocytes in vivo (12). BMPs significantly induce chondrocyte differentiation and promote the expression of cartilage-specific genes in primary cultures of chondrocytes, as well as in cell lines such as ATDC5 and C3H10T1/2 (13–15). In a previous study, the BMP-2-dependent onset of chondrogenic differentiation in the pluripotent murine mesenchymal stem cell line (C3H10T1/2) was reported to be accompanied by the immediate upregulation of FGFR3 (16). Overexpression of FGFR3 in C3H10T1/2 cells is sufficient for chondrogenic differentiation, indicating an important role for FGF signaling during the manifestation of the chondrogenic lineage in this cell line.
Despite extensive studies of FGFR3, the transcription factors that interact with the FGFR3 promoter in the process of chondrogenesis have yet to be identified and functionally characterized. As previously reported, the sequence between –220 and –27 bp (upstream of the transcription start site) of the FGFR3 gene, conferred a 20–40-fold increase in transcriptional activity upon a promoter-less vector (17), and may serve as the core promoter. In this study, we examined the order of events occurring on the core FGFR3 promoter in vivo during BMP-2-dependent transcriptional activation, using nuclease digestion assays and chromatin immunoprecipitations (ChIP). Following BMP-2 induction, the remodeling complex SWI/SNF is assembled on the proximal promoter and functions by targeting a specifically positioned nucleosome (Nuc +1) that masks the start site of transcription and Sp1-binding sites. We also revealed a cascade of molecular recruitment events following assembly of the remodeling complex, beginning with Sp1 binding to the promoter, followed immediately by p300 recruitment, resulting in an alteration in modification of the histone tail, which suggests that BMP-2 upregulates FGFR3 expression through controlling the core promoter structure.
MATERIALS AND METHODS
Materials
C3H10T1/2 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Antibodies were purchased from the following companies: Sp1 (SC-59G), p300 (SC-584), Brg1 (SC-10 768) and RNA polymerase II (SC-9001) from Santa Cruz (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); tetra-acetyl-H4 (06-866) and diacetyl-H3 (07-593) from Upstate (Charlottesville, VA, USA); dimethyl-H3-K9 (7312) and trimethyl-H3-K4 (8580) from Abcam (Abcam, Inc., Cambridge, MA, USA); p38MAPK (9212) and phospho-p38 (9215) from Cell Signaling Technology (Cell Signaling Technology, Inc., Boston, MA, USA); p38 inhibitor SB203580 was purchased from Calbiochem (Calbiochem Inc., San Diego, CA, USA); cycloheximide (CHX) was obtained from Sigma (Sigma-Aldrich, St Louis, MO, USA) and BMP-2 was from R&D systems (R&D Systems, Inc., Minneapolis, MN, USA).
Cell culture
To examine the effects of BMP-2 on FGFR3 expression, C3H10T1/2 cells were incubated for 1 day with indicated amount of BMP-2 or cells were incubated in the presence or absence of 200 ng/ml BMP-2 for indicated period of time, in the medium with 5% FBS. Pre-incubation with 10-μM CHX (inhibitor of protein synthesis) or 10 μM SB203580 (blocking the p38 MAPK pathway) was performed for 30 min, followed by the addition of BMP-2 and further incubation for 1 day in the continued presence of CHX or SB203580 (18,19).
Reverse transcription PCR (RT–PCR)
Total RNA was extracted and purified using an RNeasy Kit (Qiagen, Inc., Valencia, CA, USA). Reaction mixture (25 μl) containing 2 μg of total RNA was reverse transcribed to cDNA using SuperScript II RT-polymerase (Invitrogen Corp., Carlsbad, CA, USA). PCR was performed on the cDNA using primers specific for FGFR3 (5′-ttggcatgcggtgccttcac-3′ and 5′-gaggtccaagtactcgtcgg-3′) and 18s rRNA (5′-cctggataccgcagctagga-3′and 5′-gcggcgcaatacgaatg cccc-3′). The PCR reaction mix for quantitative RT–PCR (qRT–PCR) was prepared using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and the PCR conditions for quantitative RT-PCR were as follows: activation of enzyme at 95°C for 5 min, 45 cycles of denaturation at 95°C for 15 s, annealing at 57°C for 5 s, and extension at 72°C for 25 s. qRT–PCR was carried out using a 7300 Real Time PCR System (Applied Biosystems). The fluorescence of each sample was determined after every cycle, and denaturation curves of the PCR products were determined by increasing the temperature at the rate of 0.1°C/min from 55°C to 95°C. The fluorescence of samples was continuously traced during this period. All the melting curves of PCR products gave a single peak. Agarose gel electrophoresis of representative reactions was used to confirm the amplification of unique fragments of predicted lengths. Relative FGFR3 expression levels were calculated as ratios of FGFR3 mRNA levels normalized against those of 18s rRNA. The data from qRT–PCR were analyzed by the ΔΔCt method, and the ΔCt value was determined by subtracting the 18s rRNA Ct value from the target gene Ct value. The ΔCt of the stimulated cells (ΔCts) was subtracted from the ΔCt of the untreated cells (ΔCtu) (ΔΔCt = ΔCts – ΔCtu), and the expression level for a target gene in the stimulated cells compared with the level in the untreated cells was calculated as follows: x-fold of unstimulated control = 2−ΔΔCt. All results are expressed as the means ± standard deviations of three independent experiments.
Nuclease hypersensitivity assays
In brief, nuclear pellets were resuspended in 3 ml of buffer A (10 mM Tris [tris (hydroxymethyl) aminomethane], pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3 M sucrose). Aliquots of nuclei were digested with increasing concentrations of DNase I (0, 1, 3, 5 U/ml) (Worthington Biochemical Corp., Freehold, NJ, USA) in a 400-μl final volume of buffer A with 1 mM CaCl2 for 5 min at room temperature. The reaction was stopped by adding 400 μl of Stop buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 50 mM EDTA, 0.3% sodium dodecyl sulfate [SDS]) and incubated with 10 U of RNase at 37°C for 1 h. The samples were extracted once with phenol–chloroform–isoamyl alcohol (25:24:1) and once with chloroform–isoamyl alcohol (24:1). Nucleic acids were precipitated with 0.7 volumes of isopropanol, washed with 70% ethanol and resuspended in 1× TE buffer (10 mM Tris–HCl [pH 7.5], 1 mM EDTA). Sensitivity to micrococcal nuclease (MNase) was determined using a freshly prepared suspension of nuclei in MNase digestion buffer (15 mM Tris–HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 3 mM MgCl2, 20% glycerol, 15 mM 2-mercaptoethanol). DNA concentrations were estimated by absorption at 260 nm. Equal amounts of nuclei were digested with increasing concentrations of MNase I (0, 0.2, 0.4, 0.6 U/ml) (Worthington Biochemical Corp., Lakewood, N.J., USA) in a 400 μl final volume for 5 min at 20°C. The reactions were then stopped, and DNA was extracted as described above. For the restriction enzyme accessibility assays, nuclei were isolated as described above and digested with 25–150 U of restriction enzyme in 300 μl of the corresponding reaction buffer (New England Biolabs, Beverly, MA, USA) for 30 min at 37°C. Reactions were terminated, and DNA was extracted as described above. After purification, DNA (15 μg) was digested to completion with Hind III or Nco I. The products were resolved on agarose gel and detected by Southern blotting using the indirect end-labeling method.
Analysis of chromatin by indirect end labeling
Southern blotting was performed essentially as previously described (20). DNA (15 μg) was digested to completion with Hind III or Nco I, then electrophoresed in 0.8% or 1.5% agarose gel in 1× TAE buffer (40 mM Tris–acetate, 1 mM EDTA; pH 8.0). The digested DNA was then transferred to Hybond N+ membrane and FGFR3 gene fragments were detected by hybridization with radioactive probes obtained by random priming. The following primer sets were used to amplify probes anchored at Hind III or Nco I sites: HindIII-F, 5′-GAATTCTCCGATGGGACACTTGAATG-3′; HindIII-R, 5′-TCTAGACCCAGGTTAATGAACCACTG-3′; NcoI-F, 5′-GAATTCGAACCCAGGACTGGATTCTA-3′; NcoI-R, 5′-TCTAGAGGACTACCATGGCTCCAGAG-3′. After double digestion with EcoR I and Xba I, the PCR products were cloned into pUC19. Both probes were extracted from agarose gel after restriction enzyme digestion of plasmid pUC19 DNA by EcoR I and Xba I for purification; the probes were then labeled with 32P by random priming. Prehybridization and hybridization were performed at 65°C, using 5× SSC (10× SSC is 1.5 M NaCl, 0.15 M sodium citrate; pH 7.4), 0.7% SDS, and 5× Denhardt solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), followed by three sequential washes under stringent conditions. The intensities of the bands were determined using a molecular imager System (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the percentage of digestion was quantified as the fraction of the signal of the band compared with the total signal of the bands within a given lane on the gel. Size markers were generated by amplification of a series of 100 bp DNA ladders with a fixed 5′ end, using plasmid pcDNA3.1 as a template. PCR products were mixed together and electrophoresed with the nuclear samples, then transferred and hybridized with [32P]dATP-radiolabeled probes spanning the ladders.
ChIPs and Re-ChIPs
Cells were incubated in 1% formaldehyde for 10 min at 22°C and the reaction then stopped by addition of glycine to a final concentration of 0.125 M. The cells were then washed with cold phosphate-buffered saline and lysed in buffer. Nuclei were sonicated to shear DNA, and the lysates were pelleted and precleared. Sonicated chromatin was incubated with 4 μg of antibodies. The protein–DNA complexes were incubated with protein A beads and eluted in 1% SDS/0.1 M NaHCO3 and cross-links were reversed at 65°C. DNA was recovered by phenol–chloroform extraction and ethanol precipitation, then subjected to semi-quantitative PCR analysis. In the Re-ChIP experiments, complexes were eluted by incubation for 30 min at 37°C in 25 ml 10 mM DTT. After centrifugation, the supernatant was diluted 20 times with Re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, [pH 8.1]) and subjected again to the ChIP procedure.
Real-time PCR reactions were performed in triplicate with 2 μl of precipitated DNA. DNA recovered from samples containing an antibody was compared with no-antibody negative controls performed on aliquots from the same chromatin preparation. Additionally, uniformity of the chromatin preparations was confirmed by analyzing the background level of an unrelated genomic locus by real-time PCR. Data are presented as the amount of DNA recovered relative to the appropriate negative control antibody. In this way, differences between chromatin preparations are normalized. The results were expressed as the means ± standard deviations of three independent experiments. The PCR primers were as follows: –3k region (Forward: 5′-GCCTTTACATAGATAGAGTG-3′; Reverse: 5′-TGTACCGTGTGCTTCTAGAC-3′); +1 region (Forward: 5′-AGCTCGGCATTGCTTCAAG-3′; Reverse: 5′-TGCCACAGTGCACTGAACT-3′); +6k (Forward: 5′-GTTCCAGGGCCTGAACCTAG-3′; Reverse: 5′-TAGCACTTGCAGCCTCTGAG-3′); 3′UTR (Forward: 5′-GCTTAACACTTCCTATGCAG-3′; Reverse: 5′-AAACCACATCTGCCAGTGTG-3′).
Luciferase assay
Plasmid DNA was transiently transfected into C3H10T1/2 cells using Lipofectamine 2000 (Invitrogen Corp.). phRL-TK (Promega, Madison, WI, USA) was co-transfected as an internal control for transfection efficiency. After further cultivation under serum-deprived conditions for 24 h, the transfected cells were harvested, lysed, centrifuged and the pellet subjected to luciferase assay. Luciferase activity was measured as chemiluminescence in a luminometer (PerkinElmer Life Sciences, Boston, MA, USA) using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. All transfections were performed in triplicate, and the results were expressed as the means ± standard deviations of three independent experiments.
Construction of reporter plasmids and mutagenes
PCR was performed using a pair of oligonucleotide primers (Forward: 5′-ATGCCTTTACATA GATAGAGTG-3′; Reverse: 5′-ATGGTACCATCTCGAGTGTACCGTGTGCTTCTAGAC-3′) specific for the mouse FGFR3 promoter corresponding to the –224/+15 fragment, of which the forward primer was Kpn I-site-linked and the reverse primer Xho I-site-linked. C3H10T1/2 genomic DNA was used as the template. These PCR products were digested with Kpn I and Xho I, and subcloned into the Kpn I/Xho I sites of the pGL3 Basic vector (Promega), yielding the reporter plasmid pGLP. Substitution mutation constructs were generated using a PCR-based site-directed mutagenesis kit (TaKaRa Co. Ltd., Tokyo, Japan), using the pGLP plasmid as the template. For six-base substitutions, we introduced the EcoR I restriction site GAATTC into the substitution sites. Oligonucleotides containing three repeats of BMP-2 responsive element (3× GCCGCGCCGGCTCCAGGGCAGGAGCGGC) were synthesized and inserted into the pGL3 promoter vector (Promega) after digestion with Kpn I and Xho I, yielding the positive reporter pGL3BMRE.
Statistical analysis
Statistical evaluations were conducted using the t-test. P-values <0.05 were considered to be statistically significant.
RESULTS
BMP-2 stimulates FGFR-3 expression in C3H10T1/2 cells through the p38 pathway independent of protein synthesis
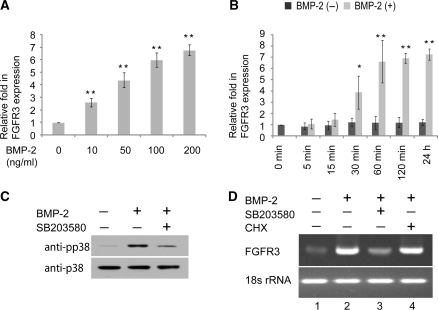
The effect of BMP-2 on the FGFR-3 mRNA expression level in C3H10T1/2 was determined by quantitative RT-PCR. As shown in Figure 1A, FGFR-3 expression was dose-dependently induced by 10–200 ng/ml of BMP-2. FGFR-3 mRNA induction appeared as early as 30 min following BMP-2 stimulation, reached a high level at 1 h, then continued to increase slowly (Figure 1B). To determine whether protein synthesis was required for BMP-2 induced FGFR3 expression, CHX, a protein synthesis inhibitor, was applied. Pretreatment with CHX did not inhibit the BMP-2-induced increase in FGFR3 expression as analyzed by qRT–PCR analysis (Figure 1D, lane 2 versus lane 4), suggesting that the upregulation of FGFR3 expression is independent of protein synthesis in C3H10T1/2 cells.
Figure 1.
Upregulation of FGFR3 expression by BMP-2 treatment in C3H10T1/2 cells. (A, B) C3H10T1/2 cells were incubated with the indicated dosage of BMP-2 with 5% FBS for 1 day (A), or for the indicated periods of time in the presence of 200 ng/ml BMP-2 with 5% FBS (B) and total RNA was extracted for measurement of FGFR3 mRNA expression level by semi-quantitative RT–PCR. *P < 0.05; **P < 0.01 versus the corresponding untreated cells. All the results are the means of three independent experiments ± standard deviation. (C) C3H10T1/2 cells were pretreated with control or SB203580 (10 μM) for 30 min and cultured for another 30 min in the presence or absence of BMP-2 (200 ng/ml). The whole lysates were examined by immunoblotting with antibodies against phosphorylated p38 and total p38. (D) C3H10T1/2 cells were pretreated with SB203580 (10 μM) or CHX (10 μM) for 30 min, followed by addition of BMP-2 (200 ng/ml) for 1 day in the continued presence of CHX or SB203580. FGFR3 mRNA expression level was examined by semi-quantitative RT–PCR. The PCR products were electrophoresed on agarose gel and stained with ethidium bromide.
In a previous study, it was determined that the BMP-2/Smads pathway was not responsible for immediate stimulation of FGFR3 expression (16). Another pathway that may play a role in FGFR3 expression is the BMP/p38 pathway. The importance of the BMP/p38 pathway was examined by pretreatment of C3H10T1/2 cells with SB203580, a specific inhibitor of p38 kinase. Pre-incubation of 10 μM SB203580 for 30 min partially blocked BMP-2-induced p38 phosphorylation (Figure 1C) and greatly inhibited the increase in FGFR3 expression elicited by BMP-2 (Figure 1D, lane 2 versus lane 3). Together, these data suggest that the p38 kinase pathway contributes to BMP2-mediated immediate upregulation of FGFR3 expression in C3H10T1/2 cells.
Open chromatin structure around the transcription start site
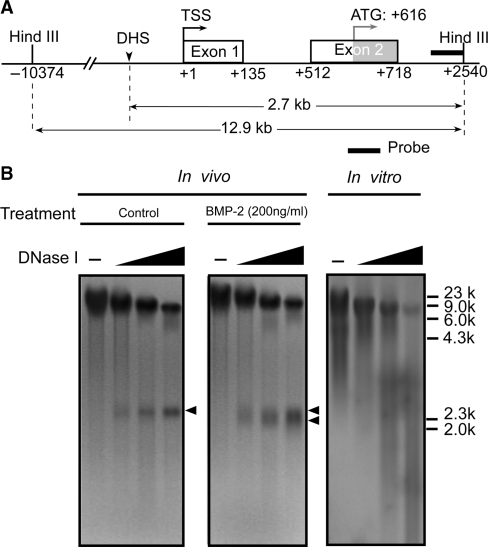
It is generally accepted that the local chromatin structure affects gene transcription. We performed DNase I hypersensitivity assays in an attempt to reveal regions of open chromatin around the FGFR3 gene promoter upon BMP-2 exposure, in order to provide cues in the search for the regulation sites of BMP-2 signaling at the promoter level. In these experiments, C3H10T1/2 cells were either left untreated or treated with BMP-2 for 1 day. Nuclei were isolated and subjected to digestion with increasing concentrations of DNase I, and the genomic fragments between –10 374 bp and +2540 bp (relative to the transcription start site) were analyzed. As shown in Figure 2, in untreated conditions, a band gradually appeared in a DNase I concentration-dependent manner, representing a single DNase I hypersensitive site (DHS) that was adjacent to the transcription start site of the FGFR3 promoter and ∼2.7 kb upstream of the Hind III site at +2540 bp. Importantly, on stimulation with BMP-2 (200 ng/ml), the hypersensitive band appeared wider than that of the untreated cells; thus, after screening a large range of ∼13 kb around the transcription start site, we found a single DNase I hypersensitive site in the proximal promoter region, which was associated with both the basal and BMP-induced transcription of FGFR3 in C3H10T1/2 cells.
Figure 2.
Inducible DNase I hypersensitivity within the FGFR3 promoter. (A) Schematic representation of the probe used to map the DNase-hypersensitive sites within the FGFR3 promoter by the indirect end-labeling technique. (B) C3H10T1/2 cells were treated with or without 200 ng/ml BMP-2 with 5% FBS for 1 day. Nuclei were then purified and digested with increasing concentrations of DNase I (0, 1, 3, 5 U/ml). DNA was purified and cleaved with Hind III. After Southern blotting, the filter was hybridized with the 32P-labeled probe. The DNase I-hypersensitive sites are indicated by arrowheads. As a control, purified genomic DNA from C3H10T1/2 cells was digested in vitro with increasing concentrations of DNase I (0, 0.05, 0.1, 0.2 U/ml).
Additionally, we performed control digestion of naked genomic DNA in vitro with increasing concentrations of DNase I. No preferential cutting was observed in conditions that generated levels of digestion comparable to those obtained in the in vivo experiments, confirming that the DNase I hypersensitivity observed was a consequence of chromatin organization and not secondary to sequence-directed cleavage preference by DNase I.
BMP-2-induced nucleosome remodeling at the proximal promoter of the FGFR3 gene
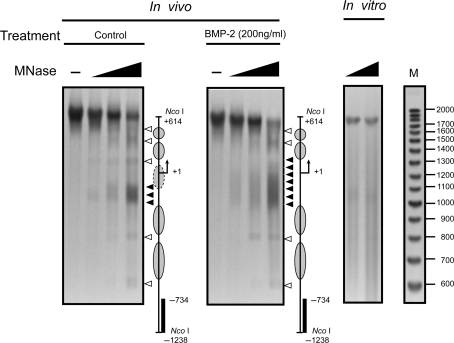
We next sought to map the hypersensitive site more precisely and to determine the nucleosomal organization within the FGFR3 proximal promoter region. MNase preferentially digests nucleosome-free DNA and linker DNA, and can therefore be used to determine nucleosome remodeling. Digestion of a nucleosomal array using limited amounts of MNase produces a ladder of DNA fragments corresponding to multiples of the nucleosome particle (∼160 bp). Nuclei were isolated from C3H10T1/2 cells and digested with increasing amounts of MNase. A discrete band pattern (Figure 3A) was revealed in untreated C3H10T1/2 cells. The overall digestion pattern indicated a pattern consistent with precise nucleosome positioning. Particularly, in untreated cells we detected a strong and specific band ∼1 kb from the Nco I site, at ∼−200 bp relative to the transcription start site. Bmp-2 treatment did not alter the whole band pattern, and the signal of the major band at –200 bp did not change as compared with that of untreated cells with the same amount of MNase. However, the 3′ boundary extended further into the next linker region in BMP-2-treated cells, which indicated that the major band in untreated cells correlates with the basal transcription of FGFR3, and the neighboring nucleosome may be removed from the local region in response to BMP-2 treatment.
Figure 3.
Mapping of the nucleosomes within the FGFR3 promoter upon BMP-2 treatment. C3H10T1/2 cells were either incubated with medium alone or stimulated with BMP-2 (200 ng/ml) with 5% FBS for 24 h. Nuclei were then purified and digested with increasing concentrations of MNase (0, 0.2, 0.4, 0.6 U/ml). The major MNase hypersensitive sites (MHSs) are indicated by the solid arrowheads; other MNase cutting sites are marked by hollow arrowheads. Increase in MNase hypersensitivity on BMP-2 stimulation is indicated by an extension of the hypersensitive site. As a control, purified genomic DNA from C3H10T1/2 cells was digested in vitro with increasing concentrations of MNase. Size markers were generated as described in ‘Material and Methods’ section. The diagrams at the right of each panel indicate the positions of putative nucleosomes (circles and ovals) and the probes used for hybridization (solid bars); the arrows indicate the transcription start site.
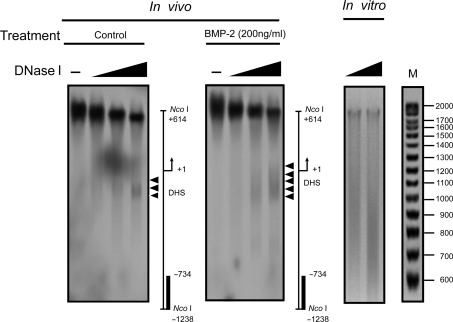
In order to confirm the relationship between the MNase hypersensitive site (the major band at –200 bp) and the DNase I hypersensitive site, we performed the DHS assay again using the same restriction enzyme, Nco I, for DNA digestion and the same probe for Southern blot as in the MNase digestion experiment. As shown in Figure 4, there is a remarkable congruence in the positions of the DNase I and MNase hypersensitive sites in the proximal promoter. Interestingly, the range of the DHS may be expanded by BMP-2 addition, suggesting that the region immediately downstream of the DHS is resistant to nuclease digestion under basal conditions, but becomes sensitive to nuclease after stimulation with BMP-2.
Figure 4.
Mapping of the BMP-2-induced DNase I hypersensitivity within the proximal FGFR3 promoter. C3H10T1/2 cells were treated with or without 200 ng/ml BMP-2 with 5% FBS for 1 day. Nuclei were then purified and digested with increasing concentrations of DNase I (0, 1, 3, 5 U/ml) and DNA was purified and cleaved with Nco I. After Southern blotting, the filter was hybridized with the 32P-labeled probe used in the MNase digestion assay. The DNase I-hypersensitive sites are indicated by arrowheads. As a control, purified genomic DNA from C3H10T1/2 cells was digested in vitro with increasing concentrations of DNase I. Size markers were generated as described in ‘Material and Methods’ section. The diagrams at the right of each panel indicate the positions of the transcription start site and the probes used for hybridization (solid bars). The large signal near +1 in the figure on the left is an autoradiograph artifact.
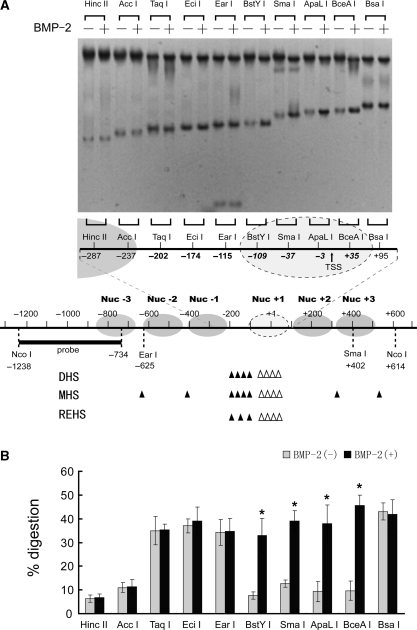
To analyze more precisely the locations and boundaries of these chromatin changes, a restriction enzyme accessibility assay was employed. Nuclei isolated from BMP-2-treated or untreated C3H10T1/2 cells were subjected to limited digestion with a panel of restriction enzymes with recognition sites at different locations around the nuclease hypersensitive site (NHS). As shown in Figure 5, under basal conditions, restriction sites of Hinc II/–287, Acc I/−237, BstY I/−109, Sma I/−37, ApaL I/−3 and BceA I/+35 were resistant to nuclease digestion, indicating their coverage by nucleosomes. In contrast, stronger cleavage was detected for Taq I/−202, Eci I/−174, Ear I/−115 and Bsa I/+95, suggesting that these sites were in two linker regions, with greater accessibility to restriction enzymes. BMP-2-dependent restriction enzyme cleavages were apparent when the nuclei were incubated with BstY I, Sma I, ApaL I and BceA I (Figure 5B); in contrast, BMP-2-induced cleavages were not observed for other restriction sites, including Taq I/−202, Eci I/−174 and Ear I/−115 (Figure 5B). These results suggest that the region in which BMP-2-induced changes in chromatin structure take place extends from at least −109 bp (BstY I site) to at least +35 bp (BceA I site). Because digestion with enzymes cutting just upstream (Ear I/−115) and downstream (Bsa I/+95) of the region remain unchanged upon BMP-2 exposure, we more precisely mapped the boundaries of the region undergoing chromatin remodeling on BMP-2 induction to between nt −115 and −109 in 5′ and nt +35 and +95 in 3′. The size of this region, which ranges from 144 to 210 bp, corresponds to the length of DNA protected by a nucleosome, suggesting the selective remodeling of this nucleosome on FGFR3 transcriptional activation.
Figure 5.
Alteration in the restriction enzyme accessibility at the FGFR3 proximal promoter induced by BMP-2 treatment. (A) C3H10T1/2 cells were incubated in the presence or absence of 200 ng/ml BMP-2 with 5% FBS for 1 day. Nuclei were then purified and digested with 25–200 U of restriction enzyme/ml, and purified DNA was digested to completion with Nco I. Products were detected by Southern blotting (top panel). A tentative assignment of the nucleosome positions in this region based on nuclease digestion is shown and is aligned with the nuclease-hypersensitive sites. Sites of cutting by DNase I and MNase and restriction enzyme are depicted by solid bars for basal conditions. Because MNase preferentially digests DNA in linker regions between nucleosomes, it is possible to locate nucleosomes within the FGFR3 proximal promoter. Hollow arrow heads indicate increases in the nuclease hypersensitivity upon BMP-2 exposure. (B) The relative nuclease sensitivity of restriction enzyme sites was quantitated, and the intensities of the radioactive bands were used to calculate the percentage of DNA digested. All the results are the means of three independent experiments ± standard deviation. *P < 0.05 versus the untreated control cells.
Among the restriction enzymes applied in our experiments, Sma I and BceA I are sensitive to DNA methylation, however the proximal promoter DNA of FGFR3 is hypomethylated even in untreated cells while the level of the basal FGFR3 transcription (shown in Figure 1D) indicated the relative active state of the FGFR3 promoter in the absence of BMP-2 treatment.
Based on the chromatin structure analysis of the FGFR3 promoter region by means of DNase I, MNase and restriction enzyme studies, we established a tentative map of positioned nucleosomes within the FGFR3 regulatory regions (Figure 5, bottom panel). Although the low-resolution methods used in our study do not pinpoint nucleosome positions on the nucleotide, the results showed that at least six nucleosomes are positioned across the core promoter. In untreated cells, the nuclease hypersensitive site is located in a histone-free region between two nucleosomes (Nuc+1 and Nuc–1). Upon BMP-2 treatment, the nucleosome (Nuc+1) just downstream of the nuclease hypersensitive site is removed, resulting in a promoter region with greater accessibility. Thus, we suppose that the remodeling of the nucleosome may contribute to BMP-2-induced FGFR3 expression.
Kinetics of FGFR3 promoter chromatin remodeling
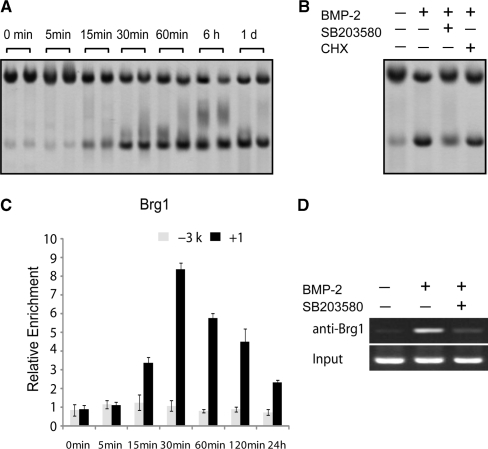
The kinetics of chromatin remodeling was next monitored. C3H10T1/2 cells were treated with BMP-2 for the indicated periods of time followed by restriction enzyme hypersensitivity analysis. An increase in nuclease accessibility was observed with ApaL I digestion as early as 15 min and became evident 30 min after BMP-2 addition. The high level of chromatin accessibility was sustained for the duration of the assay (Figure 6A). It was noticeable that chromatin remodeling slightly preceded the increase in FGFR3 mRNA (Figure 1B), suggesting that chromatin remodeling is a primary event in the transcriptional activation of FGFR3 gene expression.
Figure 6.
Kinetics of FGFR3 gene chromatin remodeling in C3H10T1/2. (A, B) C3H10T1/2 cells were incubated with 200 ng/ml BMP-2 with 5% FBS for the indicated periods of time (A), or pretreated with SB203580 (10 μM) and CHX (10 μM) respectively for 30 min, then cultured for 1 day in the presence of BMP-2 (200 ng/ml) (B). Nuclei were then purified and digested with 80 U of ApaL I, and purified DNA was digested to completion with Nco I. Products were detected by Southern blotting. (C) Chromatin from C3H10T1/2 was harvested 0, 5, 15, 30, 60, 120 min and 24 h after stimulation with 200 ng/ml BMP-2. The chromatin was precipitated with anti-Brg1 antibodies. After DNA recovery, the precipitates were evaluated by real-time PCR for the level of enrichment over the negative control antibody with the primers shown in Figure 8A. All the results are the means of three independent experiments ± standard deviation. (D) C3H10T1/2 cells were pretreated with control or SB203580 (10 μM) for 30 min and cultured for another 30 min in the presence or absence of BMP-2 (200 ng/ml). The chromatin was precipitated with anti-Brg1 antibodies, and the +1 region (shown in Figure 8A) was examined for binding of Brg1.
As BMP-2-induced immediate FGFR3 expression is independent of protein synthesis, we further analyzed the requirement for new protein synthesis in nucleosome remodeling. After pretreatment of C3H10T1/2 cells with the translation inhibitor CHX for 30 min before BMP-2 addition, the increase in restriction enzyme cleavage remained unchanged (Figure 6B). These results indicate that, new protein synthesis is not necessary for either FGFR3 transcription (Figure 1C) or for BMP-2-elicited chromatin remodeling at the FGFR3 promoter.
As shown in Figure 1, p38 MAPK is necessary for BMP-induced FGFR3 gene expression in C3H10T1/2 cells. To determine whether the inhibition of the p38 MAPK pathway has any effect on BMP-2-elicited nucleosome remodeling across the FGFR3 promoter, C3H10T1/2 cells were pretreated with SB203580 before stimulation with BMP-2 and subsequent digestion with ApaL I. The inhibition of the p38 MAPK pathway significantly abrogated the increase in restriction enzyme cleavage by BMP-2 (Figure 6B), indicating that the activation of the p38 pathway is essential for chromatin remodeling by BMP-2.
Changes in the accessibility of nucleosome-packaged DNA can be mediated by the SWI–SNF remodeling complex. To investigate whether the core SWI–SNF component Brg1 associates with the FGFR3 promoter during BMP-2-induced differentiation, ChIP was used. Two regions of the FGFR3 gene were evaluated by real-time PCR in order to determine the binding of the remodeling complex: the +1 region corresponding to both the transcription start site and the BMP-2-induced nuclease hypersensitive site, and the –3 kb region as a control (Figure 8A). Brg1 was observed at the +1 region as early as 15 min after BMP-2 addition, with peak detection occurring at 30 min, after which it began to decline but was still above the basal level 24 h after BMP-2 addition (Figure 6C). During the whole process, the remodeling complex was undetectable at the –3 kb region. These results suggest that the SWI–SNF remodeling complex may contribute to BMP-2-induced nucleosome remodeling at the FGFR3 promoter. Following on from the results shown in Figure 6B, we studied whether phosphorylation of p38 is responsible for the recruitment of the remodeling complex to the proximal promoter using the ChIP assay, and the resulting data demonstrated that SB203580 did indeed significantly inhibit binding of Brg1 to the FGFR3 promoter (Figure 6D).
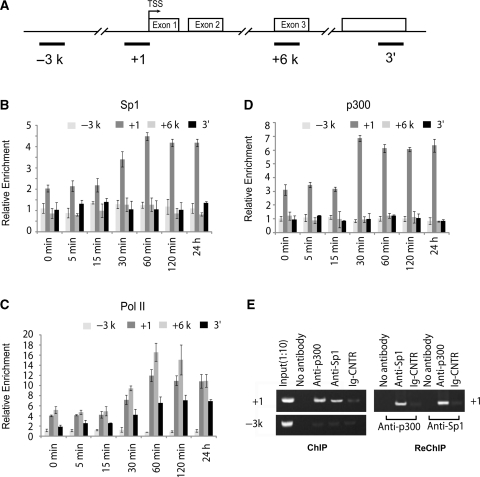
Figure 8.
Coactivator recruitment to the FGFR3 promoter. (A) The locations of the real-time PCR primer sets used for ChIP assays are shown (B, C and D). Chromatin from C3H10T1/2 cells was harvested 0, 5, 15, 30, 60, 120 min and 24 h after stimulation with 200 ng/ml BMP-2 with 5% FBS and precipitated with anti-Sp1 (B), anti-Pol II (C), and anti-p300 (D) antibodies. After DNA recovery, the precipitates were evaluated by real-time PCR for the level of enrichment over the negative control antibody. All the results are the means of three independent experiments ± standard deviation. (E) ChIP and Re-ChIP experiments performed with anti-Sp1 and anti-p300 antibodies on C3H10T1/2 cells with 200 ng/ml BMP-2 for 24 h.
BMP-2-induced binding of Sp1 to the nuclease hypersensitive site
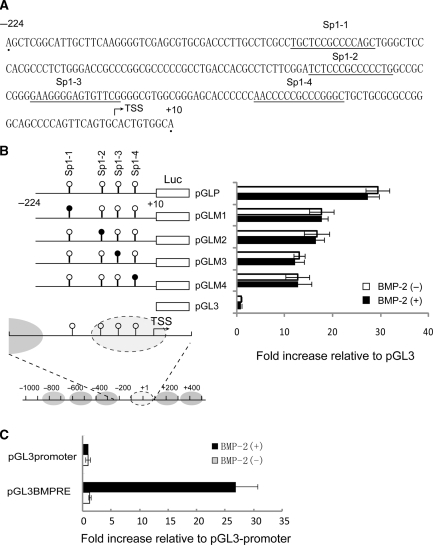
The existence of a nuclease hypersensitive site strongly indicates the binding of transcription factors as transcriptional regulators, prompting us to examine the potential role of the cis elements at this region. A search for transcription factor-binding sites using an online software program (MatInspector, http://www.genomatix.de/online_help/help_matinspector/matinspector_help.html) revealed four Sp1-binding sites immediately upstream of the transcription start site (Figure 7A). We found that, among the four Sp1 sites, Sp1-1 overlapped with the NHS under basal conditions, whereas Sp1-2, Sp1-3 and Sp1-4 were located in the BMP-2-induced NHS (Figure 7B bottom panel).
Figure 7.
Sp1-binding sites contribute to the basal activation of the FGFR3 gene promoter. (A) Location of putative Sp1-binding sites in the FGFR3 proximal promoter. (B) Transient transfection of C3H10T1/2 in the presence or absence of BMP-2 (200 ng/ml) with 5% FBS for 24 h was used to determine the transcriptional activity of chimeric construct pGLP, which included nts –224 and +10, as well as a series of 6-bp substitutions that were made within the context of pGLP (right panel). The diagram shows the structure of the FGFR3 promoter construct: the four putative Sp1-binding sites are indicated by circles; mutated Sp1 sites are shown as black circles (left panel). As a positive control, pGL3BMPRE was transiently transfected into C3H10T1/2 in the presence or absence of BMP-2 (200 ng/ml) for 24 h, followed by determination of luciferase activity (C). All the results are the means of three independent experiments ± standard deviation.
So, we determined the regulatory roles of these Sp1-binding sites by constructing a luciferase reporter containing the DNA fragment from the region spanning –224 to +10 (pGLP). After transient transfection of the reporter construct into C3H10T1/2 cells in the presence or absence of 200 ng/ml BMP-2, luciferase activities were determined. About a 30-fold increase in luciferase activity was observed with the –224/+10 fragment as compared with the control vector pGL3 (Figure 7B) in untreated cells. In order to verify the contribution of each Sp1-binding site to the overall activation, we generated mutant reporter plasmids in which the Sp1-binding sites were mutated by inserting the EcoR I restriction site GAATTC. Upon transient transfection into C3H10T1/2 cells, luciferase activities were determined, and it was found that mutation of the Sp1 sites suppressed the basal activity by 53% (Sp1-1), 55% (Sp1-2), 39% (Sp1-3) and 35% (Sp1-4) as compared with the wild-type construct (Figure 7B). These data indicate that all the Sp1-binding sites contribute to the basal promoter activity. Surprisingly, no apparent inducible activity by BMP-2 treatment was detected in any of the constructs. As a positive control, three tandem repeats of a standard BMP-2 responsive element (BMPRE) (GCCGCGCCGGCTCCAGGGCAGGAGCGGC) were inserted into the multiple cloning sites upstream of a core promoter and the luciferase gene in a pGL3-promoter vector (pGL3BMRE). Smad1 and Smad5 directly bound to the BMPRE and stimulated its transactivity. After transient transfection into C3H10T1/2 followed by BMP-2 induction for 24 h, the expression level of luciferase increased significantly as compared with that of untreated cells, suggesting that our system works well.
The interaction between Sp1 and the FGFR3 core promoter in vivo was subsequently determined by ChIP analysis using the antibody against Sp1. Although the distance between the Sp1-1 and Sp1-4 sites is smaller than the average chromatin fragments produced during sonication (300–1000 bp), and no suitable primer is available to discriminate the binding of Sp1 protein to different binding sites, primers correlating with the +1 region were used to detect Sp1 binding to all the sites. Additionally, the –3k region and another two primer sets were used for real-time PCR in order to evaluate the results of the ChIP: +6k corresponds to an area in the third exon; the 3′ region is a part of the 3′UTR contained in the last exon (Figure 8A). As shown in Figure 8B, in untreated C3H10T1/2 cells, Sp1 protein is present at the +1 region, strongly suggesting its role in regulating basal transcription of FGFR3. In contrast with the results of the luciferase reporter assay (Figure 7B), the ChIP results showed that 30 min after BMP-2 induction, binding of Sp1 protein began to increase with the peak level reached at 60 min, and remained stably bound to the promoter (Figure 8B).
Collectively, our data suggest a possible mechanism whereby Sp1 regulates FGFR3 expression: under basal conditions, the Sp1-1 site is highly accessible to Sp1 protein, resulting in basal transcription; upon BMP-2 treatment, the nucleosome occupying the +1 region is removed, thus exposing the previously masked Sp1-2, 3 and 4 sites, and the initiation site; more Sp1 proteins are recruited to the core promoter, leading to the upregulation of FGFR3 expression level.
Ordered recruitment of histone acetyltransferase and basal transcription factors to the FGFR3 core promoter in vivo
As the removal of the nuc +1 from the chromatin exposed the TSS, we performed a ChIP assay to confirm BMP-2-enhanced recruitment of RNA polymerase II to the start site. Consistent with the kinetics of FGFR3 transcription, RNA polymerase was observed under basal conditions, and enrichment began to increase 30 min after BMP-2 treatment, peaking at 1 h, and was maintained at a high level throughout the remaining period, although a moderate decrease was detected. Association of Pol II appeared to be maximal in the +1 and +6 kb regions and undetectable in the –3 kb region at any point (Figure 8C).
It has been previously reported that the HAT p300 enzyme binds Sp1 target promoters, and that its binding correlates with the presence of highly acetylated histones (21). From the results obtained thus far, we hypothesized that BMP-enhanced Sp1 binding by remodeling of the nucleosome might control the transcription of the FGFR3 gene through local recruitment of histone-modifying enzymes. Therefore, we performed experiments to verify whether Sp1 might recruit the p300 enzyme onto FGFR3 promoter sequences and consequently allow specific recruitment of acetylated histones. The ChIP results showed that p300 is present in the +1 region of the FGFR3 gene in untreated C3H10T1/2 cells (Figure 8D). An increase was observed only 30 min after BMP-2 induction, and at the same time, BMP-2 enhanced the binding of Sp1 to the same region in vivo (Figure 8D and 7B). Additionally, p300 and Sp1 shared a similar kinetic pattern of FGFR3 promoter association, indicating the local recruitment of p300 by Sp1 protein under both basal and BMP-2-induced conditions.
Next, we performed a Re-ChIP assay in order to confirm whether Sp1 and p300 are co-recruited onto the proximal promoter. In Re-ChIP analysis, we immunoprecipitated the Sp1- or p300-containing complexes with antibodies against p300 or Sp1, respectively, and only those DNA sequences that are simultaneously bound by both proteins would be amplified in the subsequent PCR. Our results showed that p300 and Sp1 were co-recruited on the same DNA fragments (Figure 8E), suggesting that the two factors co-occupy common target loci. These results strongly support a direct role of Sp1 in p300 recruitment to the FGFR3 promoter, leading to the activation of epigenetic markers at the same location, as described in the next paragraph.
Dynamics of histone modifications at the FGFR3 gene
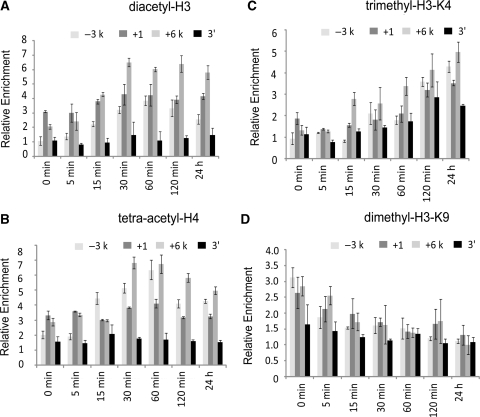
Acetylation has long been known to be associated with actively transcribed genes and open chromatin configurations. Antibodies specific to diacetylated H3 (K9 and K14) and tetraacetylated H4 (K5, K8, K12 and K16) were used in ChIP analysis to determine the pattern of histone modification upon BMP-2 exposure. In untreated cells, both H3 and H4 were acetylated at all the testing areas, with the lowest level in the 3′ extreme region. Upon treatment, histone acetylation began to modestly increase at 15 min post-induction and reached a high level at 30 min, which was sustained throughout the remaining time (Figure 9A and B). Interestingly, the histone acetylation was preferentially restricted to the 5′ region of the gene, whereas the 3′ extreme region showed a small induction of acetylation. As for histone methylation, trimethylH3K4 (a euchromatic marker) was not apparently scored under basal conditions, and increased somewhat slowly, reaching a peak at 1 day (Figure 9C). As for the heterochromatin marker dimethylH3K9, which serves as a signal for chromatin silencing by recruiting the HP1 protein (heterochromatin protein 1) and is mutually exclusive with H3–K9 acetylation, it was measurable before BMP-2 treatment, then began to drop slowly and moderately to background levels at 24 h (Figure 9D). Taken together, these results indicate that transcription activation of FGFR3 is correlated with an increase in the local recruitment of euchromatic markers and a decrease in that of the heterochromatic marker.
Figure 9.
Changes in histone acetylation and methylation upon BMP-2 treatment. Kinetic ChIP analysis of C3H10T1/2 cells as in Figure 8, with antibodies directed against diacetyl–H3 (A), tetra–acetyl–H4 (B), trimethyl–H3–K4 (C) and dimethyl–H3–K9 (D). The results shown are expressed as enrichment over the negative control antibody. All the results are the means of three independent experiments ± standard deviation.
DISCUSSION
In our study, we first identified the mechanism by which BMP-2 regulates the expression of FGFR3. The novel insight provided is 3-fold: first, Sp1-binding sites contribute to both the basal and BMP-2-enhanced activation of the FGFR3 promoter in C3H10T1/2 cells; second, BMP-2-induced dynamic alteration of nucleosome positioning in the proximal promoter was determined and third, the ordered recruitment of various nuclear factors and the covalent modification of histones in the FGFR3 promoter in response to BMP-2 treatment were examined in detail.
By comparing the timing of recruitment of Brg1, Sp1, p300 and Pol II with histone modification and chromatin remodeling, we are able to provide a nearly complete picture of the molecular mechanism that determines BMP-2-induced FGFR3 expression. Under basal conditions, several nucleosomes are found in the promoter, except in an area localized in the linker region between Nuc–1 and Nuc+1. The association of Sp1 protein with its binding site (Sp1-1) contained in this nucleosome-free region may result in the recruitment of histone acetyltransferases (such as p300) to the promoter, leading to basal acetylation of the histone tail and maintaining the open configuration of the local chromatin. Furthermore, such events occurring at the proximal promoter provide chromatin accessibility to polymerase II to initiate the transcription of the FGFR3 gene in a persistent manner, although at a relatively low level.
Upon BMP-2 treatment, the SWI/SNF complex was first found to be associated specifically with the proximal promoter region after only 15 min of induction. Although such binding is in a transient manner—reaching a peak at 30 min, then declining quickly—it is sufficient to yield a state of open configuration of the local chromatin through releasing Nuc+1 from the transcription start site. This primes for a rapid increase in the FGFR3 transcription level by exposing the Sp1-binding sites (Sp1-2, Sp1-3 and Sp1-4). An increase in Sp1 binding to the promoter is apparent at 30 min after BMP-2 treatment, and it can further recruit p300. The putative p300 and Sp1 complex may upregulate the H3 and H4 acetylation levels that are responsible for the initiation and maintenance of enhanced local chromatin accessibility to the general transcription apparatus, upregulating the FGFR3 expression level.
SWI/SNF complexes have been shown to have important functions in development and differentiation in many tissues, organs and cells (22), but their functions in osteogenic or chondrogenic differentiation are not well understood. In a previous study (23), the expression of several components of the SWI/SNF complex was reported to be upregulated during BMP-2-mediated osteogenesis in C2C12 cells. These findings also demonstrate that initiation of expression of several BMP-2-induced, Runx2-dependent skeletal genes requires SWI/SNF complexes. In another work (24), Villagra et al. reported that transcription of osteocalcin (a bone-specific gene that is expressed in the late stages of osteoblastic differentiation) involves an active chromatin-remodeling process mediated by ATP-dependent SWI/SNF activity that is specifically recruited to the proximal promoter region. In our study, we found that BMP-2 is capable of increasing the binding of the remodeling complex to the FGFR3 promoter in C3H10T1/2 cells. This recruitment of the SWI/SNF complex is a rapid response to BMP-2 induction, beginning at 15 min and reaching a peak at 30 min, consistent with the fast kinetics of FGFR3 expression upon induction (Figure 6C). But the mechanism whereby BMP-2 plays its role in this process remains to be elucidated. Our data suggest that at least two features may ensure the rapid recruitment of the remodeling complex. First, different from the study reported by Yong et al. (23), BMP-2-induced binding of the complex is independent of the upregulation of the expression level of the remodeling complex or other proteins, as this process is insensitive to CHX treatment. Second, it is well established that acetylated chromatin is the preferred substrate for SWI/SNF recruitment. Brg1 bears a bromodomain, which can interact with acetylated histone N-termini, suggesting that histone acetylation provides a higher-affinity surface for interaction with the SWI/SNF complex, thus leading to more stable nucleosomal binding. Under basal conditions, histones surrounding the proximal promoter of the FGFR3 gene are acetylated to a certain degree. We believe that basal acetylation may ‘pre-activate’ the histones to be ready for binding the remodeling complex, enabling the rapid binding of the SWI/SNF complex to the promoter, shortening the time taken to react to BMP-2 induction.
Another key issue is how the SWI/SNF complex is targeted to the specific promoter. Our results demonstrate that recruitment of the complex is dependent on p38 activation (inhibited by SB203580), which can phosphorylate several downstream targets (Figure 1D, 6B and D). The simplest possibility is that activated p38 may phosphorylate a sequence-specific transcription factor required for the cooperative recruitment of the SWI/SNF complex to the FGFR3 proximal promoter. SWI/SNF components have been shown to interact with several transcriptional activators, including nuclear steroid receptors (25), human heat shock factor 1 (26), EKLF (27), c-Myc (28), C/EBPβ (29) and C/EBPα (30); however, only a few transcription factors have been demonstrated to target the SWI/SNF complex to specific cellular promoters (31,32). We do not think that Sp1 is the candidate transcription factor, as BMP-2-induced Sp1 recruitment is preceded by Brg1 recruitment; however, the binding sites for other transcription factors not yet identified in the FGFR3 promoter region may contribute to BMP/p38-dependent recruitment of a partner of the SWI/SNF complex.
Interestingly, the remodeling complex is recruited to the promoter in a transient way, beginning at 15 min and decreasing to nearly background levels; in contrast, the BMP-2-induced FGFR3 transcription level may be maintained for at least 6 days (data not shown). So, we conclude that once the nucleosome has been removed from the proximal promoter, the remodeling complex is no longer required. Instead, other mechanisms such as chromatin modification may be responsible for maintaining the local chromatin accessibility. We observed an increase in the local recruitment of euchromatic markers and a decrease in that of the heterochromatic marker. The acetylation of the histone tails disrupts and interferes with the higher order chromatin folding, promotes the solubility of chromatin at physiological ionic strength and maintains the unfolded structure of the transcribed nucleosome, allowing transcription factor binding (33,34). At 30 min after BMP-2 induction, dramatic increases in H3 and H4 acetylation level, as well as Sp1 and p300 recruitment, were observed, indicating the possibility that p300 is responsible for histone acetylation (Figure 7B, 9A and B). The increase in trimethylation of H3-K4 (an euchromatic marker) lagged behind that of histone acetylation, reaching the peak level at 1 day (Figure 9C). We believe that this slow dynamic pattern may partially compensate for the decrease in H3 and H4 acetylation level at a late stage to maintain the unfolded chromatin structure.
In our experiment performed in order to analyze histone modification, two points should be noted:
Histone acetylation began to increase modestly at 15 min post-induction, preceding the recruitment of p300, suggesting that histone acetylation, at least at the early stage, is independent of p300. A previous report (35) indicated that p38 MAPK, which is strongly activated by LPS, is responsible for the phosphoacetylation of histone H3 on promoters of a subset of stimulus-induced cytokine and chemokine genes by enhancing the accessibility of the NF-κB-binding sites. So, we hypothesized that activation of the BMP/p38 pathway could improve the local histone acetylation level, which may facilitate the binding of the SWI/SNF complex, as discussed above. Our further study will focus on the relationships between p38, the SWI/SNF complex and histone acetylation at the FGFR3 proximal promoter region.
The +1 region that we expected to be the most prominent area in histone modification showed only modest alteration, and the strongest positivity was found in the +6k region. This is surprising, as the +1 region was where the remodeling complex, as well as Sp1 and p300, bound to chromatin and contributed most to the promoter activity. One important possibility cannot be ruled out: BMP-2-induced depletion of Nuc+1 from the +1 region may be responsible for the decreased level of histone modification, as previously reported (36,37).
The nuclear coactivator p300 is a transcriptional adaptor for many DNA-binding activators; it possesses intrinsic acetyltransferase activity, which, by chemically modifying histone tails, affects the nucleosomal environment and transcription (38). The general transcription factor Sp1, which binds and interacts with GC boxes in the promoter regions of target genes, is capable of activating the transcription of some genes. It has previously been reported the Sp1 and p300 cooperate to transactivate the expression of the embryonic globin gene (39), 12(s)-lipoxygenase (21), p21 (40) and TβRII (41). Hung et al. reported that Sp1 physically interacts with p300, and recruits p300 to the target promoter, leading to a chromatin modification that induces the transcription activity of the 12(s)-lipoxygenase gene (21); Suzuki et al. demonstrated physical and functional interaction between the acetyltransferase region of p300 and the DNA-binding domain (DBD) of the transcription factor Sp1 (42); and in another work, p300 was found to indirectly interact with Sp1 through progesterone receptors (43). In our study, the ChIP results demonstrated that p300 and Sp1 share a similar kinetic pattern of FGFR3 promoter association, and both are co-recruited on the same DNA fragments, strongly supporting a direct role of Sp1 in p300 recruitment to the FGFR3 promoter. However, is it the case that p300 directly interacts with Sp1, or that they indirectly interact through a common multi-protein complex? Further study is needed in order to gain a detailed picture of the cooperation of p300 and Sp1 uncovered in our study.
Furthermore, apart from its HAT activity, the p300 protein may act as a bridging factor to connect sequence-specific transcription factors to the basal transcription machinery (41). Sp1 is also known to interact with the N terminus of the TATA-binding protein-associated factor dTAF110 in the TFIID complex, and this interaction may account for the ability of Sp1 to assemble the pre-initiation complex to activate transcription from TATA-less promoters (44). Therefore, we believe that both Sp1 and p300 are responsible for the recruitment of Pol II to the FGFR3 promoter to activate its expression under basal and BMP2-induced conditions.
In this study, when we confined the BMP-2-upregulated nuclease hypersensitive site to a more precise region, a conventional method, luciferase reporter assay, was used to delineate the promoter: after construction and transient transfection of the luciferase reporter containing the proximal promoter, the reporter plasmids showed the proper basal promoter function, but exhibited no inducible activity upon BMP-2 treatment. We think that, unlike stably transfected plasmids or episomal constructs, the chromatin structure of transiently transfected plasmids is usually incomplete or aberrant, and may vary with cell type and the transfection method used. We postulate that in our transient transfection experiment, the proximal promoter region had not been packaged into a chromatin-like structure, and may even have been in a ‘naked’ state, and as a result, Sp1-2, 3 and 4-binding sites and the TSS may not have been occupied by nucleosomes, leading to high accessibility to various nuclear factors, including the basal transcription apparatus. Hence, BMP-2 induction cannot exert its regulatory role through remodeling the chromatin structure, and upregulated promoter activity was undetectable in our luciferase reporter assay. Therefore, it is important that the promoter activity is studied in its native chromatin context.
In conclusion, in this work we demonstrated that BMP-2 upregulates the FGFR3 expression level by controlling the local chromatin structure through the recruitment of the remodeling complex, which in turn initiates a cascade of molecular events to increase the chromatin accessibility for the general transcription apparatus.
FUNDING
National Natural Science Foundation of China (grant numbers 30400270, 30600328 and 30740030); the National 973 Program (grant number 2001CB510106); the Natural Science Foundation of Guangdong (grant number 32204018). Funding for open access charge: National 973 Program of China (grant number 2001CB510106) and the Third Phase of the National 211 Project.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Sarah Bangs (University of Oxford) for critically reading the article.
REFERENCES
- 1.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat. Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J. Clin. Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwata T, Chen L, Li C, Ovchinnikov DA, Behringer RR, Francomano CA, Deng CX. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum. Mol. Genet. 2000;9:1603–1613. doi: 10.1093/hmg/9.11.1603. [DOI] [PubMed] [Google Scholar]
- 4.Segev O, Chumakov I, Nevo Z, Givol D, Madar-Shapiro L, Sheinin Y, Weinreb M, Yayon A. Restrained chondrocyte proliferation and maturation with abnormal growth plate vascularization and ossification in human FGFR-3(G380R) transgenic mice. Hum. Mol. Genet. 2000;9:249–258. doi: 10.1093/hmg/9.2.249. [DOI] [PubMed] [Google Scholar]
- 5.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 6.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 7.Davidson D, Blanc A, Filion D, Wang H, Plut P, Pfeffer G, Buschmann MD, Henderson JE. Fibroblast growth factor (FGF) 18 signals through FGF receptor 3 to promote chondrogenesis. J. Biol. Chem. 2005;280:20509–20515. doi: 10.1074/jbc.M410148200. [DOI] [PubMed] [Google Scholar]
- 8.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 9.Ellsworth JL, Berry J, Bukowski T, Claus J, Feldhaus A, Holderman S, Holdren MS, Lum KD, Moore EE, Raymond F, et al. Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthr. Cartilage. 2002;10:308–320. doi: 10.1053/joca.2002.0514. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Green RP, Zhao G, Ornitz DM. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development. 2001;128:3867–3876. doi: 10.1242/dev.128.19.3867. [DOI] [PubMed] [Google Scholar]
- 11.Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev. Cell. 2002;3:439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 12.De Luca F, Barnes KM, Uyeda JA, De-Levi S, Abad V, Palese T, Mericq V, Baron J. Regulation of growth plate chondrogenesis by bone morphogenetic protein-2. Endocrinology. 2001;142:430–436. doi: 10.1210/endo.142.1.7901. [DOI] [PubMed] [Google Scholar]
- 13.Shukunami C, Ohta Y, Sakuda M, Hiraki Y. Sequential progression of the differentiation program by bone morphogenetic protein-2 in chondrogenic cell line ATDC5. Exp. Cell Res. 1998;241:1–11. doi: 10.1006/excr.1998.4045. [DOI] [PubMed] [Google Scholar]
- 14.Wang EA, Israel DI, Kelly S, Luxenberg DP. Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors. 1993;9:57–71. doi: 10.3109/08977199308991582. [DOI] [PubMed] [Google Scholar]
- 15.Haas AR, Tuan RS. Murine C3H10T1/2 multipotential cells as an in vitro model of mesenchymal chondrogenesis. Methods Mol. Biol. 2000;137:383–389. doi: 10.1385/1-59259-066-7:383. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann A, Czichos S, Kaps C, Bachner D, Mayer H, Kurkalli BG, Zilberman Y, Turgeman G, Pelled G, Gross G, et al. The T-box transcription factor Brachyury mediates cartilage development in mesenchymal stem cell line C3H10T1/2. J. Cell Sci. 2002;115:769–781. doi: 10.1242/jcs.115.4.769. [DOI] [PubMed] [Google Scholar]
- 17.McEwen DG, Ornitz DM. Regulation of the fibroblast growth factor receptor 3 promoter and intron I enhancer by Sp1 family transcription factors. J. Biol. Chem. 1998;273:5349–5357. doi: 10.1074/jbc.273.9.5349. [DOI] [PubMed] [Google Scholar]
- 18.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl Acad. Sci. USA. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden JM, Strong DD, Baylink DJ, Powell DR, Sampath TK, Mohan S. Osteogenic protein-1 stimulates production of insulin-like growth factor binding protein-3 nuclear transcripts in human osteosarcoma cells. Endocrinology. 1997;138:4240–4247. doi: 10.1210/endo.138.10.5457. [DOI] [PubMed] [Google Scholar]
- 20.Escamilla-Del-Arenal M, Recillas-Targa F. GATA-1 modulates the chromatin structure and activity of the chicken alpha-globin 3' enhancer. Mol. Cell Biol. 2008;28:575–586. doi: 10.1128/MCB.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 23.Young DW, Pratap J, Javed A, Weiner B, Ohkawa Y, van Wijnen A, Montecino M, Stein GS, Stein JL, Imbalzano AN, et al. SWI/SNF chromatin remodeling complex is obligatory for BMP2-induced, Runx2-dependent skeletal gene expression that controls osteoblast differentiation. J. Cell Biochem. 2005;94:720–730. doi: 10.1002/jcb.20332. [DOI] [PubMed] [Google Scholar]
- 24.Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, et al. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. J. Biol. Chem. 2006;281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- 25.Ostlund Farrants AK, Blomquist P, Kwon H, Wrange O. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol. Cell Biol. 2001;21:5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 28.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 29.Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 32.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 33.Spencer VA, Davie JR. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 34.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 36.Donati G, Imbriano C, Mantovani R. Dynamic recruitment of transcription factors and epigenetic changes on the ER stress response gene promoters. Nucleic Acids Res. 2006;34:3116–3127. doi: 10.1093/nar/gkl304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim A, Song SH, Brand M, Dean A. Nucleosome and transcription activator antagonism at human beta-globin locus control region DNase I hypersensitive sites. Nucleic Acids Res. 2007;35:5831–5838. doi: 10.1093/nar/gkm620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 39.Dempsey NJ, Ojalvo LS, Wu DW, Little JA. Induction of an embryonic globin gene promoter by short-chain fatty acids. Blood. 2003;102:4214–4222. doi: 10.1182/blood-2002-12-3766. [DOI] [PubMed] [Google Scholar]
- 40.Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
- 41.Huang W, Zhao S, Ammanamanchi S, Brattain M, Venkatasubbarao K, Freeman JW. Trichostatin A induces transforming growth factor beta type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1.NF-Y complex. J. Biol. Chem. 2005;280:10047–10054. doi: 10.1074/jbc.M408680200. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki T, Kimura A, Nagai R, Horikoshi M. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells. 2000;5:29–41. doi: 10.1046/j.1365-2443.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- 43.Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 44.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl Acad. Sci. USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]