Abstract
Rad51C is a central component of two complexes formed by five Rad51 paralogs in vertebrates. These complexes are involved in repairing DNA double-strand breaks through homologous recombination. Despite accumulating evidence suggesting that the paralogs may prevent aneuploidy by controlling centrosome integrity, Rad51C's role in maintaining chromosome stability remains unclear. Here we demonstrate that Rad51C deficiency leads to both centrosome aberrations in an ATR-Chk1-dependent manner and increased aneuploidy in human cells. While it was reported that Rad51C deficiency did not cause centrosome aberrations in interphase in hamster cells, such aberrations were observed in interphase in HCT116 cells with Rad51C dysfunction. Caffeine treatment and down-regulation of ATR, but not that of ATM, reduced the frequency of centrosome aberrations in the mutant cells. Silencing of Rad51C by RNA interference in HT1080 cells resulted in similar aberrations. Treatment with a Chk1 inhibitor and silencing of Chk1 also reduced the frequency in HCT116 mutants. Accumulation of Chk1 at the centrosome and nuclear foci of γH2AX were increased in the mutants. Moreover, the mutant cells had a higher frequency of aneuploidy. These findings indicate that the ATR-Chk1 pathway plays a role in increased centrosome aberrations induced by Rad51C dysfunction.
INTRODUCTION
Homologous recombination, along with nonhomologous end-joining, plays a major role in the repair of DNA double-stranded breaks (DSBs) (1). Rad51 is a key player in homologous recombination by exerting homologous pairing and strand exchange activities. Rad51 paralogs are assumed to be involved in the early stages of homologous recombination by assisting Rad51 function (2). Five members of the Rad51 paralog family constitute two protein complexes: Rad51B-Rad51C-Rad51D-XRCC2 (BCDX2) and Rad51C-XRCC3 (3,4). Thus, Rad51C is a central component among five members in vertebrates.
The centrosome serves as the microtubule-organizing center, ensuring correct chromosome segregation to prevent aneuploidy (5). Accumulating evidence suggests that centrosome dysfunction, typically represented by abnormal numbers of centrosomes, is involved in human diseases, particularly in cancers (6). More than 100 proteins have been reported to be localized in the centrosome (7). Deletion of these proteins often leads to centrosome aberrations.
Mutations of XRCC2, XRCC3, Rad51B and Rad51D were shown to increase centrosome fragmentation and aneuploidy (8–10). Despite these observations, the role of Rad51C in the maintenance of centrosome integrity and chromosome stability remains unclear. Initially, Rad51C-deficient Chinese hamster ovary (CHO) cells, CL-V4B, were shown to exhibit no increase in centrosome aberrations (11). A recent study, however, demonstrated that centrosome numbers were increased only in mitosis and not in interphase in CL-V4B cells (12). Moreover, although increased numbers of centrosomes are assumed to generate aneuploidy, no studies using mammalian cells have demonstrated that Rad51C deficiency leads to increased aneuploidy.
The mechanisms underlying centrosome aberrations observed in cells with a defect in homologous recombination are controversial. In chicken DT40 cells with a conditional mutation of Rad51, the ATM-dependent checkpoint pathway was proposed to be responsible for centrosome amplification at the G2 phase (13). However, the results of a study using CHO cells with the dominant-negative Rad51 protein argued against this result (14). The hereditary breast cancer susceptibility protein BRCA1 is also involved in homologous recombination. Recent evidence suggests that HMMR, encoding the hyaluronan-mediated motility receptor, is a substrate of BRCA1-BARD1 E3 ubiquitin ligase activity and plays a role in centrosomal function (15).
Supernumerary centrosomes induced by ionizing radiation were shown to be caused by the Chk1-mediated pathway, indicating that the DNA damage response signal is involved in centrosome amplification (16). Treatment with caffeine, an inhibitor of ATM and ATR kinases, reduced centrosome amplification induced by ionizing radiation, suggesting that either or both kinases may be involved in centrosome amplification. However, caffeine treatment in ATM- or ATR-deficient cells also reduced centrosome amplification. Thus, the roles of ATM and ATR in promoting centrosome amplification induced by ionizing radiation appear to be complementary.
To investigate Rad51C's role in the maintenance of chromosome stability, we knocked out the gene in the human colon cancer cell line HCT116. We also silenced the gene by RNA interference in the human fibrosarcoma cell line HT1080. Supernumerary centrosomes in these cells with Rad51C dysfunction were increased at both interphase and metaphase in an ATR-Chk1-dependent manner. Consistent with this observation, aneuploidy was increased in HCT116 cells with Rad51C dysfunction. Our observations suggest that the ATR-Chk1 pathway plays a role in increased centrosome aberrations induced by Rad51C dysfunction in human cancer cells.
MATERIALS AND METHODS
Cell culture
HCT116 cells were cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS). HT1080 cells were cultured in the minimum essential medium Eagle (MEM) supplemented with 10% FBS. These cells were obtained from the American Type Culture Collection. 2-Morpholin-4-yl-6-thianthren-1-yl-pyran-4-one (KU55933) and 7-hydroxystaurosporine (UCN-01) were purchased from Merck Calbiochem.
Generation of Rad51C mutant cells
The 8-kb left arm and 1.5-kb right arm of the RAD51C genomic fragments were isolated from the EMBL3 SP6/T7 human genomic library (Clontech) and inserted into pBluescript SK for neomycin and blasticidin selection. The shorter genomic fragment was isolated from the genomic DNA of HCT116 by PCR amplification for puromycin selection. The 2.4-kb left arm was amplified using primers 5′-TTTGGCTGCTCCGGGGTTA-3′ and 5′-CAGAGTTTCTAAGGCTTCTGC-3′. The 1.5-kb right arm was amplified using primers 5′-CTCGAGTGGAGTGCCCTTAAT-3′ and 5′-CTCGAGTCAACAGAAAGATGAC-3′. Promoterless neomycin-, blasticidin- and puromycin-resistant genes were inserted into the Dra I site in exon 2. Gene targeting in HCT116 was carried out as previously described (17).
Complementation experiments
The RAD51C cDNA was amplified from normal human cDNA using primers 5′-TTTGGCTGCTCCGGGGTTA-3′ and 5′-CATTCATGCCATAGTGTG-3′, and cloned into pCR2.1 (Invitrogen) by the TA cloning method. After the sequence was confirmed, the cDNA was inserted into an expression vector under the control of the MSV enhancer and the MMTV promoter. The generation of the XRCC3 expression vector has been described earlier (18).
Sensitivity to DNA-damaging agents
After 1 × 103 cells were cultured in a 60 mm dish for 24 h, they were exposed to mitomycin C (MMC) (Kyowa Hakko Kogyo) for 10 min, hydroxyurea (HU) (Sigma-Aldrich) for 24 h or γ-ray irradiation. They were further cultured for 9 days, and the colonies were counted.
Sister chromatid exchange (SCE) and gene targeting frequency
SCEs were analyzed essentially as previously described (17). Cells were cultured in 16 μM 5-bromodeoxyuridine (BrdU) for 36 h (wild type), 42 h (RAD51C+/−/−/−) or 41 h (RAD51C+/−/−/− + cDNA), and pulsed with 0.1 μg/ml colcemid for the last 1 h. To examine the effect of MMC, cells were incubated in the presence of 0.8 μg/ml MMC for 8 h before harvest. Harvested cells were treated with 75 mM KCl:1% sodium citrate (4:1) for 20 min and fixed with methanol:acetic acid (3:1). Cells were fixed on slides and incubated with 12.8 μg/ml Hoechst 33258 for 20 min. The slides were irradiated with ultraviolet for 1 h at 60°C and stained with Giemsa solution. Gene targeting frequencies at the RAD54B locus were examined as previously described (17).
Antibodies
Anti-ATM (5C2), ATR (N-19), Chk1 (G-4 and DCS-310) and Cdk2 (M2) antibodies were from Santa Cruz Biotechnology. Anti-γ-tubulin (T3559 and T6557) and β-actin (A1978) antibodies were from Sigma-Aldrich. The anti-XRCC3 antibody (NB100-165) was from Novus Biologicals. Anti-phospho-H2AX Ser 139 (07-164) and Chk2 (05-649) antibodies were from Upstate Biotechnology. The anti-Chk2 antibody (DCS-273) was from Medical & Biological Laboratories. The anti-Rad51B antibody was obtained from rabbits immunized with the recombinant Rad51B protein (19). The anti-Rad51C antibody (ab3669) was from Abcam.
Immunofluorescence
Cells grown on coverslips were fixed for 10 min in 4% paraformaldehyde. Then, they were permeabilized with 0.5% Triton X-100 and 0.1% SDS/PBS for 5 min. Cells were blocked with 10% horse serum at 4°C for 30 min and incubated with primary antibodies at 37°C for 1 h and with secondary antibodies for 30 min. Finally, cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted. Slides were analyzed using Olympus BX51-DP70, Carl Zeiss AxioImagerZ1/Apotome and Nikon Confocal Laser Microscope system A1.
Fluorescence in situ hybridization analysis
Chromosome-specific centromeric probes were obtained from Vysis. DNA in cells was denatured in 70% formamide/2× SSC at 73°C for 5 min. Hybridization was performed in a CEP hybridization buffer (Vysis) at 42°C for 1 h. Slides were washed in 0.4× SSC/0.3% NP40 at 73°C for 2 min and in 2× SSC/0.1% NP40 at room temperature for 1 min. Cellular DNA was stained with DAPI.
RNA interference
siCONTROL, Rad51C siGENOME set of 4, ATM SMARTpool, ATR SMARTpool, Chk1 SMARTpool and Chk2 SMARTpool small interference RNAs (siRNAs) were purchased from Dharmacon. Cells were transfected with 100 nM siRNA and Dharmafect4 (Dharmacon) according to the manufacturer's instructions. Cells were harvested at 72 h post-transfection.
Cell-cycle analysis
Cells treated with siRNAs were harvested 72 h after transfection. EPICS XL (Beckman Coulter) was used for cell-cycle analysis.
Kinase assay
Cells were dissolved into a lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Nonidet P40, 10% glycerol and 2 mM EDTA) containing phosphatase inhibitors (50 nM cantharidin, 5 nM microcystin and 25 mM bromotetramisole oxalate), 2 mg/ml aprotinin and 0.5 mM PMSF for 15 min on ice and lysed by sonication. After centrifugation (12 000 rpm, 5 min), the supernatant was collected and precleared by incubation with normal IgG for 1 h and Protein G for 30 min. Precleared lysate was incubated with anti-Chk1 or anti-Chk2 antibody for 1 h. After incubation with Protein G, immunoprecipitates were washed three times each with the lysis buffer and 25 mM Hepes. The washed precipitate was incubated in a kinase reaction buffer (25 mM Hepes pH 7.4, 15 mM MgCl2, 80 mM EGTA and 1 mM DTT) with 0.1 mM ATP, 3 μCi[γ-32P]ATP and 10 μg glutathione S-transferase (GST)-Cdc25C (200–256) at 30°C for 30 min. The reaction was stopped by the addition of the sample buffer and subjected to SDS-PAGE. GST-Cdc25C (200–256) was prepared as previously described (20).
RESULTS
Rad51C is involved in homologous recombination in human cells
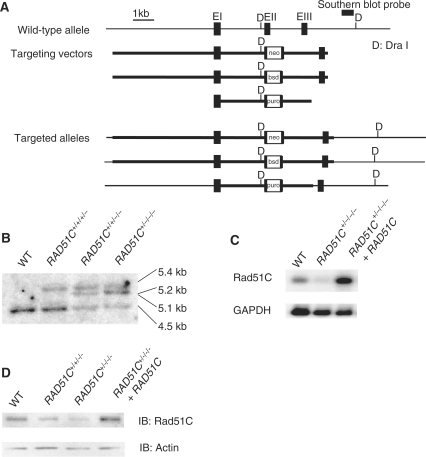
To investigate the role of Rad51C in human cells, the gene was sequentially knocked out in HCT116 cells (Figure 1A). In accordance with the finding that RAD51C was frequently amplified in human cancer cells, Southern blot analysis demonstrated that this cell line harbors four alleles of the gene (Figure 1B) (21,22). Two independent RAD51C+/−/−/− cell lines (#1 and #2) were isolated. The levels of Rad51C expression in proportion to targeting events were confirmed by northern and western blot analyses (Figure 1C and D). These cells were used in function analysis as Rad51C mutant cells because clear phenotypes indicating defective homologous recombination in comparison with wild-type HCT116 cells were observed. Complementation experiments were performed using cells transfected with the exogenous Rad51C expression vector.
Figure 1.
Generation of Rad51C mutant cells by gene targeting. (A) Strategy for gene targeting at the RAD51C locus. The targeting vectors, restriction sites and the position of the probe used for the Southern blot analysis are shown. (B) Southern blot analysis of DNA digested with Dra I. The size of the wild-type allele is 4.5 kb. Insertion of drug-resistance genes changes the size to 5.1, 5.2 and 5.4 kb. (C) Northern blot analysis of polyadenylated RNAs hybridized with the full-length RAD51C cDNA. (D) Western blot analysis. The Rad51C protein was immunoprecipitated with the rabbit polyclonal antibody and visualized by western blot analysis using the mouse monoclonal antibody.
The mean of the frequency of SCE induced by MMC per cell was 5.8 in the Rad51C mutant cells, a value that was significantly lower than 8.3, the mean value for wild-type cells (Table 1). The SCE frequency increased to a level comparable to that of the wild-type cells in cells transfected with the Rad51C expression vector. Gene targeting frequency was examined at the RAD54B locus, because it was assumed that Rad54B is not functionally related to Rad51C (17). The frequency in the Rad51C mutant cells was 1.16%, which was significantly lower than 12.0%, the wild-type value. These results indicate that Rad51C plays a role in homologous recombination, a finding that is in accord with the observations in DT40, CHO and HeLa cells (11,12,23,24).
Table 1.
Sister chromatid exchanges and targeting frequency
| Cell line | SCEs per cella |
Targeted integrationb | |
|---|---|---|---|
| Spontaneous | MMC- induced | RAD54B- hygro | |
| RAD51C+/+/+/+ | 4.2 ± 0.2 | 8.3 ± 0.4 | 12.0% (13/108) |
| RAD51C+/−/−/− | 4.3 ± 0.3 | 5.8 ± 0.3c | 1.16% (1/86)d |
| RAD51C+/−/−/−+cDNA | 5.0 ± 0.2 | 8.2 ± 0.4 | N.D.e |
aNumber of SCEs per cells represents the mean ± SEM from 100 mitotic cells.
bThe frequency of targeted integration is shown as a percentage of the targeted clones relative to the total number of hygromycin resistant clones; the numbers of clones are given in parentheses.
cP < 1.0 × 10−4, based on Mann–Whitney U-tests, comparing wild-type cells with RAD51C+/−/−/− cells.
dP = 0.0025, based on Fisher's exact tests, comparing wild-type cells with RAD51C+/−/−/− cells.
eN.D., not determined.
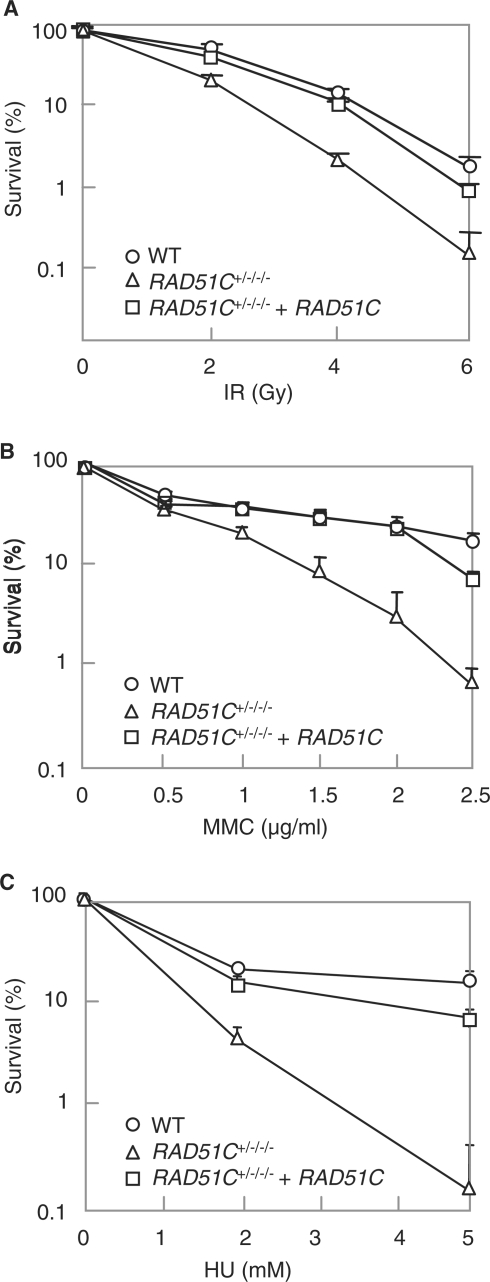
Homologous recombination is required for the repair of DSBs induced by DNA-damaging agents. The Rad51C mutant cells were hypersensitive to ionizing radiation, MMC and HU, which was complemented by the exogenous expression of the gene, indicating that Rad51C plays a role in DSB repair (Figure 2A–C). A defect in homologous recombination leads to an increase in chromosome aberrations such as gaps and breaks. Giemsa staining using a metaphase spread revealed that both chromosome- and chromatid-type aberrations were increased in Rad51C mutant cells, indicating that spontaneously arising DSBs were not properly repaired (Table 2). These observations suggest that Rad51C plays a role in the repair of DSB through homologous recombination in human cells.
Figure 2.
Sensitivity to DNA-damaging agents. (A) Sensitivity to ionizing radiation. (B) Sensitivity to MMC. (C) Sensitivity to HU. In (A), (B) and (C), the results represent the means ± SD of three experiments.
Table 2.
Spontaneous chromosomal aberrations
| Cell line | Chromatid type | Chromosome type | Abnormal cells |
|---|---|---|---|
| RAD51C+/+/+/+ | 2 | 3 | 3 |
| RAD51C+/−/−/− | 12 | 7 | 17 |
| RAD51C+/−/−/−+cDNA | 4 | 4 | 7 |
A total of 100 cells were scored for each cell line.
Rad51C dysfunction leads to increased centrosome aberrations
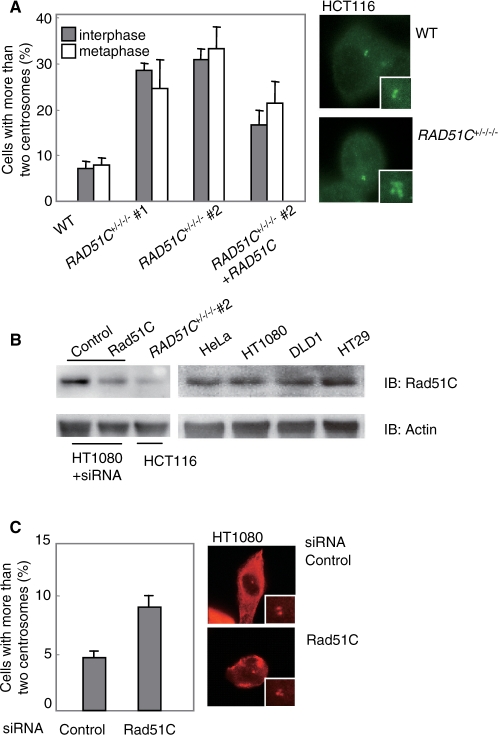
Since Rad51C's role in maintaining centrosome integrity has been controversial (11,12), we examined centrosome aberrations by immunostaining γ-tubulin. Cells with more than two centrosomes are observed more frequently in two independent Rad51C mutant clones than in wild-type cells (Figure 3A). The frequency of aberrant numbers of interphase centrosomes (more than two) was 7.0% in wild-type cells, whereas it was 28.0% (#1) and 30.3% (#2) in the Rad51C mutant cells. RAD51C cDNA expression in the mutant cells reduced the frequency to 16.3% (#2). In mitosis, the frequency was 8.0% in wild-type cells, and 24.0% (#1) and 32.8% (#2) in the mutant cells. cDNA expression in the mutant cells reduced the frequency to 21.5% (#2). Thus, unlike CHO cells with Rad51C mutation, in which centrosome aberrations were observed only at mitosis, the corresponding HCT116 cells demonstrated centrosome aberrations at both interphase and mitosis.
Figure 3.
Centrosome aberrations. (A) Centrosome aberrations in HCT116 cells. For each clone at interface and metaphase, 100 cells and 50 cells, respectively, stained with the anti-γ-tubulin antibody by immunofluorescence were counted. (B) Western blot analysis showing the expression of Rad51C in HT1080 cells treated with either control or Rad51C siRNA. Rad51C levels in human cells are also shown as controls. (C) Centrosome aberrations in HT1080 cells. Cells were treated with either control or Rad51C siRNA and stained with the anti-γ-tubulin antibody. For each siRNA treatment at interface, 100 cells were counted. In (A) and (C), the results represent the means ± SEM of three independent experiments.
To confirm that a reduction in Rad51C levels leads to centrosome aberrations in other human cells, we silenced the gene in HT1080 cells by RNA interference. Western blot analysis showed that Rad51C expression was not altered in wild-type HT1080 cells and was reduced in the cells transfected with Rad51C siRNA (Figure 3B). The frequency of aberrant numbers of interphase centrosomes was 4.6% in cells transfected with control siRNA, whereas it was 9.1% in cells transfected with Rad51C siRNA (Figure 3C). Thus, supernumerary centrosomes were also increased in HT1080 cells with Rad51C dysfunction.
Rad51C affects XRCC3 levels
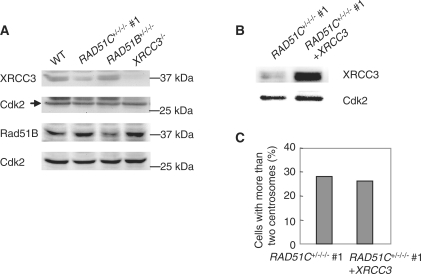
Since Rad51C is a central component of protein complexes formed by Rad51 paralogs, we investigated the effect of Rad51C dysfunction on the BCDX2 and Rad51C-XRCC3 complexes by measuring levels of Rad51B and XRCC3. Western blot analysis revealed that XRCC3 levels but not Rad51B levels were reduced in the mutant cells (Figure 4A). The previous report that XRCC3 was destabilized in Rad51C-deficient human cells is in accord with this finding (24). Next, we investigated the effect of reduced XRCC3 levels on centrosome aberrations by expressing the XRCC3 cDNA in the Rad51C mutant cells (Figure 4B and C). The frequency of centrosome aberrations was not changed by stable expression of XRCC3. Thus, centrosome aberrations induced by Rad51C dysfunction are unlikely to result from the destabilization of other Rad51 paralogs.
Figure 4.
Effect of Rad51C dysfunction on levels of other Rad51 paralogs. (A) Expression of XRCC3 and Rad51B in Rad51C mutant cells. (B) Exogenous expression of XRCC3 in the mutant cells. (C) Effect of complementation of XRCC3 on centrosome aberrations in the mutant cells. A total of 200 cells were counted.
ATR is involved in promoting centrosome aberrations
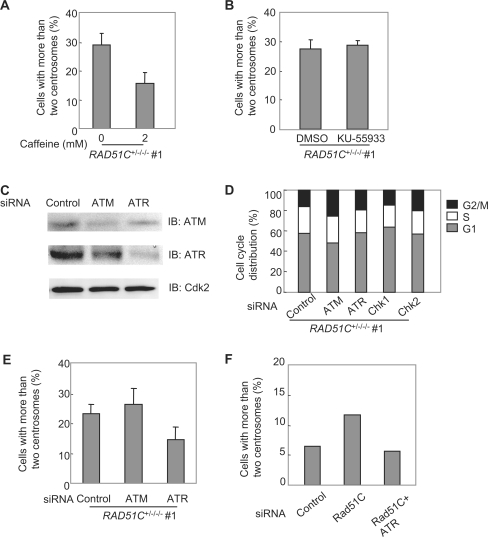
We next examined the roles of ATM and ATR in centrosome aberrations in Rad51C mutant cells by a treatment with 2 mM caffeine (Figure 5A). FACS analysis confirmed that caffeine treatment at this concentration did not affect cell-cycle progression (data not shown). This treatment reduced the frequency of centrosome aberrations in the mutant from 29.0% to 15.8% (P < 0.01, paired t-test), indicating that ATM and/or ATR may be involved in the generation of supernumerary centrosomes.
Figure 5.
ATR-dependent centrosome aberrations. (A) Effect of caffeine treatment on centrosome aberrations in the Rad51C mutant cells. The cells were treated with 2 mM caffeine for 40 h. (B) Effect of an ATM inhibitor on centrosome aberrations in the Rad51C mutant cells. The cells were treated with either DMSO alone or 10 μM KU55933 dissolved in DMSO for 24 h. (C) Western blot analysis showing the expression of ATM and ATR in cells treated with siRNAs. (D) Cell-cycle distribution of the mutant cells treated with siRNAs. (E) Effect of silencing of ATM and ATR on centrosome aberrations in the Rad51C mutant cells. The frequencies of centrosome aberrations were examined in the cells treated with control, ATM or ATR siRNA. In (A), (B) and (E), a total of 100 cells were counted for each measurement. The results represent the means ± SEM of three independent experiments. (F) Effect of ATR silencing on centrosome aberrations in HT1080 cells treated with Rad51C siRNA. A total of 300 cells were counted.
ATM's effect on the centrosomes was examined by treatment with the ATM-specific inhibitor, KU55933 (Figure 5B). This treatment did not change the frequency of supernumerary centrosomes, indicating that ATM was not involved in the damage-dependent increase in supernumerary centrosomes. Next, the effects of ATM and ATR on the centrosomes were examined by using RNA interference to silence the genes. Western blot analysis demonstrated that each protein was down-regulated by RNA interference (Figure 5C). Treatment with ATM siRNA slightly affected the cell-cycle distribution of the mutant cells, whereas treatment with ATR siRNA did not affect the distribution (Figure 5D). In accord with the finding using the ATM inhibitor, the silencing of ATM did not affect the frequency, whereas the silencing of ATR reduced the frequency from 23.3% to 14.7% (P < 0.01) (Figure 5E). This observation was also confirmed in HT1080 cells with Rad51C dysfunction. The silencing of ATR reduced the frequency from 11.8% to 5.8% (P < 0.01) (Figure 5F). These results indicate that ATR but not ATM was involved in the generation of supernumerary centrosomes. Because treatment with ATR siRNA rescued centrosome aberrations related to Rad51C depletion, ATR is likely to promote the generation of supernumerary centrosomes in the damage response signaling pathway rather than in a pathway directly associated with Rad51C.
Chk1 is involved in promoting centrosome aberrations
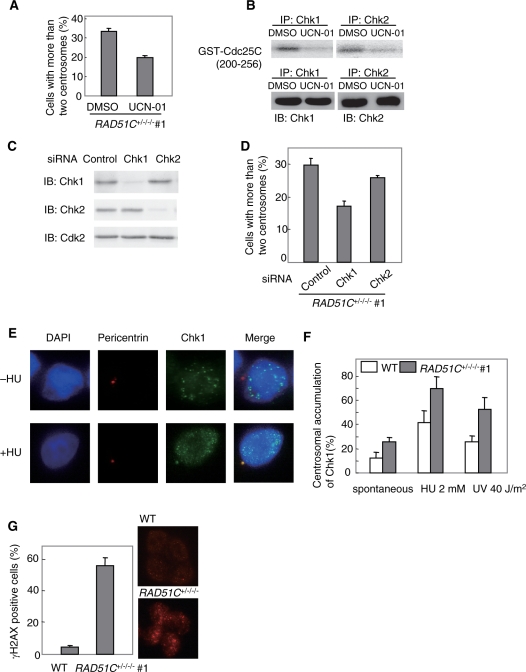
Since Chk1 is activated by ATR in response to DNA damage, we examined the effect of Chk1 inhibition on centrosome aberrations by treatment with the Chk1 inhibitor UCN-01 at the concentration of 100 nM in the mutant cells. This treatment reduced the frequency of centrosome aberrations from 33.0% to 19.7% (P < 0.01) (Figure 6A). Although UCN-01 was originally shown to inhibit Chk1 rather than Chk2, a subsequent study using GST-Cdc25C (200–256) as a substrate showed that the drug inhibited the immunoprecipitated kinase activities of Chk1 and Chk2 from wild-type HCT116 cells after exposure to ionizing radiation (25). We examined inhibition of both kinases by the drug using GST-Cdc25C (200–256) as a substrate in the mutant cells. In accord with the previous result, UCN-01 inhibited both kinases (Figure 6B). To determine which kinase is involved in the generation of supernumerary centrosomes, we examined the effect of RNA interference of the kinases on the centrosome aberrations (Figure 6C). Treatment with Chk1 or Chk2 siRNA did not affect the cell-cycle distribution of the mutant cells (Figure 5D). Silencing of Chk1 by RNA interference reduced the frequency from 30.0% to 17.3%, whereas silencing of Chk2 did not affect the frequency (Figure 6D). These results indicate that the ATR-Chk1 pathway was involved in the generation of supernumerary centrosomes.
Figure 6.
Chk1-dependent centrosome aberrations. (A) Effect of a Chk1 inhibitor on centrosome aberrations in the Rad51C mutant cells. The cells were treated with either DMSO alone or 100 nM UCN-01 dissolved in DMSO for 48 h. (B) Effect of UCN-01 on Chk1 and Chk2 kinase activities in the mutant cells. The cells were treated with 100 nM UCN-01 for 1 h. (C) Western blot analysis showing the expression of Chk1 and Chk2 in cells treated with siRNAs. (D) Effect of silencing of Chk1 and Chk2 on centrosome aberrations in the Rad51C mutant cells. The frequencies of centrosome aberrations were examined in the cells treated with control, Chk1 or Chk2 siRNA. (E) Damage-induced centrosomal accumulation of Chk1. HU treatment induces the accumulation of Chk1 at the centrosome. (F) Accumulation of Chk1 at the centrosome. (G) Focus formation of γH2AX in the nucleus. Immunofluorescence was carried out using the anti-γH2AX antibody. In (A), (D), (F) and (G), a total of 100 cells were counted for each measurement. The results represent the means ± SEM of three independent experiments.
Rad51C dysfunction promotes centrosomal accumulation of Chk1
Chk1 was shown to localize to centrosomes in response to the DNA damage induced by ultraviolet irradiation or HU treatment (26). This observation suggested that accumulation of Chk1 at centrosomes contributes to its checkpoint functions. We examined centrosomal localization of Chk1 by double immunofluorescence of Chk1 and pericentrin in HCT116 cells (Figure 6E and F). In accord with the previous report, ultraviolet irradiation or hydroxyurea treatment promoted Chk1's centrosomal localization in HCT116 cells. The frequency of cells in which Chk1 localizes to centrosomes was 12.0% in wild-type cells, whereas it was 25.8% in the mutant cells (P < 0.01). We next examined focus formation of phosphorylated form of histone H2AX (γH2AX) in the nucleus by immunofluorescence of the protein. γH2AX is recruited to DSBs in response to DNA damage. γH2AX-positive cells were defined as cells harboring the discrete large-sized foci of the protein. The frequency of γH2AX-positive cells was 4.3% in wild-type cells, whereas it was increased to 55.7% in the mutant cells (Figure 6G). These observations indicate that Chk1 accumulates at centrosomes in response to unrepaired DNA damage caused by Rad51C dysfunction in the absence of exogenous insults. In addition, localization of Chk1 at centrosomes in response to DNA damage induced by ultraviolet irradiation or hydroxyurea treatment was promoted by Rad51C dysfunction. Together with our observation that the Rad51C mutant cells were hypersensitive to HU (Figure 2C), these results suggest that Rad51C dysfunction may lead to prolonged replication fork arrest.
Rad51C dysfunction leads to increased aneuploidy
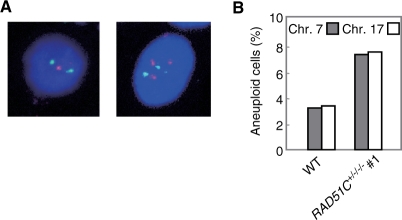
Supernumerary centrosomes are assumed to play a role in the generation of aneuploidy by inducing aberrant chromosome segregation. The frequency of aneuploidy, characterized by one or three chromosomes, was investigated by FISH using two independent chromosome-specific centromere probes (Figure 7A). The frequencies of aneuploidy at chromosomes 7 and 17 were 3.3% and 3.4%, respectively, in wild-type cells, whereas they were increased to 7.4% and 7.6% in the Rad51C mutant cells (P < 0.01, Fisher's exact test) (Figure 7B). This indicates that Rad51C, like other Rad51 paralogs, maintains chromosome stability.
Figure 7.
Increased aneuploidy. (A) FISH on Rad51C mutant cells using a probe for chromosome 7 (orange) and a probe for chromosome 17 (green). (B) Frequency of aneuploidy. For each chromosome-specific FISH, 500 cells were counted.
DISCUSSION
We have shown here that Rad51C dysfunction leads to increases in supernumerary centrosomes in an ATR-Chk1-dependent manner and to aneuploidy in human cells. This observation indicates that the DNA damage-response signal activated by Rad51C dysfunction causes chromosome instability by affecting centrosome integrity.
In contrast to previous studies using CL-V4B cells (11,12), the present study demonstrated that centrosome aberrations were induced by Rad51C dysfunction at both mitosis and interphase in two independent human cell lines. Since centrosome aberrations in interphase are likely to be generated at the S and G2 phases, such a difference may be explained by a mechanism that regulates centrosome integrity at these phases of the cell cycle. Given that the DNA damage response is involved in the maintenance of centrosome integrity, the absence of a protein involved in the signaling in response to unrepaired DNA damage may explain the lack of centrosome aberrations in interphase in CL-V4B cells.
A defect in the mismatch repair pathway has been shown to interfere with the DNA damage response (27). Since MLH1, a mismatch repair protein, is mutated in HCT116 cells, some phenotypes observed in the derivatives of this cell line are likely to be affected by a defect in mismatch repair. However, ATR-dependent centrosome aberrations in interphase were also observed in mismatch-proficient HT1080 cells, thus excluding this possibility.
The present results, together with those of previous studies (8–10,12), strengthen the role of Rad51 paralogs in the maintenance of centrosome integrity. However, it is unlikely that Rad51 paralogs directly regulate centrosome functions. There is no evidence to suggest that any member of the Rad51 paralog family localizes to the centrosome (7). We confirmed that Rad51C does not localize to the centrosome by exogenous expression of the RAD51C cDNA fused with the green fluorescent protein in Rad51C mutant cells (data not shown). Since Rad51C participates in the formation of two protein complexes consisting of five members of the Rad51 paralog family, these complexes are unlikely to localize to the centrosome.
Evidence that the DNA damage response plays a role in centrosome functions has been accumulating. ATM, ATR, ATRIP, Chk1 and Chk2 were shown to localize to the centrosome in HeLa cells (28). Supernumerary centrosomes in human lymphoblastoid cells exposed to ionizing radiation were cancelled by treatment with 2 mM caffeine or by the depletion of Chk1, suggesting that the ATM/ATR-Chk1 pathway may be involved in promoting centrosome amplification induced by DNA damage (16). The ATR-Chk1 pathway's role in regulating centrosome functions has been proposed from the identification of mutations in pericentrin, a centrosomal protein, in Seckel syndrome as well as in Majewski osteodysplastic primordial dwarfism type II (MOPD II) syndrome (29,30). These rare genetic disorders share dwarfism and microcephaly. Since ATR mutations are responsible for Seckel syndrome (31), the ATR-dependent DNA damage response signaling pathway might be associated with the pericentrin-dependent pathway. Although the exact role of ATR in centrosome functions remains to be elucidated, these results support the present finding that ATR is involved in increased supernumerary centrosomes. It is also noteworthy that caffeine treatment in Rad51C mutant cells did not reduce the frequency of centrosome aberrations to the wild-type levels, suggesting that an ATR-independent pathway may play a minor role in promoting centrosome aberrations in the mutant cells.
Defective homologous recombination causes prolonged replication fork arrest. Increased centrosome numbers that arise spontaneously in Rad51C mutant cells are likely to be attributable to prolonged arrest at replication forks or at the G2/M boundary. Furthermore, the present finding that ATR or Chk1 depletion reduced centrosome aberrations suggests that prolonged checkpoint arrest may cause supernumerary centrosomes. The previous finding that centrosome amplification induced by Rad51 deletion was caused during the prolonged G2 phase in an ATM-dependent manner supports this model (13). However, ATM is unlikely to be involved in the DNA damage response that increases centrosome numbers in Rad51C mutant cells. Although the interplay between ATM and ATR was shown to function in the DNA damage response (32,33), Rad51C dysfunction appears to activate the ATR-specific pathway. The DNA damage response signaling pathway activated by Rad51C dysfunction may be different from the pathway activated by depletion of Rad51. Rad51C may play a role in the DNA repair process that does not depend on Rad51. Rad51C or its associated proteins have been proposed to have a resolvase activity for Holliday junctions, suggesting that Rad51C may be involved in the late phase of homologous recombination (34). Thus, discrimination in the choice of the DNA damage response signal in their mutant cells may reflect functional differences between Rad51 and Rad51C in DSB repair through homologous recombination.
FUNDING
Japanese Society for Promoting Science (18310037 to K.M.); the Ministry of Education, Culture, Sports, Science and Technology (18012012 to K.M.); the Ministry of Health, Labor and Welfare of Japan (19140401 to K.M.); Hiroshima University 21st Century Center of Excellence Program (M.K. and T.Y.). Funding for open access charge: Japanese Society for Promoting Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Masson J-Y, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu N, Schild D, Thelen MP, Thompson LH. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 7.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 9.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 10.Date O, Katsura M, Ishida M, Yoshihara T, Kinomura A, Sueda T, Miyagawa K. Haploinsufficiency of RAD51B causes centrosome fragmentation and aneuploidy in human cells. Cancer Res. 2006;66:6018–6024. doi: 10.1158/0008-5472.CAN-05-2803. [DOI] [PubMed] [Google Scholar]
- 11.Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PPW, Kanaar R, Zdzienicka MZ. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 2002;30:2172–2182. doi: 10.1093/nar/30.10.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindh AR, Schultz N, Saleh-Gohari N, Helleday T. RAD51C (RAD51L2) is involved in maintaining centrosome number in mitosis. Cytogenet. Genome Res. 2007;116:38–45. doi: 10.1159/000097416. [DOI] [PubMed] [Google Scholar]
- 13.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daboussi F, Thacker J, Lopez BS. Genetic interactions between RAD51 and its paralogues for centrosome fragmentation and ploidy control, independently of the sensitivity to genotoxic stresses. Oncogene. 2005;24:3691–3696. doi: 10.1038/sj.onc.1208438. [DOI] [PubMed] [Google Scholar]
- 15.Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 16.Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyagawa K, Tsuruga T, Kinomura A, Usui K, Katsura M, Tashiro S, Mishima H, Tanaka K. A role for RAD54B in homologous recombination in human cells. EMBO J. 2002;21:175–180. doi: 10.1093/emboj/21.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara T, Ishida M, Kinomura A, Katsura M, Tsuruga T, Tashiro S, Asahara T, Miaygawa K. XRCC3 deficiency results in a defect in recombination and increased endoreduplication in human cells. EMBO J. 2004;23:670–680. doi: 10.1038/sj.emboj.7600087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoyama H, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]
- 20.Hiyama T, Katsura M, Yoshihara T, Ishida M, Kinomura A, Tonda T, Asahara T, Miyagawa K. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promote rereplication in human cells. Nucleic Acids Res. 2006;34:880–892. doi: 10.1093/nar/gkj495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bärlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O-P, Kallioniemi A. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 22.Wu G-J, Sinclair CS, Paape J, Ingle JN, Roche PC, James CD, Couch FJ. 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGMA1B genes. Cancer Res. 2000;60:5371–5375. [PubMed] [Google Scholar]
- 23.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lio Y-C, Schild D, Brenneman MA, Redpath JL, Chen DJ. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J. Biol. Chem. 2004;279:42313–42320. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- 25.Yu Q, La Rose J, Zhang H, Takemura H, Kohn KW, Pommier Y. UCN-01 inhibits p53 up-regulation and abrogates γ-radiation-induced G2-M checkpoint independently of p53 by targeting both of the checkpoint kinases, Chk2 and Chk1. Cancer Res. 2002;62:5743–5748. [PubMed] [Google Scholar]
- 26.Löffler H, Bochtler T, Fritz B, Tews B, Ho AD, Lukas J, Bartek J, Krämer A. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 2007;6:2541–2548. doi: 10.4161/cc.6.20.4810. [DOI] [PubMed] [Google Scholar]
- 27.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J. Cell Biochem. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 29.Griffith E, Walker S, Martin C-A, Vagnarelli P, Stiff T, Vernay B, Sanna NA, Saggar A, Hamel B, Earnshaw WC, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 31.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 32.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GCM, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 33.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Masson J-Y, Shah R, O’Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]