Abstract
Catalytic RNA molecules possess simultaneously a genotype and a phenotype. However, a single RNA genotype has the potential to adopt two or perhaps more distinct phenotypes as a result of differential folding and/or catalytic activity. Such multifunctionality would be particularly significant if the phenotypes were functionally inter-related in a common biochemical pathway. Here, this phenomenon is demonstrated by the ability of the Azoarcus group I ribozyme to function when its canonical internal guide sequence (GUG) has been removed from the 5′ end of the molecule, and added back exogenously in trans. The presence of GUG triplets in non-covalent fragments of the ribozyme allow trans-splicing to occur in both a reverse splicing assay and a covalent self-assembly assay in which the internal guide sequence (IGS)-less ribozyme can put itself together from two of its component pieces. Analysis of these reactions indicates that a single RNA fragment can perform up to three distinct roles in a reaction: behaving as a portion of a catalyst, behaving as a substrate, and providing an exogenous IGS. This property of RNA to be multifunctional in a single reaction pathway bolsters the probability that a system of self-replicating molecules could have existed in an RNA world during the origins of life on the Earth.
INTRODUCTION
Group I introns catalyze RNA phosphoester transfer reactions at specific splice sites both in vivo and in vitro. The splice site selection is precise, and relies on base-pairing interactions between the 5′ portion in P1 of the catalytic intron and a pseudo-complementary 3–6-nt region of the 3′ portion of the 5′ exon (1,2). The former has been termed the internal guide sequence (IGS) and the latter can be referred to as the IGS complement. Base-pairing between the IGS and its complement depends on Watson–Crick pairing at most positions; however the 3′ nt of the IGS complement always forms a G•U wobble pair with the IGS to define precisely the 5′ splice site (1–4). For example, the IGS of the group I intron in the tRNAIle transcript from the purple bacterium Azoarcus can be shortened in vitro to 5′-GUG-3′, and this pairs with the complement 5′-CAU-3′ to effect splicing after the terminal U in the complement (5–7) (Figure 1A).
Figure 1.
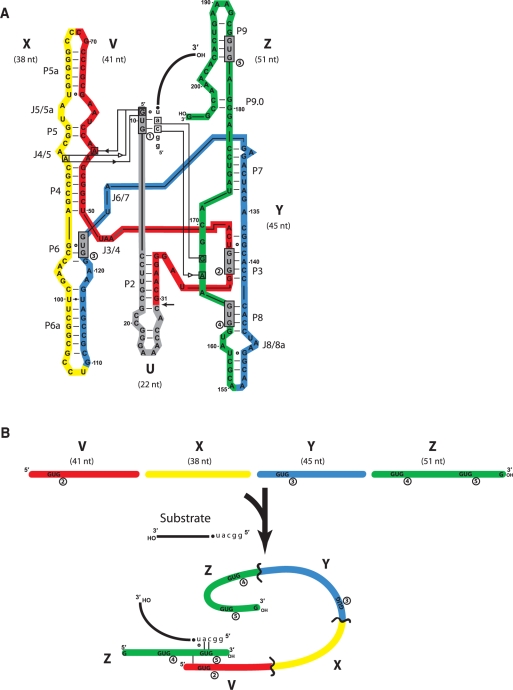
Schematic of the partitioning of the Azoarcus ribozyme into fragments. (A) The 197-nt source molecule was partitioned into five fragments (U, gray; V, red; X; yellow; Y, blue; Z, green) such that catalytic activity in trans, plus covalent self-assembly could be assayed. Removal of the U fragment from the system at the location indicated by the arrow leaves an L–30 construct which can then be divided into four fragments that can exhibit activity when 5-nt head groups (h = GGCAU) are appended to the 5′ portions of X, Y and Z (see text). A substrate oligomer (lower-case letters = head; black line = tail) binds via a 5′-CAU-3′ to the IGS (GUG) of the ribozyme, which catalyzes phosphotransfer of the tail to its own 3′ end. The native IGS of the ribozyme is denoted by GUG1, while the four exogenous GUG triplets that occur naturally in the remainder of the ribozyme are gray boxes denoted 2–5 (in circles). The tertiary interactions that hold the IGS and the IGS complement into the active site as determined by X-ray crystallography (24) are denoted using the hydrogen-bonding symbolism of Leontis et al. (41). (B) Schematic of how a ribozyme complex (either as a covalently contiguous molecule or as several fragments cooperating in trans) can perform catalysis in the absence of the U fragment that contains the native IGS. Here, the use of an exogenous IGS (GUG5) present in the h•Z fragment to bind to the IGS complement CAU is depicted.
In vitro, group I introns can behave as true enzymes, exhibiting accurate substrate specificity and multiple turnover. Notably, these enzymes often remain active when broken into non-covalent trans complexes. The work of Inoue and others has shown that the Tetrahymena, sunY, Azoarcus, Synechococcus, phage T4 td and other group I introns can retain catalytic efficiency when fragmented into several non-covalent pieces and then reconstituted in trans via base-pairing and tertiary interactions (8–16). These studies have isolated the regions of the ribozyme that are necessary to carry out the various catalytic events of self-splicing, a multi-step process that involves two transesterification reactions plus often a hydrolytic cleavage reaction. While trans-catalysis by reconstructed group I introns—including that from Azoarcus—is possible, complete removal of the IGS or the catalytic core will abolish splicing activity. For group I introns, and the Azoarcus ribozyme in particular, deletion of the P1 and P2 elements, along with the IGS, results in a molecule that cannot self-splice (and thus recombine exogenous RNAs), even though site-specific hydrolysis at the 3′ splice site is still possible because P1 (and hence the IGS) is only required for 5′ splice site selection (17,18).
Here, we have used the Azoarcus group I intron, with 5′-GUG-3′ as its IGS as a model system to show that if the canonical IGS is missing from the ribozyme, another RNA fragment containing GUG can act in trans to rescue transesterification activity. In fact, we show that one RNA fragment can play as many as three distinct roles in the covalent self-assembly of a self-replicating ribozyme. These results have bearing on models of the origins of genetic information in which multiple RNA oligonucleotides may have had to cooperate in order to allow construction of a catalytic entity.
MATERIALS AND METHODS
RNA preparation
RNAs were either purchased from IDT (Coralville, IA) or prepared by run-off transcription from double-stranded DNA templates constructed through recursive gene synthesis. RNAs were gel purified and desalted prior to use. Salts and buffers were made from the highest purity available (Sigma-Aldrich) and all water used was nuclease free (Ambion). Barrier pipette tips and other strict contamination controls, including the use of dedicated rooms and isolation hoods to purify primers and set up gene synthesis and PCR reactions, were always used to ensure correct sequence identities of all RNAs employed in experimentation.
Trans-splicing assays
The ability of Azoarcus ribozyme constructs to perform trans-splicing was assayed by incubating 2 μM of an enzyme (intron only) complex with 2 μM of a substrate oligonucleotide with or without the 5′-GUG-3′ IGS triplet somewhere within its sequence. Assays were performed at 48°C in 100 mM MgCl2 and 30 mM EPPS buffer (pH 7.5). Reactions were carried out for 0–60 min in 200 μl or 600 μl microcentrifuge tubes and then quenched by the addition of an equal volume of gel-loading solution containing 8 M urea, SDS, 200 mM EDTA and bromphenol blue dye. The RNAs were heat denatured at 80°C for 4 min, and then immediately electrophoresed through 8% polyacrylamide/8 M urea gels. In most cases, the V (or W) fragment was 5′-end-labeled with γ[32P]•ATP and OptiKinase (USB) and then gel-purified (19) prior to use. This allowed visualization of the products via phosphorimaging with a Typhoon 9200 instrument (GE Healthcare). In other cases the 3′ portion of the substrate molecule was radiolabeled using the method of Huang and Szostak (20). In some cases no radioactivity was employed, and all oligomers were visualized by SYBR Green II staining, although when quantification was needed, this technique was not used.
Self-assembly experiments
Ribozyme covalent self-assembly from two or three oligomers was performed as described previously (21–23). Briefly, RNA oligomers were incubated together at 42–48°C at a final concentration of ≤2 μM each. All reactions contained a final concentration of 100 mM MgCl2 and 30 mM EPPS buffer (pH 7.5) unless otherwise indicated. Reactions were carried out for 1–6 h with visualization methods as described above.
Genotyping
Full-length RNA covalent constructs were identified by comparison to a bona fide Azoarcus RNAs (with or without the 5′ U section) run as size controls. The bands corresponding to self-assembled RNAs were carefully excised from the gel and subjected to reverse transcription using the primer T20a (5′-CCGGTTTGTGTGACTTTCGCC-3′), which targets the 3′ portion of the Z fragment. One-twentieth of these reactions was used to seed PCR reactions employing T20a and W3′-h (5′-TAATACGACTCACTATAGCAAGGGATGGTG-3′) primers, the latter being specific for the 5′ portion of V. The PCR products were cloned into the vector pJET1.2 (Fermentas) and transformed into Escherichia coli. Individual colonies were picked as templates for colony PCR reactions employing the primers pJET1.2-F and pJET1.2-R (Fermentas), which generate products of ∼310 bp [= the insert size (∼192 bp) plus about 120 bp]. Products of the correct size were genotyped using BigDye (v.3) cycle sequencing chemistry and a Prism 3100 (ABI) instrument.
RESULTS AND DISCUSSION
A group I intron without an IGS
To create an IGS-less (‘blind’) version, of the L–8 Azoarcus ribozyme, the first 30 nt of the naturally occurring form of the 205-nt intron were removed, resulting in an L–30 construct lacking the first occurrence of the GUG triplet (termed GUG1), which is the canonical IGS for the ribozyme. Without this triplet, transesterification activity should be abolished. Previously it has been shown that the Azoarcus ribozyme can covalently self-assembly through recursive and autocatalytic recombination reactions after having been fragmented into four roughly equally sized pieces termed W, X, Y and Z (21). The removal of the first 22 nt in W results in a new fragment, here termed V (Figure 1A). Of interest is that the remaining 175 nt of the wild-type version of this ribozyme contain four additional GUG triplets, denoted GUG2, GUG3, GUG4 and GUG5 (Figure 1A and B). By chance, any given triplet such as GUG should appear fewer than three times in 175 nt. Regardless, these ‘exogenous’ GUGs could potentially complement the missing canonical IGS if they were to work in trans. Such activity would be similar to the manner in which fragments of the Tetrahymena ribozyme can non-covalently assemble to restore activity, although with each RNA presumably only adopting one role (9,10).
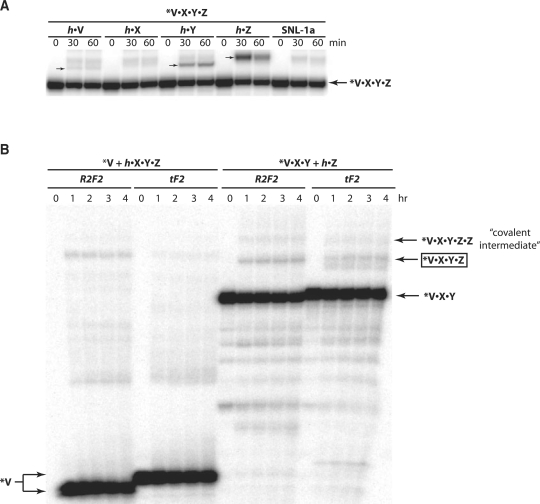
The efficacy of an exogenous IGS to rescue a blind Azoarcus ribozyme was first tested using an assay in which the L–30 ribozyme V•X•Y•Z was challenged to perform trans-splicing on an external substrate, where ‘•’ signifies a covalent phosphoester bond (Figure 2A). In these reactions, specific recognition of the substrate at its IGS complement CAU would require the articulation of a GUG triplet into the active site of the V•X•Y•Z construct. Five substrates were assayed, three of which possess at least one GUG sequence (h•V, h•Y and h•Z, where h refers to a 5-nt head, GGCAU), while two of which do not (h•X, and SNL-1a = 5′-GGCAU•AAAUAAAUAAAUAAAUA-3′). Measurable trans-splicing activity was observed only for the three GUG-containing substrates, with Z, possessing two such sequences, exhibiting by far the most activity (Figure 2A). This activity diminishes when the GUGs are mutated to CUG in a dosage dependent manner (Supplementary Figure S1). While it is conceivable that the V•X•Y•Z ribozyme itself could be supplying the exogenous IGS by partially unfolding in partnership with a folded and catalytic complex, the differential response to exogenous substrates suggests that this is not occurring under these assay conditions. The more likely scenario involves the utilization of a GUG triplet in one molecule of the substrate itself to direct splicing at the splice site of another molecule of the substrate. The case in which GUG5 of Z is performing this role is diagrammed in Figure 1B. The product sizes observed during gel electrophoresis (Figure 2A) are those expected should transesterification occur following the single CAU sequence present in each substrate, and these were confirmed by RT–PCR using reverse primers specific for each substrate.
Figure 2.
Activity assays of ribozymes lacking a native IGS. (A) Splicing activity of the Azoarcus L–30 construct V•X•Y•Z when supplied with an exogenous substrate that either contains the IGS sequence GUG (h•V, h•Y and h•Z) or that does not (h•X and SNL-1a). Upon splicing, the 5′-radiolabeled ribozyme is expected to append the 3′ portion of the substrate to its own 3′ end, producing a product of the sizes indicated by the small arrows; no splicing is visible with h•X or SNL-1a, which should give products of lower molecular weight than the product with h•V. (B) Bipartate covalent self-assembly reactions. In each set, the V-containing fragment is 5′ radiolabeled. The R2F2 reactions were designed with splice junctions (V–X or Y–Z) that can only form a covalent bond via a two-step recombination reaction, while the tF2 reactions were designed with splice junctions that can form a covalent bond through either a two-step recombination or through a one-step recombination (22). This accounts for the slight size difference between the reactants or products in each system. The covalent self-assembly product V•X•Y•Z (∼175 nt) is indicated.
Covalent self-assembly of a blind ribozyme by multifunctional RNA fragments
Next, the ability of a single RNA fragment to assume multiple roles in an RNA reaction network was investigated using covalent self-assembly systems. Here, two pieces of the Azoarcus L–30 ribozyme were co-incubated at 2 μM concentrations. Covalent self-assembly requires that non-covalent assemblages initially catalyze a series of recombination reactions leading to the ‘full-length’ covalent ribozyme, which can then feed back in an autocatalytic fashion (21,23). In the current experiments, an additional requirement exists that one fragment behave as an exogenous IGS, with the consequence that the overall yield of covalent ribozyme is expected to be low. Nonetheless, assembly was observed in a variety of systems. When V was incubated with h•X•Y•Z, where h refers to the 5-nt sequence GGCAU, self-construction of the V•X•Y•Z ribozyme was observed in 0.1–1.3% yield, and when V•X•Y was incubated with h•Z the yield was 1.0–3.5% (Figure 2B). Trace amount of product was also seen when V•X was incubated with h•Y•Z (data not shown). The higher yields seen when the Z fragment was used in isolation is consistent with this molecule's possession of two GUG triplets, such that either one could be serving as an exogenous IGS. Highest self-assembly yields were obtained at 42°C, as opposed to the 48°C optimum for self-assembly with the U fragment in place (21), suggesting that a multifunctional RNA fragment containing an exogenous IGS is binding to the catalytic core through a set of relatively weak hydrogen-bonding interactions.
Mechanism of covalent self-assembly
Recombination in these self-assembly reactions can proceed through a one-step (tF2) or a two-step (R2F2) mechanism (22), and the junctions between the two fragments were designed here in two alternative ways to favor one or the other mechanism. However, because the tF2 mechanism requires a pre-formed RNA duplex to bind to the IGS, rather than a single RNA strand as in the R2F2 mechanism, the scheme as portrayed in Figure 1B that depends on an exogenous IGS seemed less plausible with the tF2 mechanism. To test this hypothesis, the assembled RNAs at the size indicated by the arrow in Figure 2B were excised, converted to DNA by RT–PCR, and then cloned and subjected to nucleotide sequence analysis. The two mechanisms can be distinguished by the fact that the tF2 mechanism leaves a characteristic 3–4-nt insertion, while the R2F2 does not (21,22). All clones analyzed in all experiments failed to exhibit insertions, and thus it can be concluded that two sequential recombination reactions using single-stranded substrates (R2F2 mechanism) are required when an exogenous IGS is utilized, consistent with the depiction in Figure 1B. In fact, most clones possessed deletions of 3–12 nt at the splice junction, suggesting that splicing is often sloppy, occurring a few positions 3′ of the canonical CAU target site. Some of these products are even visible in the gel shown in Figure 2B, and are further evidenced when the 5′ splicing products are examined (Supplementary Figure S2).
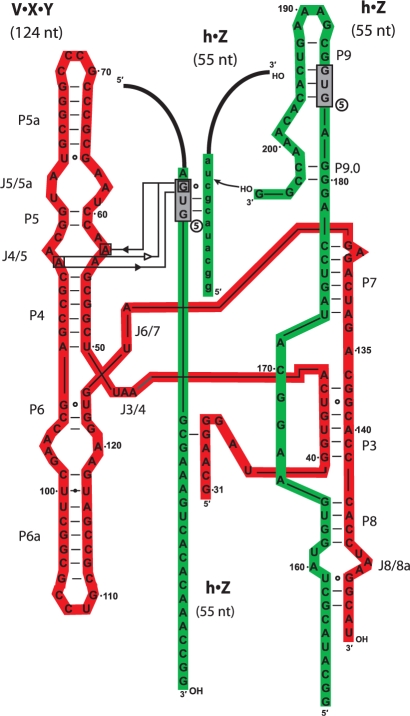
One manner in which mispairing can occur under the high salt conditions employed in self-assembly (100 mM MgCl2) is when the IGS pairs with a single-nucleotide variant of CAU (22). In Figure 3, an example of this is diagrammed, whereby the exogenous IGS is pairing with the triplet CGC of the substrate, leading to a 4-nt deletion that was commonly detected in the clones. It should be noted that the pairing between the 5′ G of the IGS and the 3′ C in this triplet is likely not a canonical cis Watson–Crick pair because the base-paring surface of this guanosine of the IGS is participating in tertiary interactions with the J4/5 bulge (Figure 1A), at least in the wild-type Azoarcus model deduced from X-ray crystallography (24). In any event, use of an exogenous IGS leads to somewhat promiscuous splicing, as can be seen with the Tetrahymena ribozyme (8). It also can lead to promiscuous self-assembly events, with the consequence that should autocatalytic feedback be significant in this system, then the RNA denoted at the arrow in Figure 2B should be the result of a selection process for catalytically proficient molecules. As a rudimentary test of this notion, three clones of self-assembled ribozymes (with deletions of 3, 4 or 8 nt) were tested for splicing activity and all were found to be active for transesterification (Table 1). Of interest is that the shortest of these clones, one that exhibits an 8-nt deletion at the Y–Z junction is clearly the most active and would be expected to emerge from a selection process. The deletion in this clone disrupts the tetraloop receptor in P8 (Figure 1A), but in the absence of the P2 tetraloop that normally docks at that location, this deletion becomes less deleterious.
Figure 3.
Model of utilization of an exogenous IGS. In this example, the h•Z RNA molecule is multifunctional, performing three discrete roles: acting as the 3′ end of the ribozyme, acting as a substrate for transesterification, and acting as an exogenous IGS. In the last case, the h•Z molecule must be held in place in the catalytic core of the ribozyme complex via tertiary hydrogen-bonding interactions. Some potential examples of these are indicated in the diagram, although no solid evidence exists for any of these interactions. In addition, the loose interaction between the fragment supplying the exogenous IGS and the remainder of the ribozyme apparently can lead to mis-pairing between the IGS and the IGS complement (see text); here GUG5 is depicted as acting as the exogenous IGS and binding to CGC instead of CAU, which would lead to a 4-nt deletion in the splicing product.
Table 1.
Activity assays of three clones derived from ribozymes self-assembled from RNAs lacking the canonical IGS
| Clone | Sequence at Y–Z junction | Relative yield using h•Z substrate (%) |
|---|---|---|
| L–30 wild type | … CUAAGGCAU•ACGCUAUGGUGAAGG … | 100 |
| 1398: Δ4nt | … CUAAGGCAU•[ ]UAUGGUGAAGG … | 32 |
| 1401: Δ8nt | … CUAAGGCA[ ]CGGUGAAGG … | 63 |
| 1403: Δ3nt | … CUAAGGCACAU•[ ]UAUGGUGAAGG … | 60 |
Table values are relative transesterification yields after 1 h with the indicated substrate, normalized to that of the V•X•Y•Z construct, set at 100%. Brackets indicate a deletion relative to the wild type.
Multifunctionality in catalytic RNAs
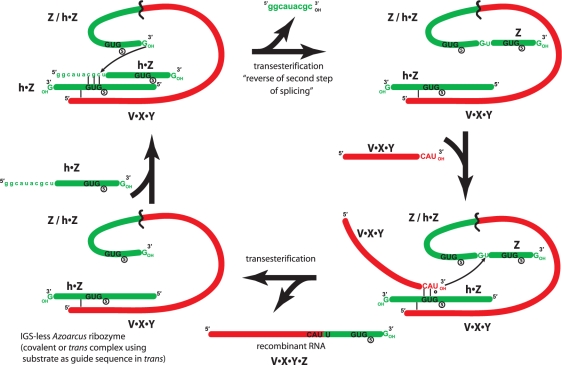
Together, these data allow a model in which RNA multifunctionality is a requisite aspect of the construction of self-replicating catalytic RNA from it component fragments (Figure 4). In this model, one RNA sequence (h•Z in this case) is actually performing three distinct roles. First, it is operating as part of a catalytic recombinase ribozyme, either in trans as a non-covalent partner with the remainder of the catalyst, or in cis, as a covalent element within a ribozyme. Second, it is behaving as an exogenous IGS, bringing a requisite GUG into the catalytic core of a ribozyme complex. The multifunctional fragment containing the exogenous IGS is held likely in place by tertiary interactions (13) such as those postulated in Figure 3. And third, it is being utilized as a substrate for recombination, in which its CAU sequence (or a low-error variant) is targeted by an IGS for transesterification to another RNA molecule.
Figure 4.
Model in which a recombinase ribozyme can covalently self-assemble from two RNA fragments despite the lack of a native IGS in the catalytic complex. Here, this self-assembly requires the h•Z fragment to be multifunctional and exhibit three distinct phenotypes.
A fundamental confirmation of the notion that h•Z is playing three distinct roles was obtained when the stoichiometry was varied between V•X•Y and h•Z in self-assembly reactions (Table 2). When the concentration of h•Z was kept constant at 1 μM, the optimum ratio of V•X•Y to h•Z was in fact 1:3, which gave a significantly better yield of V•X•Y•Z after 4 h than either lower (1:2) or higher (1:5) amounts of h•Z (one-tailed t-tests; P = 0.03 and 0.002, respectively).
Table 2.
Covalent self-assembly yields in different ratios of V•X•Y and h•Z after 4 h
| V•X•Y : h•Z | Percent yield of V•X•Y•Z |
|---|---|
| 1:1 | 1.11 ± 0.01 |
| 1:2 | 1.24 ± 0.10 |
| 1:3 | 1.50 ± 0.06 |
| 1:5 | 1.17 ± 0.03 |
Data based on results from three independent trials with [h•Z] fixed at 1 μM.
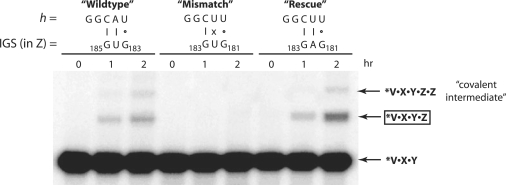
Another test of this model was provided when either the IGS, or its complement in the h portion of substrates, was mutated and loss-of-activity assays were performed. One approach was to track covalent self-assembly between V•X•Y and h•Z with matched and mis-matched pairings between potential exogenous IGS triplets in the Z portion of h•Z and the IGS complement in the h portion of h•Z (Figure 5). When h was changed from GGCAU to GGCUU, self-assembly was completely abolished. However, this required a second mutation at position 182 to alter a natural GAG to GUG to destroy a fortuitous IGS that would function with CUU. This A182U mutation by itself does not abolish activity (data not shown). Notably, when this second mutation is not made, the self-assembly activity is fully rescued, presumably by the GAG functioning as the IGS and pairing with the CUU in the h of h•Z (Figure 5).
Figure 5.
Test of the ability of nucleotide triplets in h•Z to perform as exogenous IGSs. Here, covalent self-assembly was assayed when 2 μM V•X•Y was incubated with 2 μM h•Z for 2 h at 42°C. In the ‘wild-type’ scenario, neither the h nor the Z portions of h•Z contained any mutations, as in Figure 2B. Self-assembly is achieved when an exogenous IGS in the Z portion is utilized, such as GUG5 at positions 183–185 (depicted). In the ‘mismatch’ scenario, the h was mutated from GGCAU to GGCUU, while an A182U mutation was simultaneously made to destroy a fortuitous exogenous IGS triplet of GAG. Self-assembly was abolished, but could be fully restored in the ‘rescue’ scenario when this second mutation was not present.
A second approach was simply to mutate exogenous GUG triplets, and test for loss-of-function with the canonical GGCAU in h (Table 3). Triplets 2, 4 and 5 were mutated to GUC, CUG and CUG, respectively. GUG3 was not mutated because it lies in J6/7 and has been shown previously to be essential for catalytic activity (25). When GUG2, GUG4, or GUG5 was mutated, the ability of the ribozyme to self-assemble covalently from two fragments was abolished, although it is not clear if this is a consequence of the effect this mutation has on the ribozyme as a whole. However, mutations in GUG4 and GUG5 are not inhibitory to the wild-type (W•X•Y•Z) Azoarcus ribozyme, and have minimal effect on the L–30 construct (V•X•Y•Z). Yet these two mutations severely inhibit both the covalent self-assembly reaction (Table 3) and transesterification (Supplementary Figure S1), implicating them as operating as exogenous IGS triplets in a cooperative RNA–RNA interaction.
Table 3.
Activity assays of reaction systems in which mutations were engineered into the exogenous IGS triplets
| IGS mutations | Relative yield in V•X•Y + h•Z self-assembly reaction (%) | Relative activity in W•X•Y•Z ribozyme (%) | Relative activity in V•X•Y•Z ribozyme (%) |
|---|---|---|---|
| (wild type) | 100 | 100 | 100 |
| GUG2 | 0 | 59 | 2.5 |
| GUG4 | 0 | 125 | 60 |
| GUG5 | 0 | 102 | 71 |
| GUG2/GUG4 | 0 | 1.5 | 5.0 |
| GUG2/GUG5 | 0 | Trace | 2.4 |
| GUG4/GUG5 | 0 | 4.6 | 45 |
| GUG2/GUG4/GUG5 | 0 | Trace | 8.8 |
Table values are relative bipartite covalent self-assembly yields after 3 h (second column) or splicing assays using an exogenous substrate after 1 h (third and fourth columns), normalized to that of the unmutated construct, set at 100%. Trace = less than 1% product detected. 0% = no V•X•Y•Z product detected.
Evolutionary implications
The RNA World scenario posits the existence at some point in time on the prebiotic Earth the existence of one or a few RNA-like molecules with the capacity to carry out all catalytic and information transfer events needed for life (26–29). To envision catalytic RNAs that are self-replicating requires an evolutionary scenario by which larger and more complex ribozymes develop from smaller and simpler structural motifs (30). In the case of group I introns, which can perform RNA recombination reactions that could have been of evolutionary importance (31–33), piecing together the entire catalytic complex may have required physical separation of key structural elements, including the IGS.
The trans-splicing activity of the blind Azoarcus ribozyme is in accordance with a previous study that demonstrated that if the IGS were disarticulated from the catalytic core on a separate fragment, and then an effectively full-length ribozyme could polymerize oligonucleotide sequences on its 3′ end (13). In that study an exogenous IGS (termed the external guide sequence, or EGS) required the GUG from P1 plus an additional 7 nt from P10 of the natural bacterial intron. The observations in the current study that the IGS can not only be externalized but also embodied in another portion of the ribozyme itself and shortened to GUG beg the question of what the minimum possible IGS/EGS could be. It will be interesting to test whether the trinucleotide GUG itself would impart any splicing activity to a blind ribozyme. The minimum length is unlikely to be <3 nt, because then almost all specificity would be lost. Provocatively, the IGS complement used by GUG in this system is CAU, which is the biochemical precursor to the L(lysidine)AU anticodon that is used to specify isoleucine in bacterial translation (34). The embedding of the Azoarcus self-splicing intron in an isoleucyl-tRNA gene evokes a direct relationship between an IGS/EGS and the origin of the genetic code, as alluded to by Shub upon its original discovery (5).
The ability of RNA molecules to act multifunctionally facilitates our ability to envision the origins of complex catalytic function and lends support to the RNA World scenario. RNA molecules are excellent candidates for this, as they clearly can fold variably, often leading to inhomogeneous populations of three-dimensional shapes (35–37). In fact, Bartel and colleagues have previously described a single RNA sequence that can fold into two different and essentially unrelated ribozymes (38), while here we report a single sequence with three distinct functions in the same reaction pathway. Primordial RNA molecules may have had to interact in a cooperative fashion to participate in hypercycles (39) or autocatalytic sets (40) in which the system as a whole benefits from the multiplicity of interactions among its components. The advent of such diversity would have its probability heightened if each component could play multiple roles such that fewer molecular types were required initially.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Aeronautics and Space Administration (NNX07AU05G to N.L.). Funding for open access charge: National Aeronautics and Space Administration.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank A. Burton and E. Hayden for useful discussions during the preparation of this manuscript.
REFERENCES
- 1.Davies RW, Waring RB, Ray JA, Brown TA, Scazzochio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982;300:719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. Conserved sequences and structures of group I introns: building an active site for RNA catalysis – a review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 3.Been MD, Cech TR. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986;47:207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- 4.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 5.Reinhold-Hurek B, Shub DA. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 6.Tanner MA, Cech TR. Activity and thermostability of the small self-splicing group I intron in the pre-tRNAIle of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo LY, Davidson LA, Pico S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta. 1999;1489:281–292. doi: 10.1016/s0167-4781(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 8.Doudna JA, Cormack BP, Szostak JW. RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc. Natl Acad. Sci. USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudry AA, Joyce GF. Minimum secondary structure requirements for catalytic activity of a self-splicing group I intron. Biochemistry. 1990;29:6534–6539. doi: 10.1021/bi00479a027. [DOI] [PubMed] [Google Scholar]
- 10.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl Acad. Sci. USA. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doudna JA, Couture S, Szostak JW. A multisubunit ribozyme that is a catalyst of and template for complementary strand RNA synthesis. Science. 1991;251:1605–1608. doi: 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- 12.Doudna J, Cech TR. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA. 1995;1:36–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Chowrira BM, Berzal-Herranz A, Burke JM. Novel system for analysis of group I 3′ splice site reactions based on functional trans-interaction of the P1/Pl0 reaction helix with the ribozyme's catalytic core. Nucleic Acids Res. 1995;23:849–855. doi: 10.1093/nar/23.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanczyc M, Dorit RL. Experimental evolution of complexity: in vitro emergence of intermolecular ribozyme interactions. RNA. 1998;4:268–275. [PMC free article] [PubMed] [Google Scholar]
- 15.Ikawa Y, Shiraishi H, Inoue T. Trans-activation of the Tetrahymena group I intron ribozyme via a non-native RNA-RNA interaction. Nucleic Acids Res. 1999;27:1650–1655. doi: 10.1093/nar/27.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikawa Y, Shiraishi H, Inoue T. Minimal catalytic domain of a group I self-splicing intron RNA. Nature Struct. Biol. 2000;7:1032–1035. doi: 10.1038/80947. [DOI] [PubMed] [Google Scholar]
- 17.van der Horst G, Inoue T. Requirements of a group I intron for reactions at the 3′ splice site. J. Mol. Biol. 1993;229:685–694. doi: 10.1006/jmbi.1993.1072. [DOI] [PubMed] [Google Scholar]
- 18.Ikawa Y, Naito D, Shiraishi H, Inoue T. Structure-function relationships of two closely related group IC3 intron ribozymes from Azoarcus and Synechococcus pre-tRNA. Nucleic Acids Res. 2000;28:3269–3277. doi: 10.1093/nar/28.17.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton AS, Madix RA, Vaidya N, Riley CA, Hayden EJ, Chepetan A, Díaz Arenas C, Larson BC, Lehman N. A simple method for gel-purification of radiolabeled nucleic acids via phosphorimaging: “Dip-N-Dot”. Anal. Biochem. 2009;388:351–352. doi: 10.1016/j.ab.2009.02.010. doi:10.1016/j.ab.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Huang Z, Szostak JW. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden EJ, Lehman N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem. Biol. 2006;13:909–918. doi: 10.1016/j.chembiol.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Draper WE, Hayden EJ, Lehman N. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res. 2008;36:520–531. doi: 10.1093/nar/gkm1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayden EJ, von Kiedrowski G, Lehman N. Systems chemistry on ribozyme self-construction: evidence for anabolic autocatalysis in a recombination network. Angew. Chem. Int. Ed. 2008;47:8424–8428. doi: 10.1002/anie.200802177. [DOI] [PubMed] [Google Scholar]
- 24.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 25.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helicies and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl Acad. Sci. USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crick FHC. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 27.Orgel LE. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- 29.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 30.Levy M, Ellington AD. The descent of polymerization. Nat. Struct. Biol. 2001;8:580–582. doi: 10.1038/89601. [DOI] [PubMed] [Google Scholar]
- 31.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 32.Riley CA, Lehman N. Generalized RNA-directed recombination of RNA. Chem. Biol. 2003;10:1233–1243. doi: 10.1016/j.chembiol.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Lehman N. A recombination-based model for the origin and early evolution of genetic information. Chem. Biodivers. 2008;5:1707–1717. doi: 10.1002/cbdv.200890159. [DOI] [PubMed] [Google Scholar]
- 34.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for the evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:1–4. [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt T, Lehman N. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem. Biol. 1999;6:857–869. doi: 10.1016/s1074-5521(00)80005-8. [DOI] [PubMed] [Google Scholar]
- 37.Ancel LW, Fontana W. Placticity, evolvability, and modularity in RNA. J. Exp. Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 38.Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 39.Eigen M, Schuster P. The hypercycle: a principle of natural self-organization. Part A: emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 40.Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. New York: Oxford University Press; 1993. NY. [Google Scholar]
- 41.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matricies. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.