Abstract
In the presence of Mn2+, an activity in a preparation of purified Bacillus subtilis RecN degrades single-stranded (ss) DNA with a 3′ → 5′ polarity. This activity is not associated with RecN itself, because RecN purified from cells lacking polynucleotide phosphorylase (PNPase) does not show the exonuclease activity. We show here that, in the presence of Mn2+ and low-level inorganic phosphate (Pi), PNPase degrades ssDNA. The limited end-processing of DNA is regulated by ATP and is inactive in the presence of Mg2+ or high-level Pi. In contrast, the RNase activity of PNPase requires Mg2+ and Pi, suggesting that PNPase degradation of RNA and ssDNA occur by mutually exclusive mechanisms. A null pnpA mutation (ΔpnpA) is not epistatic with ΔrecA, but is epistatic with ΔrecN and Δku, which by themselves are non-epistatic. The addA5, ΔrecO, ΔrecQ (ΔrecJ), ΔrecU and ΔrecG mutations (representative of different epistatic groups), in the context of ΔpnpA, demonstrate gain- or loss-of-function by inactivation of repair-by-recombination, depending on acute or chronic exposure to the damaging agent and the nature of the DNA lesion. Our data suggest that PNPase is involved in various nucleic acid metabolic pathways, and its limited ssDNA exonuclease activity plays an important role in RecA-dependent and RecA-independent repair pathways.
INTRODUCTION
Genome stability is dependent on numerous DNA metabolic proteins, which ensure that replication, repair and recombination occur with high fidelity. The rapid detection and subsequent repair of DNA double-strand breaks (DSBs) is critical for the survival of all organisms. DNA damage causes perturbations in DNA replication, arrests cell cycle progression and triggers a transcriptional response that increases the chances of survival (1–4). Among the first responders to DSBs are the bacterial RecN or the eukaryotic Mre11, Rad50 and Xrs2 (in budding yeast) or Nbs1 (in mammals) complex, MRX(N) (5,6). In Bacillus subtilis, a prototype of the Firmicutes phylum, introduction of site-specific or randomly induced DSBs results in re-localization of RecN from a diffuse distribution to a discrete RecN focus (7–9). DSBs are mainly repaired by error-free homologous recombination (HR), but the presence of template- and RecA-independent repair by non-homologous end joining (NHEJ) is not ruled out (5,10–13). HR initiates with 5′ → 3′ end processing by the RecJ exonuclease in concert with a RecQ-like helicase (RecQ or RecS) or by the AddAB nuclease–helicase complex (counterpart of Escherichia coli RecBCD) (5,10,11,14). The resulting 3′ single-stranded (ss) tails are bound by SsbA (homologous to E. coli SSB), with RecN binding to the 3′-OH ends (14,15). RecN facilitates tethering of DNA ends and promotes the relocalization to a discrete repair centre (RC) (8,9). RecO, RecR and RecA, and, later, RecF and RecU co-localize with RecN at the RC (7,15–18). The absence of resection (addAB ΔrecJ cells) prevents the formation of a discrete RecN focus and RecA localization to RecN-promoted RC (8,9). RecA, with the help of RecA mediators, polymerizes onto SsbA-coated 3′-tails, and catalyzes DNA strand invasion with an intact homologous duplex (D-loop intermediate) and strand transfer, leading to Holliday junction (HJ) formation (17–21). The D-loop acts as a target for loading the primosome that then primes DNA synthesis, and the HJ acts as a target for loading the branch-migration translocases, RecG or RuvAB (22,23). RuvAB promotes loading of the RecU HJ resolvase to the RecN-promoted RC (16) and RecU (ortholog of E. coli RuvC) in concert with RuvAB, or perhaps RecG, cleaves the HJ, which is sealed by DNA ligase. The repair of single strand gaps (SSGs) by daughter strand gap repair is a poorly understood process in B. subtlis cells.
Recent studies revealed that archaeal Mre11/Rad50 or the yeast MRX-Sae2 complex catalyzes a limited 3′-end resection, which is rapidly processed by either the Sgs1–Dna2 complex or Exo1 to yield a duplex with a 3′-ssDNA tail (24–26). In vitro analyses indicate that both eukaryotic Rad50 and bacterial RecN, which are members of the structural maintenance of chromosome family of proteins, bridge and tether DNA ends (15,27). Eukaryotic Mre11 degrades 3′ ends in an Mn2+-dependent manner, and Rad50 and Xrs2 (Nbs1) stimulate such activity, which is modulated by ATP (28,29). We may ask whether there is a ssDNA exonuclease linked to RecN in B. subtilis cells. The functions involved in the processing of the 5′-end are conserved between E. coli and B. subtilis cells. While E. coli encodes at least four ssDNA exodeoxyribonucleases with a 3′ → 5′ polarity (ExoI, ExoVII, ExoX and ExoXI [TatD]) (30,31), no functional equivalent to any of these ssDNA exonucleases has been reported in B. subtilis (see http://www.genolist.pasteur.fr/SubtiList/). These observations prompted us to search for a 3′ → 5′ ssDNA exonuclease activity associated with RecN protein. We show here that an Mn2+-dependent ssDNA exodeoxyribonuclease co-purifies with RecN and is attributed to PNPase. PNPase is capable of limited 3′-end resection that is likely necessary to remove blocked 3′-ends or to generate blunt-ends.
PNPase is a non-essential multifunctional enzyme responsible for Mg2+- and inorganic phosphate (Pi)-dependent 3′ → 5′ processive exoribonuclease activity (32,33). PNPase can convert ribonucleoside-5′-monophosphates (NMPs) into ribonucleoside-5′-diphosphates (NDPs) and can synthesize RNA polymers using NDP substrates (32–37) or DNA polymers using dNDP substrates, in the absence of DNA template, when MgCl2 is replaced by FeC13 (38). A B. subtilis ΔpnpA strain shows a number of phenotypes including cold sensitivity, competence deficiency, tetracycline sensitivity and long multiseptate growth. All of these phenotypes are presumably linked to mRNA turnover and/or recycling of NDPs (39–41).
To shed light on the role of PNPase in Mn2+-dependent degradation of ssDNA, genetic and biochemical experiments were performed.
MATERIALS AND METHODS
Bacterial strains and survival studies
E. coli BL21(DE3)[pLysS] and B. subtilis BG214 strains were described previously (14). The pCB422-borne recN gene, under the control of the recA promoter, in B. subtilis strain BG214 was used to over-express RecN upon mitomycin C (MMC) induction, as described in (14). The pCB730-borne recN-His gene was used to over-express the recN-His gene under the control of the recA promoter, where a DNA segment coding for a hexa-histidine followed by a stop codon was used to replace the stop codon of the recN wild type (wt) gene. The pNP21-borne pnpA gene in E. coli strain EG763 was used to over-express the PNPase protein (42). Chromosomal DNA isolated from the kanamycin-resistant (KmR) ΔpnpA prophage-containing strain (40) was used to transform BG214 (rec+) to give BG993 or BG125 (addA5) to give BG1007. Plasmids carrying ΔrecQ, ΔrecN, ΔrecO, ΔrecU, ΔrecG or ΔrecA constructs were used to transform BG993 as previously described (23). All B. subtilis strains used in this study were isogenic to BG214 (rec+ control) and are listed in Supplementary Table S1.
B. subtilis cells were grown to an OD560 = 0.4 at 37°C in LB broth or S7 minimal medium as indicated. To measure acute sensitivity, cells growing exponentially in LB broth were exposed to increasing concentrations of methyl methanesulfonate (MMS) or hydrogen peroxide (H2O2) for 15 min, and were plated on LB agar and incubated overnight at 37°C. To measure chronic sensitivity to H2O2, MMS, MMC or 4-nitroquinoline-1-oxide (4NQO), serial dilutions of exponential phase cells were spotted on plates containing the indicated amount of the drug and were incubated overnight at 37°C.
Image acquisition
Fluorescence microscopy was performed on an Olympus BX61 microscope. Cells were mounted on agarose pads containing S7 growth medium on object slides. Images were acquired with an Olympus DP70 color CCD camera; signal intensity and cell length were measured using the Metamorph 4.6 program. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; final concentration 0.2 μg/ml).
Reagents, protein purification and antibodies
RecN was over-expressed and purified from BG214 (wt) and BG993 (ΔpnpA) B. subtilis cells bearing pCB422 after MMC induction to >98% homogeneity as described in (14). RecN-His was over-expressed from BG214 cells and purified using a nickel-chelating column, according to the instructions of the manufacturer (Qiagen). Rabbit polyclonal anti-RecN-His antibodies were obtained using standard techniques. B. subtilis PNPase, which was purified from E. coli cells to ∼95% purity, as previously described, was free of E. coli PNPase (42). The molar extinction coefficients for RecN and PNPase were calculated to be 30 600 and 77 800 M−1 cm−1 at 280 nm, as previously described (19).
The DNA modification enzymes were supplied by Roche. Dithiothreitol (DTT), ATP, dATP, ATPγS and AMP-PNP were from Sigma. [α-32P]dATP, [α-32P]ATP, [α-32P]ddATP and [γ-32P]ATP were from Amersham Bioscience. All chemicals used were reagent grade, and nucleotides concentrations were measured spectrophotometrically using an extinction coefficient of 1.54 × 10−4 M−1 cm−1 at 260 nm. They were dissolved as concentrated stock solutions at pH 7.5.
Western blot analysis
Cells were grown to an OD560 = 0.4 at 37°C in LB. MMC (3 μM) was added and the cells were incubated for 30 min. The cells were centrifuged, re-suspended in buffer A (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 5% glycerol) and lysed by sonication. For Western blotting, extracts containing equal concentrations of protein were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). Blots were probed with the indicated rabbit polyclonal antibodies raised against RecA or RecN, which were obtained using standard techniques.
2D-PAGE and peptide fingerprint
The proteins present in the RecN sample, prior to the gel filtration step, were separated by preparative two-dimensional (2D) PAGE using a linear pH gradient in the 3–10 pH range. The protein bands were excised manually, digested with modified porcine trypsin, and the resulting peptides analyzed using a MALDI-TOF mass spectrometer as described in Supplementary Annex 1.
Assays of PNPase activities
Linear oligonucleotide ssDNA60 (5′-CTCCTATTATGCTC AACTTAAATG ACCTACTCTATAAAGCTATAG TACTGCTATCTAATC-3′) and dsDNA60, formed by annealing of ssDNA60 with its complementary 60-mer, were end-labelled at the 3′-end using [α-32P]-dATP or [α-32P]-ddATP and terminal transferase (Roche). ssDNA60, dsDNA60, SS360 (5′ AAAAAAAAAAAAAAAAAAAAA AAAAAAAAAGAATTCGAGCTCGGTA CCCGGGGATCCTCT-3′), SS460 (5′-AGAGGATCCCCGGGTACCGA GCTCG AATTCAAAAAAAAAAAAA AAAAAAAAAAAAAAAAA-3′), SS580 (5′-CGCAAGC G ACAGGAACCTCGAGGGATCCG TCCTAGCAAGGGGCTGCTACCGGA AGCTTCTCGAGG TTCCTGTCGCTTGCG-3′) and RNA (5′-CAUCCUGUUCCAUGGCCAAUU-3′) were 5′-end labelled using [γ-32P]-ATP and polynucleotide kinase (New England Biolabs). DNA concentrations were determined using molar extinction coefficients of 8780 and 6500 M−1 cm−1 at 260 nm for ssDNA and dsDNA, respectively.
The [γ32P]-DNA or [γ32P]-RNA was incubated with PNPase in buffer B (50 mM Tris–HCl [pH 7.5], 50 mM NaCl, 2 mM MnCl2, 1 mM DTT, 2% PEG-6000) or in buffer C (50 mM Tris–HCl [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 2% PEG-6000) containing 1 mM ATP, ADP, ATPγS or AMP-PNP, or no nucleotide, with or without 2 mM sodium phosphate. Where indicated a 10-fold excess of unlabelled DNA was added to the reaction mixture. The samples were separated on native (n) or 7 M Urea denaturing (d) PAGE (14). Quantitation of band shifts and of DNA degradation products was done with a PhosphorImager instrument (GE Healthcare).
RESULTS
Purified RecN shows Mn2+-dependent exonuclease activity
The B. subtilis RecN protein was purified from the soluble fraction of a B. subtilis extract to near homogeneity (∼98%) as previously described (14). RecN, as conventionally purified, contains ∼1% of GroEL, based on N-terminal sequencing (see 14). We have shown previously that incubation of RecN with a 60-nt long ssDNA (ssDNA60), in the presence of Mg2+, results in the formation of several slowly migrating RecN–DNA complexes, designated CI, CII and CIII (14,15). Indeed, RecN specifically binds and protects the radiolabelled 3′-OH termini from ExoVII degradation (14), and forms large networks in which many DNA segments are end-bridged by RecN (15).
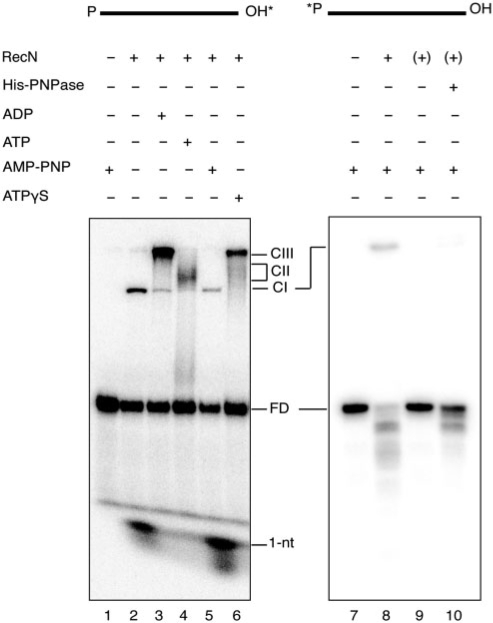
In the presence of Mn2+, without a nucleotide cofactor or with AMP-PNP present, incubation of RecN with 3′-end-labelled ssDNA60 led to the accumulation of a ∼1-nt long product (Figure 1, lanes 2 and 5), in addition to the slowly migrating complexes. In the presence of ADP·Mn2+, ATP·Mn2+ or ATPγS·Mn2+, the ∼1-nt product was not observed (Figure 1, lanes 3, 4 and 6). Degradation of the ssDNA was also observed when RecN was incubated with 5′-end-labelled ssDNA60 in the presence of AMP-PNP·Mn2+ (Figure 1, lane 8). The pattern of decay products from the 5′-end-labelled substrate suggested that limited degradation was occurring from the 3′ end. This would explain accumulation of the labelled 1-nt product from the 3′-end-labelled substrate (lanes 2 and 5). Degradation of ssDNA60 was not observed under any condition when Mg2+ was present instead of Mn2+ (data not shown, and see below, Figure 3).
Figure 1.
3′-to-5′ ssDNA exonuclease activity in the RecN sample. Linear ssDNA60 (1 nM), labelled (*) at the 3′-end (lanes 1–6) or the 5′-end (lanes 7–10) was incubated with RecN (10 nM) purified from wt cells or from the ΔpnpA strain [denoted as (+)] in buffer B for 10 min at 37°C. Where indicated, ADP, ATP, AMP-PNP or ATPγS (1 mM) was added and incubation continued for 30 min at 37° C. For the reaction shown in lane 10, His-PNPase (0.1 nM) was added after 10 min to the pre-formed RecN·ssDNA60 complex. Incubation products were separated in 10% nPAGE and visualized by autoradiography. RecN·ssDNA complexes are labelled CI, complex I; CII, complex II; CIII, complex III (14,15); FD, free DNA. Migration of [32P]-labelled mononucleotide released by incubation with RecN is indicated (1 nt).
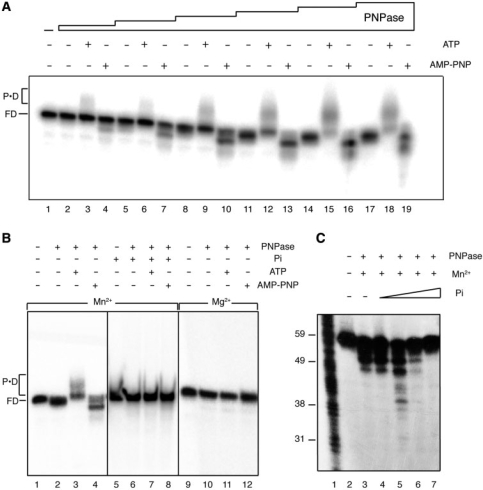
Figure 3.
Binding and degradation of ssDNA by PNPase. In (A) [γ32P]-ssDNA60 (1 nM) was incubated with increasing concentrations of PNPase (0.05, 0.1, 0.25, 0.5, 1.5 and 2 nM) for 30 min in buffer B containing 1 mM ATP, AMP-PNP or lacking a nucleotide cofactor, and the samples were separated in 10% nPAGE. FD, free DNA; PD, PNPase–DNA complexes. In (B) [γ32P]-ssDNA60 (1 nM) was incubated with PNPase (2 nM) for 30 min in buffer B (Mn2+) or buffer C (Mg2+) containing 1 mM ATP, AMP-PNP or lacking a nucleotide cofactor, and with or without added 2 mM Pi, and the samples were separated in 10% nPAGE. In (C) [γ32P]-ssDNA60 (1 nM) was incubated with PNPase (0.3 nM) for 30 min in buffer B containing 1 mM AMP-PNP and no added Pi, or increasing Pi (0.2, 2, 20 and 200 μM), and the samples were separated in 15% dPAGE.
The 3′ → 5′ ssDNA exonuclease activity is not associated with RecN itself
A genuine nuclease domain cannot be predicted from the primary sequence of RecN, which raised doubts about its ability to function as a 3′ → 5′ ssDNA exonuclease. We considered the possibility that an exodeoxyribonuclease activity co-purified with RecN. The last step of purification of conventionally purified RecN protein includes an FPLC gel filtration chromatography (7,14). This chromatographic step was used to remove the greater part of GroEL (MR = 840 000) and the traces of RecN linked to ssDNA from the MR = 520 000 RecN peak (7,14). However, under these conditions traces of GroEL (∼1%) were still present in the MR = 520 000 peak of highly enriched RecN (14).
Analysis of the polypeptides present in the RecN sample prior to gel filtration chromatography revealed that RecN and GroEL were present in comparable amounts and, to a smaller extent, four other polypeptides were detected (Supplementary Figure S1). Partial proteolysis and mass spectrometry were used to identify these polypeptides as AhpC, PncB, PNPase and YqfO proteins (Table 1 and Supplementary Annex 1).
Table 1.
Proteins present in the RecN sample and relative protein ratio in the MR = 520 000 peak
| Protein | Estimated Mass | Relative mass | Estimateda pI | Experimental pI | Relative proportionb (%) |
|---|---|---|---|---|---|
| RecN | 64.3 | 62 | 4.8–4.9 | 4.9 | ≥98 |
| GroEL | 57.2 | 60 | 4.5–4.7 | 4.6 | 1.2 ± 0.2 |
| PncBc | 56 | 44 | 5.0–5.2 | 5.1 | 0.3 ± 0.1 |
| PNPase | 77.2 | 75 | 4.8–5.0 | 4.9 | 0.2 ± 0.1 |
| Yqfo | 40.7 | 40 | 5.3–5.4 | 5.3 | 0.1 ± 0.1 |
| AhpC | 20.4 | 19 | 4.2–4.4 | 4.1 | 0.3 ± 0.1 |
aThe estimated pIs were obtained from two different data bases (subtiList and Expasy).
bThe relative proportion of polypeptides present in the MR = 520 000 RecN peak is shown.
cPeptide fingerprinting revealed that a PncB truncated form was present in the RecN sample.
Since the protein spots could be easily separated on a 2D-PAGE, the relative amounts of the six proteins present in the highly enriched RecN fraction were estimated from spots obtained by heavily overloading a 2D-PAGE (Table 1). PNPase accounted for 0.2% of the total protein (see Supplementary Annex 1).
When RecN protein was purified from the ΔpnpA strain, the Mn2+-dependent 3′ → 5′ ssDNA exodeoxyribonuclease activity, in the presence or absence of AMP-PNP, was no longer detected (Figure 1, lane 9). The addition of partially purified, His-tagged PNPase from E. coli cells to RecN purified from B. subtilis ΔpnpA cells, at a proportion comparable to the amount present in the conventionally purified RecN protein, resulted in the accumulation of degradation products with a similar pattern to RecN purified from the wt strain (Figure 1, compare lanes 8 and 10). These results suggested that the degradation of ssDNA in the presence of purified RecN was due to the contaminating PNPase.
The proteins that co-purify with RecN are involved in nucleic-acid metabolism (Supplementary Annex 2), but pull-down experiments failed to detect any stable interaction between purified RecN and His-PNPase (data not shown).
PNPase is required for DNA repair
The suggestion that PNPase, which is the major RNA-degrading enzyme in B. subtilis cells (40,43), might contribute to ssDNA degradation in vivo was intriguing. There has been relatively little work done on the nature of the DNA lesions in vegetative growing of B. subtilis cells, but in other bacteria the lesions generated by H2O2 and MMS are mainly removed by base excision repair (BER), and the lesions generated by the ultraviolet light-mimetic agent 4NQO and MMC are removed by nucleotide excision repair (NER) (3). Encounter of a replication fork with a DNA lesion introduced by MMS or 4NQO leads mainly to the accumulation of SSGs, MMC is likely to lead to formation of SSGs and/or DSBs, while the wide variety of lesions introduced by H2O2 includes single-strand breaks, DSBs, base modifications, abasic sites and sugar modifications (3).
To examine the involvement of PNPase in repair-by-recombination, the chronic sensitivity of ΔpnpA cells to different DNA damaging agents (H2O2, MMS, 4NQO and MMC) was measured. ΔpnpA cells (see Table S1) chronically exposed to MMS, 4NQO or MMC showed increased DNA damage tolerance, but the cells were more sensitive to reactive oxygen species (ROS) generated by H2O2, as compared to wt cells (Figure 2).
Figure 2.
Survival of ΔpnpA cells exposed to a chronic dose of H2O2, MMS, 4NQO or MMC. Cells were grown to OD560 = 0.4 in LB medium and serially diluted (10−3 to 10−7) and plated on LB plates containing 1 mM H2O2, 2.7 mM MMS, 200 nM 4NQO or 180 pM MMC. − Drug, no drug added.
Mechanism of PNPase effect on DNA repair
Several hypotheses could be put forth to explain the observed alteration in DNA repair. First, mRNA degradation is an important step by which gene expression can be controlled. Since the cellular response to DNA damage involves changes in the coordinated control of gene expression, one could hypothesize that the absence of PNPase affects the processing of gene transcripts required for DNA repair and/or recombination (see 44). To test this, DNA replication was perturbed by addition of MMC (3 μM), which induces a LexA-/RecA-dependent global transcriptional response. The absence of PNPase did not alter the level of RecA induction when compared to wt cells, and RecA reached levels comparable to those observed in the ΔlexA or ΔlexA ΔpnpA strain (see Table S2). Furthermore, a genome-wide analysis of mRNA levels in wt and ΔpnpA cells revealed no significant difference in the relative amounts of transcripts of genes involved in DNA repair and/or repair-by-recombination (GD and DHB, unpublished results), suggesting that the absence of PNPase did not affect the level of mRNAs specified by DNA repair genes. PNPase might be required to process and convert mRNAs into an active form (39) or might be involved in the degradation of small noncoding (nc) RNAs, as previously suggested (45). However, we observed that the ΔpnpA strain showed a different outcome upon exposure to H2O2 and MMS (Figure 2). Since both H2O2- and MMS-induced DNA damage is specifically removed by BER, if PNPase processes specific mRNAs or degraded ncRNAs that control BER genes, exposure to either of these drugs should show a similar phenotype.
A second hypothesis to explain how the absence of PNPase causes sensitivity to H2O2 is that a reduction in mRNA turnover might cause an imbalance in DNA replication and/or DNA segregation, and such defect could reduce cell survival. Indeed, E. coli mukB (mukBEco) mutants can be suppressed by altering mRNA turnover (46), and pnpA mutants of B. subtilis grow as multiseptate filaments, perhaps suggesting a defect in cell division that is tied to chromosome segregation (40). To investigate whether the ΔpnpA strain shows a chromosomal segregation defect, wt and ΔpnpA cells were grown to mid-exponential phase, the nucleoids were stained with DAPI, and the cells were fixed and visualized by fluorescence microscopy. Anucleate cells, measured as the total or partial absence of DAPI-stained material, were rare in ΔpnpA and wt cells (Figure S2A). Furthermore, it has been shown that the absence of RecU or RecG causes an increase in anucleate cells of ∼85- and 150-fold, respectively, when compared with wt cells (23,47). The fate of chromosomal segregation was not affected in ΔrecG ΔpnpA (7.7% anucleates) or ΔrecU ΔpnpA (4.5% anucleates) cells (Figure S2B and S2C), when compared to single ΔrecG (7.3%) or ΔrecU (4.4%) controls (23,47).
A third hypothesis is that PNPase plays an active role in the accumulation of nucleoside diphosphate and specifically of the limiting CDP. It has been suggested previously that there is a link between RNA turnover and DNA replication (41,48). However, if the defect was attributed to an imbalance in pyrimidine metabolism, a decrease of survival should be independent of the type of DNA lesion in ΔpnpA cells, whereas we find that ΔpnpA cells are more sensitive only to H2O2 (Figure 2).
A fourth explanation is that PNPase might interact directly or indirectly with a DNA repair protein, and the absence of such an interaction might interfere with a DNA repair pathway. Previously, it was proposed that PNPaseEco interacts with RecAEco (49), and we have shown that RecN and PNPase co-purify (Figure S1). However, we have failed to detect any stable in vitro interaction of RecA or RecN with the PNPase protein (data not shown).
Finally, we considered the hypothesis that, rather than having an indirect effect due to alterations on gene expression, pyrimidine metabolism or protein-protein interactions, PNPase might degrade DNA, and in its absence the persistence of any unprocessed substrate alters the efficiency of DNA repair. It is possible that PNPase fulfils an antagonistic role of curbing unnecessary recombination by ensuring prompt removal of 3′-ssDNA tails (as might occur in the presence of MMS-, 4NQO- or MMC-induced damage) at SSG, and by eliminating blocked 3′-ends of an extruded chicken-foot structure (as occurs with H2O2-induced lesions) that could be extended by a DNA polymerase.
Purified PNPase binding and degradation of ssDNA
The effect of increasing concentrations of purified PNPase on ssDNA was analyzed. In the presence of Mn2+ and ATP, PNPase bound 5′-end-labelled ssDNA60 with high affinity (apparent binding constant [Kapp]) of 0.6 ± 0.2 nM) (Figure 3A, lanes 3, 6, 9, 12, 15, 18) and in a sequence-independent manner (see below). To confirm that the shifted bands were due to the PNPase·ssDNA60 complexes, rather than to the addition of AMP residues by the polymerase activity of PNPase (35), cold ssDNA60 was incubated with PNPase in the presence of Mn2+ and [α32P]-ATP. The very low level of radiolabel material incorporated into ssDNA60 (0.2% of total ssDNA60) (data not shown) cannot explain the shift in mobility of >95% of total 5′-end-labelled ssDNA60 (Figure 3A, lane 18).
The accumulation of the slow-moving PNPase·ssDNA60 complexes was not observed in the presence of 2 mM Pi (Figure 3B, compare lanes 3 and 7) or when Mn2+ was replaced by Mg2+ (Figure 3B, compare lanes 3 and 11).
In the absence of added nucleotide, accumulation of the slow-moving PNPase·ssDNA60 complexes was not observed (Figure 3A, lanes 2, 5, 8, 11, 14, 17), but a slight degradation was observed at high PNPase concentrations (Figure 3A, lanes 11, 14, 17). When AMP-PNP was present instead of ATP, degradation products from the ssDNA60 were observed (Figure 3A, lanes 4, 7, 10, 13, 16, 19). Degradation of ssDNA60 was not observed in the presence of 2 mM Pi (Figure 3B, compare lanes 4 and 8), or when Mg2+ was present instead of Mn2+ (Figure 3B, compare lanes 4 and 12).
To address the effect of Pi concentration on ssDNA degradation, experiments were performed in the absence of AMP-PNP and increasing concentrations of Pi. In the absence of Pi or with addition of Pi up to 2 μM, PNPase degradation of ssDNA was observed (Figure 3C, lanes 3–5). A partial inhibition of ssDNA degradation was observed at 20 μM Pi (Figure 3C, lane 6), and a block to ssDNA degradation was observed at the highest Pi concentration (Figure 3C, lane 7). Similar results were observed in the presence of AMP-PNP (data not shown). We conclude that the slight degradation seen in the absence of added nucleotide (e.g. Figure 4, lanes 3–7) relies on contaminating Pi, and the increased activity in the presence of AMP-PNP (e.g. Figure 4, lanes 14–19) is due to contaminating Pi in the added AMP-PNP.
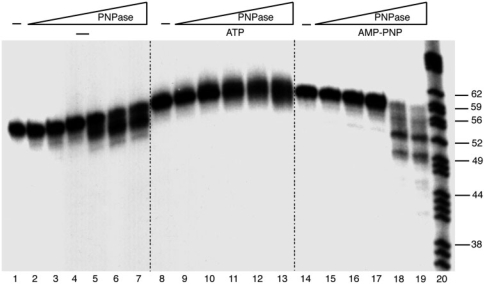
Figure 4.
PNPase promotes limited degradation of ssDNA. [γ32P]-ssDNA60 (1 nM) was incubated with increasing concentrations of PNPase (0.02, 0.04, 0.08, 0.16, 0.33 and 0.66 nM) for 30 min in buffer B containing 1 mM ATP, AMP-PNP or lacking a nucleotide cofactor, and the samples were separated in 15% dPAGE. Sizes of molecular markers (in nt) are indicated (lane 20).
The dependence of the degradation reaction on PNPase concentration under several conditions was assayed in more detail (Figure 4). In the absence of added nucleotide, 5′-end-labelled ssDNA60 was degraded slightly from the 3′ end, yielding products that were a few nucleotides shorter than the full length (Figure 4, lanes 1–7). In the presence of ATP, very little degradation was observed, except for a slight activity in the presence of the highest concentrations of PNPase (Figure 4, lanes 8–13). Alternatively, the lack of degradation could be due to the presence of contaminating concentrations of Pi in the ATP. In the presence of AMP-PNP and high concentrations of PNPase, ssDNA60 was degraded from the 3′ end to yield products that were up to ∼10 nt shorter than the full length, including several discrete bands. To determine whether these discrete degradation products were due to an inhibitory effect of any particular sequence, PNPase degradative activity was assayed using different ssDNA substrates (see ‘Materials and methods’ section). The SS360 substrate (polyA [30 As] at the 5′ end and high dC + dG at the 3′ end) and the SS460 substrate (high dC + dG at the 5′ end and polyA [30 As] at the 3′ end) were degraded with similar efficiency. Similar discrete degradation bands accumulated as observed with ssDNA60 (data not shown). These results argue against any particular sequence being involved in inhibiting PNPase processivity. It is also possible that secondary structure in the ssDNA could lead to the accumulation of discrete ssDNA bands. However, applying the Zuker m-fold version 3.2 analysis to ssDNA60 gave no significant predicted secondary structure.
PNPase was unable to linearize circular ssDNA (data not shown), confirming that the PNPase activity was exonucleolytic. Also, PNPase failed to degrade the dsDNA60 substrate, even in the presence of both Mn2+ and AMP-PNP (data not shown), demonstrating that the limited degradative activity was specific for ssDNA.
Effect of divalent cations on PNPase exoribonuclease activity
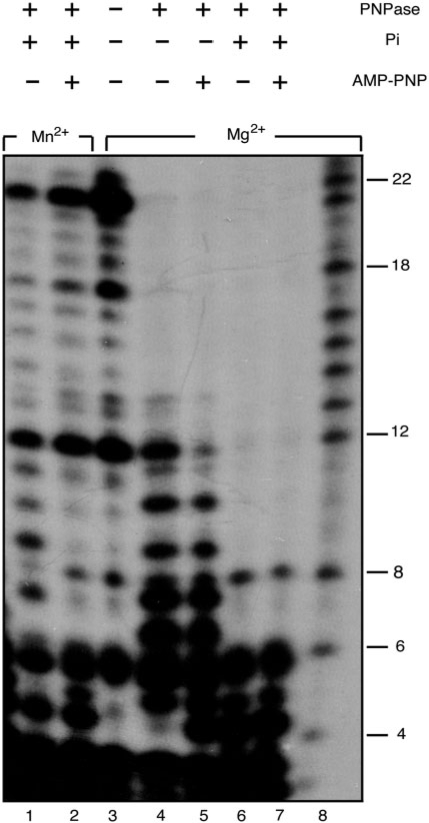
PNPase has an Mg2+- and Pi-dependent 3′ → 5′ exoribonuclease activity (48,50). To test whether Mn2+ affected the exoribonuclease activity, a 21-nt RNA was incubated with PNPase under different experimental conditions. When Mg2+ and Pi were present, PNPase actively degraded the RNA substrate (Figure 5, lanes 6 and 7). In the presence of Mn2+, PNPase showed poor RNase activity (Figure 5, compare lanes 1 and 2 to control lane 3 with no PNPase added). Addition of Mg2+ alone or with AMP-PNP gave an intermediate level of degradation (Figure 3, lanes 4 and 5), probably due to the presence of contaminating phosphate. It is likely that the exodeoxyribonuclease and exoribonuclease activities of PNPase are mutually exclusive, such that PNPase degrades ssDNA in a manner dependent on the presence of Mn2+and low Pi concentration, whereas PNPase degrades RNA in a manner dependent on the presence of Mg2+ and a range of Pi concentrations.
Figure 5.
Mn2+ inhibits the exoribonuclease activity of PNPase. [γ32P]-RNA21 (1 nM) was incubated with PNPase (0.3 nM) for 30 min in buffer B (Mn2+) or buffer C (Mg2+) containing 1 mM AMP-PNP, 2 mM Pi or no addition, and the samples were separated in 15% dPAGE. Sizes of molecular markers (in nt) are indicated (lane 8).
PNPase end processing is distributive
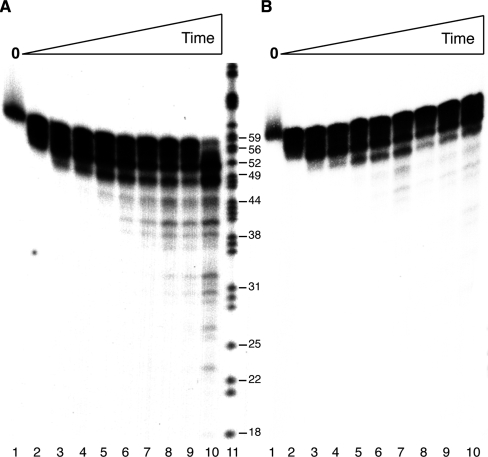
The rate of end processing was tested using limiting PNPase and 5′-labelled ssDNA60, with or without addition of an excess of cold ssDNA60. In the presence of Mn2+ and AMP-PNP, PNPase was capable of processing ssDNA60 at the earliest time point to yield ∼56 nt ssDNA (Figure 6, lane 2). With increasing times, discrete smaller ssDNA segments were observed (Figure 6A, lanes 3–9). After 120 min of incubation, 10–12% of the full-length substrate remained and ∼75% had been degraded to <50 nt (Figure 6A, lane 10). In the presence of Mn2+ and AMP-PNP and a 10-fold excess of cold ssDNA60, which was added after 1 min, incubation with PNPase led to the accumulation of at least three discrete ssDNAs of ∼56, ∼52 and ∼49 nt within the first 10 min (Figure 6B, lanes 2–5). However, after incubation for longer times degradation was markedly reduced (compare Figure 6A, lanes 8–10 vs 6B lanes 8–10), suggesting that PNPase preferentially degraded a few nucleotides at a time before releasing the substrate. The PNPase activity was not lost during incubation, because similar results were obtained when the enzyme was pre-incubated before addition of ssDNA (data not shown).
Figure 6.
PNPase activity on ssDNA is distributive. In (A) [γ32P]-ssDNA60 (1 nM) was incubated with PNPase (0.3 nM) for increasing times (1, 2, 5, 10, 20, 30, 40, 60 and 120 min) in buffer B containing 1 mM ATP, AMP-PNP or lacking a nucleotide cofactor. In (B) as in (A) except that at 1 min a 10-fold excess of cold ssDNA60 was added. The reaction was stopped by the addition of proteinase K and SDS, and the samples were separated by 15% dPAGE. Lanes marked ‘0’ were before addition of PNPase. Sizes of molecular markers (in nt) are indicated (lane 11).
We conclude that PNPase degrades ssDNA to a limited extent in the 3′ → 5′ direction via a mechanism that requires Mn2+ and low Pi, that is inhibited by ATP, and that occurs by a distributive mode of processing.
Epistasis analysis
The products of the recombination genes, other than recA, have been classified within different epistatic groups (51). Genetic interactions were used to define whether the PNPase enzyme is involved in HR. The ΔpnpA mutation was combined with mutations or null mutations in genes impaired in HR that are representatives of six different epistatic groups: α (ΔrecO and ΔrecR), β (addA5), δ (ΔrecN), ϵ (ΔrecU), ζ (ΔrecQ and ΔrecJ) and η (ΔrecG). In addition, the ΔrecA mutation was introduced into the ΔpnpA strain (Table S1). When a double mutant strain is exposed to a DNA-damaging agent, three outcomes are expected: (i) the double mutant is as sensitive as the more sensitive single mutant (interacting products), (ii) the sensitivity of the double mutant is equal to the sum of each of the single-mutant parents (non-interacting, additive effect) or (iii) the sensitivity of the double mutant is greater that the sum of each of the single-mutant parents (non-interacting, synergistic effect).
The ΔpnpA strain showed a significantly longer doubling time (∼38 min) than the wt (∼29 min), and the absence of ΔpnpA in rec-deficient cells also significantly increased the cell doubling time (Table S1). In the absence of any external DNA damage the number of viable cells, per colony or in liquid medium, for the ΔpnpA, ΔrecQ, addA5 and ΔrecN mutant strains was not significantly reduced (less than 1.3-fold) when compared with the wt strain. The number of viable cells decreased ∼5-fold in the ΔrecO, ΔrecG, ΔrecU and ΔrecA mutant strains, and 6- to 10-fold in the ΔrecO ΔpnpA, ΔrecU ΔpnpA or ΔrecA ΔpnpA strains, when compared to the wt strain (Figure S3, − Drug). However, the number of viable ΔrecG ΔpnpA cells increased slightly when compared to ΔrecG cells (Figure S3, − Drug).
The single and double mutant strains were exposed acutely to increasing concentrations of MMS or H2O2. The ΔpnpA strain was sensitive to H2O2; the lethal concentration to kill 90% (LC90) of the cells was ∼10 mM compared to wt cells with an LC90 of ∼14 mM (Table 2, Figure S4D). However, ΔpnpA cells were more tolerant of MMS (LC90 of ∼27 mM) compared to wt cells (LC90 of ∼18 mM) (Table 2, Figure S4A). Similarly, ΔpnpA cells were sensitive when exposed chronically to H2O2, but were tolerant when exposed chronically to MMS, MMC or 4NQO, compared to wt cells (Table 3, Figure S3).
Table 2.
Sensitivity of ΔpnpA and recombination-deficient strains upon acute exposure to H2O2 and MMS
| Relevant phenotype (epistatic group) | Lethal concentration to kill 90% of the cells (LC90)a |
|||
|---|---|---|---|---|
| H2O2 (in mM) |
MMS (in mM) |
|||
| pnp+ | ΔpnpA | pnp+ | ΔpnpA | |
| wt (NA) | 13.5 ± 0.1 | 10.0 ± 0.3 | 17.8 ± 0.1 | 27.1 ± 0.09 |
| ΔrecA (NA) | 0.20 ± 0.03 | 0.11 ± 0.02 | 0.41 ± 0.04 | 0.30 ± 0.02 |
| ΔrecO (α) | 0.30 ± 0.02 | 0.19 ± 0.01 | 0.73 ± 0.08 | 0.54 ± 0.10 |
| ΔrecR (α) | 0.31 ± 0.02 | 0.20 ± 0.03 | 0.72 ± 0.09 | 0.54 ± 0.08 |
| addA5 (β) | 0.35 ± 0.03 | 0.19 ± 0.02 | 7.1 ± 0.07 | 7.8 ± 0.08 |
| ΔrecN (δ) | 0.7 ± 0.04 | 5.2 ± 0.1 | 8.8 ± 0.14 | 9.8 ± 0.15 |
| ΔrecU (ϵ) | 0.31 ± 0.02 | 0.30 ± 0.04 | 0.67 ± 0.10 | 1.25 ± 0.10 |
| ΔrecQ (ζ) | 9.8 ± 0.4 | 7.0 ± 0.2 | 16.6 ± 0.08 | 31.1 ± 0.19 |
| ΔrecJ (ζ) | 9.6 ± 0.3 | 7.1 ± 0.2 | 16.7 ± 1.9 | 30.8 ± 1.2 |
| ΔrecG (η) | 0.27 ± 0.04 | 0.48 ± 0.02 | 0.63 ± 0.09 | 1.21 ± 0.14 |
| Δku (NA) | 10.2 ± 0.1 | 15.9 ± 0.3 | 16.1 ± 0.1 | 30.9 ± 0.1 |
The single and double mutant strains were exposed to increasing concentrations of the genotoxic agent for 15 min, and dilutions were plated on LB agar plates and incubated overnight at 37°C. Standard errors are indicated. Figures S4 and S5A show the primary data from where the data in Table 2 was derived. NA: not applied.
aThe LC90 results were the average of at least three independent experiments.
Table 3.
Growth of different strains on plates containing different concentrations of MMS, H2O2, MMC or 4NQO
| Strain | No drug | H2O2 (mM) | MMS (mM) | MMC (pM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.2 | 0.5 | 1 | 0.07 | 1.2 | 2.7 | 5 | 90 | 180 | |||||
| wt | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ | +++ | − | |||
| ΔpnpA | ++++ | ++++ | ++++ | (+++) | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | |||
| ΔrecO | +++ | (++) | − | − | (+++) | − | − | (+++) | − | − | |||
| ΔrecO ΔpnpA | ++ | + | − | − | (+) | − | − | (++) | − | − | |||
| addA5 | ++++ | ++++ | (++) | − | ++++ | (++) | − | ++++ | − | − | |||
| addA5 ΔpnpA | +++ | +++ | (+) | − | ++++ | (+++) | − | ++++ | (+) | − | |||
| ΔrecQ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | (++) | ++++ | +++ | − | |||
| ΔrecQ ΔpnpA | ++++ | ++++ | +++ | (++) | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | |||
| ΔrecN | ++++ | ++++ | (+++) | (+) | ++++ | +++ | (+) | ++++ | (+) | − | |||
| ΔrecN ΔpnpA | +++ | ++++ | (+++) | + | ++++ | +++ | + | ++++ | (+++) | + | |||
| ΔrecU | (+++) | (+++) | − | − | (+++) | − | − | +++ | − | − | |||
| ΔrecU ΔpnpA | (++) | (+) | − | − | − | − | − | (+) | − | − | |||
| ΔrecG | +++ | + | − | − | + | − | − | (+++) | − | − | |||
| ΔrecG ΔpnpA | (+++) | (++) | − | − | (++) | − | − | (+++) | − | ||||
| Strain | No drug | H2O2 (mM) | MMS (mM) | MMC (pM) | 4NQO (nM) | ||||||||
| 0.2 | 0.5 | 1 | 0.07 | 1.2 | 2.7 | 5 | 90 | 180 | 7.5 | 50 | 200 | ||
| wt | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ | +++ | − | ++++ | (++++) | ++ |
| ΔpnpA | ++++ | ++++ | ++++ | (+++) | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ |
| Δku | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ | +++ | − | ++++ | (++++) | ++ |
| Δku ΔpnpA | ++++ | ++++ | ++++ | (+++) | ++++ | ++++ | +++ | ++++ | +++ | ++++ | ++++ | ++++ | +++ |
| ΔrecA | ++ | (++) | − | − | ++ | − | − | (+++) | − | − | (++) | − | − |
| ΔrecA ΔpnpA | (++) | (+) | − | − | (+) | − | − | (++) | − | − | (+) | − | − |
| ΔrecA Δku | (++) | (++) | − | − | ++ | − | − | (++) | − | − | (++) | − | − |
Growth of different strains on plates containing the indicated concentrations of MMS, H2O2 or MMC. Symbols: normal growth, ++++; compromised growth, +++; compromised growth and small colony, (+++); poor growth with normal size colonies, ++; compromised growth and small colony, (++); poor growth and tiny colony, +; extremely poor growth and tiny colony, (+); no growth, −. Results are the average of at least three independent experiments. Drug concentrations that highlight the differences between strains are shown. Figures S3 and S5B show the primary data from which the data in this table were derived.
PNPase might contribute to HR in the context of the dynamic flow of recombinational repair. This process has five steps: (i) damage recognition, (ii) end processing, (iii) RecA loading onto SsbA-coated ssDNA, (iv) strand invasion and strand transfer, and (v) migration of the HJs and their resolution (5). The single rec mutants and rec mutants in combination with the pnpA deletion were used to assess the involvement of PNPase in HR. The single rec-deficient strains were moderately sensitive (ΔrecQ and ΔrecJ), sensitive (addA5 and ΔrecN), very sensitive (ΔrecO, ΔrecR, ΔrecU, ΔrecG) and extremely sensitive (ΔrecA) to both MMS and H2O2 when compared to the wt strain (Tables 2 and 3).
RecN, which is among the first responders to DSBs, recognizes the DNA damage and tethers DNA ends together (5). The absence of PNPase suppressed somewhat the ΔrecN sensitivity to acute exposure to MMS and suppressed greatly the ΔrecN sensitivity to acute exposure to H2O2 (Table 2). The absence of PNPase partially suppressed the ΔrecN sensitivity to chronic exposure to MMS, H2O2 or MMC (Table 3). It is possible that in the absence of both PNPase and RecN, an alternative DNA recombination pathway becomes operative.
The AddAB enzyme or RecJ in concert with a RecQ-like helicase (RecQ or RecS) processes the DNA ends to generate 3′ termini (9). The ΔrecQ ΔpnpA, ΔrecJ ΔpnpA and addA5 ΔpnpA cells were more sensitive to H2O2 than the single rec mutants alone, but the absence of PNPase partially suppressed the DNA repair defect of addA5, ΔrecQ and ΔrecJ cells when exposed to MMS (Table 2). A similar pattern was observed when double mutant addA5 ΔpnpA and ΔrecQ ΔpnpA (ΔrecJ ΔpnpA) strains were exposed chronically to DNA damaging agents. The double mutant strains were more sensitive than the single rec mutants to H2O2 but more tolerant to MMS and MMC (Table 3). Thus, it appears that at DNA ends, such as H2O2-mediated breaks, PNPase should be involved in the removal of blocked 3′-ends in the absence of 5′-end processing by AddAB or RecJ/RecQ. At SSGs in the template, such as MMS- or MMC-generated lesions, the removal of 3′-ends should restrict HR.
A RecA mediator (e.g. RecF, RecO and RecR) overcomes the negative effect exerted by SsbA and loads RecA onto ssDNA. RecA protein then promotes the central step of DNA pairing (17,18). The ΔrecO ΔpnpA or ΔrecR ΔpnpA cells were more sensitive to MMS or H2O2 than the ΔrecO and ΔrecR cells (Table 2). A similar degree of sensitivity was observed when the cells were exposed chronically to H2O2, MMS and MMC (Table 3). These results suggest that PNPase and the RecA mediators are non-interacting products, and deficiencies in these functions show an additive effect.
The ΔrecA ΔpnpA cells were more sensitive to acute exposure to MMS or H2O2 or chronic exposure to H2O2, MMS or MMC than the ΔrecA strain (Tables 2 and 3), suggesting that PNPase also may be affecting a step that is not RecA-dependent.
RecG or RuvAB translocate branched structures, and RecU, in concert with RuvAB, resolves the HJ (5). The absence of PNPase partially suppressed the ΔrecG sensitivity to acute exposure to both MMS and H2O2 and the ΔrecU defect to MMS, but ΔrecU ΔpnpA cells were as sensitive to low H2O2 concentrations as the single mutant strain (Table 2). However, a different outcome was observed when the cells were exposed chronically to MMS, H2O2 or MMC. Here, the absence of PNPase increased the sensitivity of ΔrecU cells to the cytotoxic agent, but ΔrecG cells were more tolerant (Table 3).
The multiseptate strands of ΔpnpA cells (Figure S2) were shorter in many of the double mutant strains (data not shown), but no correlation could be drawn between the survival of cells and the relative amount of cells in the multiseptate strand.
From these results we can hypothesize that the absence of both RecN and PNPase or RecG and PNPase might curb unnecessary recombination and/or lead to stabilization of the replication fork, by a mechanism that avoids a DNA break. However, the absence of both RecA and PNPase renders cells more sensitive to H2O2, MMS or MMC than the more sensitive single recA mutant, suggesting the presence of a RecA-independent repair mechanism (Tables 2 and 3). The presence of this repair mechanism could be explained if PNPase played a role in RNA repair (52). Alternatively, PNPase might play a role in the alternative NHEJ pathway.
A null ku mutation is epistatic with pnpA
Most bacterial DSB repair proteins appear to function exclusively in HR or NHEJ, but some proteins may influence both pathways. In B. subtilis stationary phase cells, RecA-independent DNA repair by NHEJ requires a DNA end-binding component called Ku (YkoV) (53). The potential role of PNPase in DNA repair in exponentially growing cells by NHEJ was addressed. The Δku mutation was introduced into the ΔpnpA strain, and the survival of exponentially growing cells to different type of DNA lesions was analyzed.
In the absence of DNA damage from external agents, the number of viable cells in the Δku or ΔpnpA mutant strain was not significantly reduced (less than 1.5-fold) when compared with the wt strain. However, the number of viable cells, per colony (data not shown) or in liquid medium, decreased ∼10-fold in the Δku ΔpnpA, ΔrecA, ΔrecA ΔpnpA or ΔrecA Δku strains when compared to the wt strain (Figure S5B, − Drug).
Exponentially growing Δku cells were slightly more sensitive to acute exposure to MMS or H2O2 when compared to wt cells (Table 2, Figure S5A). A similar degree of sensitivity was observed when Δku or wt cells were exposed chronically to H2O2 or MMC (Figure S5B), but the Δku strain grew slightly better than wt when exposed to MMS or 4NQO (Table 3, Figure S5B). Both ΔpnpA and Δku ΔpnpA were tolerant to acute exposure to MMS, but while Δku ΔpnpA cells were tolerant to acute exposure to a low dose of H2O2, they became as sensitive as ΔpnpA at the high dose of H2O2 (Table 2). The ΔpnpA and Δku ΔpnpA cells were tolerant to chronic exposure to MMS, 4NQO or MMC, but were sensitive to H2O2 (Table 3). The ΔrecA Δku strain showed a similar degree of sensitivity to the ΔrecA strain when exposed chronically to H2O2, MMS, 4NQO or MMC, whereas the ΔrecA ΔpnpA strain was more sensitive than the ΔrecA strain to acute exposure to MMS or H2O2 or chronic exposure to MMS, H2O2, 4NQO or MMC (Tables 2and 3, Figures S3–S5). It is likely that the H2O2-mediated DSBs with blocked 3′-OH end should be processed, perhaps by PNPase, prior to ligation by the LigD enzyme in concert with Ku. Indeed, the B. subtilis LigD enzyme contains a ligase and polymerization domain, but lacks the Mn2+-dependent 3′ → 5′ exonuclease one (13).
DISCUSSION
The multifunctional PNPase enzyme is involved in various nucleic acid metabolic pathways, in addition to its primary role in mRNA turnover (32,33,37). In the presence of ATP, B. subtilis PNPase binds to ssDNA but degradative activity is inhibited. Addition of Pi to >20 μM inhibits not only PNPase 3′ → 5′ exodeoxyribonuclease activity but also PNPase binding to ssDNA. It remains to be determined how ATP or Pi modulates negatively the exodeoxyribonuclease activity. Our results suggest that ATP and Pi exert a negative effect on PNPase by different mechanisms. It has been shown that the exoribonuclease activity of PNPaseEco, in the presence of both Mg2+ and Pi, is also inhibited by ATP (54). It is likely that the inhibition exerted by ATP on both the exoribonuclease activity (in the presence of Mg2+ and Pi) and the exodeoxyribonuclease activity (in the presence of Mn2+ and Pi) of PNPase is physiologically relevant.
Many bacteria of the Firmicutes phylum accumulate Mn2+, and high intracellular levels of Mn2+ directly or indirectly protect proteins and allow fast repair of damaged DNA after DSBs (55,56). Lactobacillus also incorporates Mn2+ as a protectant rather than as a cofactor of the superoxide dismutase (Mn-SOD) (57). Furthermore, not only PNPase but also AhpC, which is involved in the response to peroxide stress that can cause DNA damage, co-purified with RecN (Figure S1 and Supplementary Annex 2), suggesting that Mn2+ ions, ATP and/or Pi might play a role in DSB DNA repair.
PNPase is involved in homologous DNA recombination
Our data do not provide a mechanistic understanding of the role of PNPase in DNA or RNA repair. We presented in vivo evidence that the absence of PNPase increases DNA damage tolerance to MMS, 4NQO or MMC. However, the absence of PNPase increased the sensitivity to H2O2, suggesting the need for nucleolytic processing at 3′-ends. The role of the PNPase enzyme in 3′ → 5′ degradation of ssDNA in RecA-dependent repair was studied. A synergistic epistasis of ΔpnpA with any rec mutation should be expected if the sole role of PNPase is RNA repair (52); however, any of the three different outcomes should be expected if repair-by-recombination is the major role of PNPase. When rec-deficient strains in the ΔpnpA context were exposed acutely to H2O2 or MMS or chronically to H2O2, MMS or MMC, the double mutant was either more sensitive than the sensitive parent (loss-of-function) or was more resistant than the sensitive parent (gain-of-function). In the case where the absence of PNPase caused greater sensitivity, we hypothesize that, when PNPase is present, it provides 3′ → 5′ ssDNA exonuclease activity that trims a few nucleotides from the 3′ end, thereby enabling priming synthesis and enhancing cell survival. In the case where the absence of PNPase caused greater resistance, it is probable that the presence of the PNPase 3′ → 5′ ssDNA exonuclease activity plays an anti-recombinogenic role by rapidly removing ssDNA tails, as postulated for the SbcBEco enzyme (58).
Epistasis experiments suggested that the pnpA mutation was non-epistatic with mutations in functions involved in 5′-end resection (namely addA or recQ and recJ) during acute or chronic exposure to H2O2. However, the absence of PNPase showed a gain-of-function phenotype in ΔrecQ (ΔrecJ) or addA5 cells exposed to DNA lesions caused by MMS, 4NQO or MMC (Tables 2 and 3). It is likely that the DNA lesions generated by H2O2 lead to the accumulation of DNA breaks that depend on PNPase for 3′-exonucleolytic resection, whereas prompt removal of SSGs upon methylation-induced toxicity (by MMS), purine adducts (by 4NQO) or inter-strand crosslinks (by MMC) at the arrested replication fork by PNPase should curb unnecessary recombination. This is consistent with the phenotype observed when E. coli 3′ → 5′ ssDNA exonucleases (e.g. ExoI, ExoVII and ExoX) are absent (59). Alternatively, in the absence of PNPase, the 8-oxoG generated by the BER process might be recycled back to 8-oxodG for incorporation into DNA.
RecU is involved in the resolution of HJs (23,60). ΔrecU ΔpnpA cells were sensitive to different types of DNA lesions caused by chronic exposure to H2O2, MMS and MMC. However, acute exposure of recU ΔpnpA cells to H2O2 or MMS made them more resistant than the parent. It is likely that, in the ΔpnpA context, an alternative function can overcome the absence of RecU upon acute exposure to DNA-damaging agents, but not after chronic exposure.
RecN is involved in DNA damage recognition and joining DNA ends, while RecA is the central recombination enzyme (5,51). The absence of PNPase partially suppressed the ΔrecN defect, but increased the sensitivity of ΔrecA cells to DNA damage, suggesting that a pnpA mutation was epistatic with recN, but was not epistatic with recA. However, recA is epistatic with recO (epistatic groups α), addA (β), recN (δ), recU (ϵ), recQ (ζ) and recG (η) (9,23,61,62).
DNA DSB repair pathways
DNA breaks can arrest cell growth, lead to loss of genetic integrity and, if unrepaired, cause cell death (63). H2O2—directly, or indirectly via production of ROS—promotes DNA breaks. HR is the major pathway to repair DSB in exponentially growing B. subtilis cells, and the contribution of NHEJ is only apparent when HR is inactivated. Indeed, ROS-induced DSBs lead to two-ended DSBs that are processed via RecA-dependent recombination, but with a low efficiency a RecA-independent pathway(s) might become operative. We have shown that ΔpnpA and Δku mutations are non-epistatic with ΔrecA (Figure S5). However, bacteria have very little non-coding DNA and the genome is organized in operon structures; hence, an error-prone repair such as single-strand annealing or NHEJ should drastically reduce the number of viable cells per colonies. Therefore, we propose that NHEJ plays a relatively minor role in DSB repair in exponentially growing cells.
While most DSB repair proteins appear to function exclusively in HR or NHEJ, some proteins may influence both pathways, with the MRX(N) complex being one example (64–66), and RecN, perhaps in association with PNPase, being the bacterial ‘sentinels’ of damage DNA. At present we cannot rule out that PNPase also monitors RNA molecules for oxidative damage, preventing them from functioning and recruiting additional ‘factors’ to destroy damaged transcripts (52).
The responses of bacteria and eukaryotic cells to DNA DSBs show some similarities: (i) RecN, perhaps in association with PNPase, and the MRX(N) complex are among the first responders to DNA DSBs (6,7); (ii) both RecN and Rad50, which belong to the large SMC superfamily, sense DNA ends in an ATP-dependent manner and tether them (15,27); (iii) both Mre11, of the Rad50-Mre11 complex (28,29), and PNPase show a Mn2+-dependent 3′ → 5′ exonuclease activity (this work); (iv) Mre11, of the MRX(N) complex, and PNPase, perhaps in concert with RecN, remove a few nucleotides from the DNA ends to form an early intermediate, which is subsequently processed by dedicated enzymes (24,26,67, this work) and (v) the activity of the Mre11 and PNPase Mn2+-dependent nucleases are modulated by ATP (28, this work).
PNPase is a ubiquitous trimeric enzyme, in which each subunit comprises two RNase PH domains, a K-homology (KH) domain and an S1 or OB-fold-like domain (68). We propose that PNPase interacts with ssDNA through the OB-fold-like domain, as other ss-binding proteins, and the RNase PH domain degrades ssDNA from the 3′-end. Such activities might be directly involved in DNA repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants BFU2009-07167 from Ministerio de Ciencia e Innovación, Dirección General de Investigación (MCI-DGI) (to J.C.A.) and GM48804 from the US National Institutes of Health (to D.H.B.). Funding for open access charge was provided by MCI-DGI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alan Grossmann (Harvard University) for providing the AIG266 strain and to Sylvia Ayora and Gianni Dehò for critical reading of the manuscript.
REFERENCES
- 1.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 2.Begley TJ, Samson LD. Network responses to DNA damaging agents. DNA repair. 2004;3:1123–1132. doi: 10.1016/j.dnarep.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 4.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez H, Carrasco B, Ayora S, Alonso JC. Dynamics of DNA Double-Strand Break Repair in Bacillus subtilis. Norfolk, UK: Caister Academic Press; 2007. [Google Scholar]
- 6.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 8.Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 2005;170:357–366. doi: 10.1083/jcb.200412090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez H, Kidane D, Cozar MC, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J. Bacteriol. 2006;188:353–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spies M, Kowalczykowski SC. In: The Bacterial Chromosome. Higgins NP, editor. Washington, DC: ASM Press; 2005. pp. 389–403. [Google Scholar]
- 12.Mascarenhas J, Sanchez H, Tadesse S, Kidane D, Krishnamurthy M, Alonso JC, Graumann PL. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol. Biol. 2006;7:20. doi: 10.1186/1471-2199-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Ann. Rev. Microbiol. 2007;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez H, Alonso JC. Bacillus subtilis RecN binds and protects 3′-single-stranded DNA extensions in the presence of ATP. Nucleic Acids Res. 2005;33:2343–2350. doi: 10.1093/nar/gki533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez H, Cardenas PP, Yoshimura SH, Takeyasu K, Alonso JC. Dynamic structures of Bacillus subtilis RecN-DNA complexes. Nucleic Acids Res. 2008;36:110–120. doi: 10.1093/nar/gkm759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, Sharples GJ, Alonso JC. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171:873–883. doi: 10.1534/genetics.105.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrasco B, Manfredi C, Ayora S, Alonso JC. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair. 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Manfredi C, Carrasco B, Ayora S, Alonso JC. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cañas C, Carrasco B, Ayora S, Alonso JC. The RecU Holliday junction resolvase acts at early stages of homologous recombination. Nucleic Acids Res. 2008;36:5242–5249. doi: 10.1093/nar/gkn500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeeles JT, Dillingham MS. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 22.Noirot P, Polaed P, Noirot-Gros MF. Replication of the Bacillus subtilis Chromosome. Norfolk, UK: Caister Academic Press; 2007. [Google Scholar]
- 23.Sanchez H, Carrasco B, Cozar MC, Alonso JC. Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation. Mol. Microbiol. 2007;65:920–935. doi: 10.1111/j.1365-2958.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- 24.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins BB, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 28.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 29.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 30.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 32.Condon C. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 2003;67:157–174. doi: 10.1128/MMBR.67.2.157-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Ann. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 34.Ochoa S. Enzymic synthesis of polynucleotides. III. Phosphorolysis of natural and synthetic ribopolynucleotides. Arch. Biochem. Biophys. 1957;69:119–129. doi: 10.1016/0003-9861(57)90479-4. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campos-Guillen J, Bralley P, Jones GH, Bechhofer DH, Olmedo-Alvarez G. Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 2005;187:4698–4706. doi: 10.1128/JB.187.14.4698-4706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar D, Fisher PB. Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 2006;112:243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Beljanski M. De novo synthesis of DNA-like molecules by polynucleotide phosphorylase in vitro. J. Mol. Evol. 1996;42:493–499. doi: 10.1007/BF02352279. [DOI] [PubMed] [Google Scholar]
- 39.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Bechhofer DH. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danchin A. Comparison between the Escherichia coli and Bacillus subtilis genomes suggests that a major function of polynucleotide phosphorylase is to synthesize CDP. DNA Res. 1997;4:9–18. doi: 10.1093/dnares/4.1.9. [DOI] [PubMed] [Google Scholar]
- 42.Deikus G, Bechhofer DH. Initiation of decay of Bacillus subtilis trp leader RNA. J. Biol. Chem. 2007;282:20238–20244. doi: 10.1074/jbc.M702747200. [DOI] [PubMed] [Google Scholar]
- 43.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrasco B, Cozar MC, Lurz R, Alonso JC, Ayora S. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction-processing functions in chromosome segregation. J. Bacteriol. 2004;186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su C, Peregrin-Alvarez JM, Butland G, Phanse S, Fong V, Emili A, Parkinson J. Bacteriome.org-an integrated protein interaction database for E. coli. Nucleic Acids Res. 2008;36:D632–D636. doi: 10.1093/nar/gkm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitra S, Hue K, Bechhofer DH. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Molecular microbiology. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez H, Carrasco B, Ayora S, Alonso JC. Homologous recombination in low dC + dG Gram-positive bacteria. Berlin, Heidelberg: Springer Berlin/Heidelberg; 2007. [Google Scholar]
- 52.Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, Li Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48:2012–2020. doi: 10.1021/bi801752p. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d’Adda di Fagagna F, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 54.Del Favero M, Mazzantini E, Briani F, Zangrossi S, Tortora P, Deho G. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J. Biol. Chem. 2008;283:27355–27359. doi: 10.1074/jbc.C800113200. [DOI] [PubMed] [Google Scholar]
- 55.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD, Lai B, Ravel B, Li SM, Kemner KM, et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 57.Jakubovics NS, Jenkinson HF. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology. 2001;147:1709–1718. doi: 10.1099/00221287-147-7-1709. [DOI] [PubMed] [Google Scholar]
- 58.Clark AJ, Sandler SJ. Homologous genetic recombination: the pieces begin to fall into place. Critical Reviews in Microbiology. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 59.Dutra BE, Sutera VA, Jr, Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc. Natl Acad. Sci. USA. 2007;104:216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ayora S, Carrasco B, Doncel E, Lurz R, Alonso JC. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl Acad. Sci. USA. 2004;101:452–457. doi: 10.1073/pnas.2533829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez S, Sorokin A, Alonso JC. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 64.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair. 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 66.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.