Abstract
RecA, the key protein in homologous recombination, performs its actions as a helical filament on single-stranded DNA (ssDNA). ATP hydrolysis makes the RecA–ssDNA filament dynamic and is essential for successful recombination. RecA has been studied extensively by single-molecule techniques on double-stranded DNA (dsDNA). Here we directly probe the structure and kinetics of RecA interaction with its biologically most relevant substrate, long ssDNA molecules. We find that RecA ATPase activity is required for the formation of long continuous filaments on ssDNA. These filaments both nucleate and extend with a multimeric unit as indicated by the Hill coefficient of 5.4 for filament nucleation. Disassembly rates of RecA from ssDNA decrease with applied stretching force, corresponding to a mechanism where protein-induced stretching of the ssDNA aids in the disassembly. Finally, we show that RecA–ssDNA filaments can reversibly interconvert between an extended, ATP-bound, and a compressed, ADP-bound state. Taken together, our results demonstrate that ATP hydrolysis has a major influence on the structure and state of RecA filaments on ssDNA.
INTRODUCTION
RecA–single-stranded DNA (ssDNA) filaments are the catalytic core of homologous recombination (HR) in Escherichia coli and RecA has become the prototypical member of a family of proteins ubiquitous to all organisms including the human RAD51 and DMC1 recombinases (1–3). The RecA–ssDNA filament must perform multiple actions during recombinational DNA repair: first, it has to assemble on ssDNA and search for homologous duplex DNA; subsequently the RecA–ssDNA filament invades and exchanges strands with the homologous duplex DNA; and finally it dissociates to allow for further processing by polymerases. The repair of DNA double-stranded breaks by HR is vital to the cells’ survival and therefore all these steps have to be robust enough to deal with heterology or other obstacles that may hinder the process. ATP hydrolysis by RecA is essential for successful HR but many questions on how ATP hydrolysis influences the molecular structure and dynamics of RecA–ssDNA filaments remain.
The assembly of RecA filaments on ssDNA is an essential early step in HR and requires both a nucleotide cofactor (i.e. ATP) and a divalent cation (i.e. Mg2+) bound at the monomer–monomer interfaces (4,5). Filament assembly has been studied in detail on double-stranded DNA (dsDNA) where the consensus is that filament formation is dominated by a slow nucleation step, requiring a nucleation cluster of 5 or 6 monomers, followed by rapid filament extension resulting in long continuous filaments (6–9). On the biologically more relevant ssDNA substrate, nucleation also requires a multimeric unit and is orders of magnitude faster (9,10). Filament extension on ssDNA is less cooperative, possibly resulting in the formation of short filament patches as observed for RAD51 (11–13). The 3′ end is typically favored for addition of RecA monomers and disassembly occurs from the 5′ end resulting in a net 5′ to 3′ assembly direction on ssDNA (9,14). Due to the 5′ to 3′ directionality, the 3′ end is more likely to be covered with RecA resulting in more efficient pairing reactions at the 3′ end in vitro (15,16).
Disassembly of the RecA filament also is an important step in the overall recombination process. The nucleoprotein filament needs to be dismantled after strand exchange to clear the path for DNA synthesis and the completion of HR. Recent experiments suggest that strand exchange and filament disassembly are coupled, underlining the importance of disassembly kinetics (17). Disassembly of RecA is coupled to ATP hydrolysis at the monomer–monomer interface and proceeds from filament ends on both ss- and dsDNA. Since the monomer–monomer interfaces are presumably identical throughout the filament, the asymmetry in assembly and disassembly rates can only be sustained at the expense of external energy (18). The recently resolved structure of RecA complexed with ssDNA indicates that ATP binding and ATP hydrolysis indeed mediate the binding and release of RecA from DNA through allosteric coupling (5). ATP hydrolysis is, however, not the only driving factor for dissociation, as the mechanical interactions of a recombinase filament with its stretched DNA substrates also facilitate disassembly (19). Although pairing reactions proceed readily when RecA disassembly is blocked by non- or slowly hydrolysable ATP analogs, e.g. ATPγS, the delicate balance between assembly and disassembly of the RecA–ssDNA filament is likely important for strand exchange and homology search (20,21).
RecA forms a right-handed helical filament on ssDNA in the presence of ATP, extending it by 50% compared to B-form dsDNA. DNA is bound within the filament with a stoichiometry of three nucleotides (nt) or base pairs (bp) per RecA monomer and 6 monomers make up one helical turn (5,22–24). ssDNA is thus considerably extended and restricted in its conformational freedom within the RecA filament. ATP hydrolysis occurs throughout the filament and is not restricted to the disassembly end (25). The hydrolytic cycles of individual monomers are uncoupled on ssDNA in contrast to RecA bound to dsDNA where hydrolytic cycles of RecA monomers cooperatively influence each other (26–28). The structure of the ssDNA-RecA filament is also affected by the bound nucleotide cofactor and in the presence of ADP a compressed filament forms (29).
ATP hydrolysis not only drives the turnover of RecA on DNA, but is also linked to several other functions: the bypass of long stretches of heterology, fork regression, and four-strand-exchange reactions (30). All these processes seem to require the generation of torsion in the pairing strands, and several models to generate this torsion have been proposed. The simplest models link the generation of torsion to a redistribution of RecA monomers on DNA (31,32). The accompanying change in helical pitch of the dsDNA, between its canonical B-form of 10.5 bp per turn to 18.5 bp per turn when bound by RecA, could generate the required torsion. A second model postulates the presence of concerted motor action of RecA monomers to actively rotate the homologous strand around the filament (1). A third model links the generation of torsion to a conformational change of the RecA filament upon ATP hydrolysis (33).
To describe RecA filament dynamics and how the mechanical changes of the filament are coupled to ATP hydrolysis, we measured RecA filament assembly and disassembly on single ssDNA molecules. We found that ATP hydrolysis by RecA was essential for the formation of long uninterrupted filaments on ssDNA. By fitting filament assembly data to Monte Carlo (MC) simulations, we concluded that both nucleation and extension of these filaments occurred with multimers. RecA disassembly is facilitated by the energy stored in the bound and stretched ssDNA. Dissociation from ssDNA is more sensitive to force than dissociation of recombinases from dsDNA. Interestingly, we show that the mechanical properties of RecA–ssDNA filaments change drastically upon ATP hydrolysis and that this process is fast and reversible, implying a rapid exchange of ADP from the filament.
MATERIALS AND METHODS
DNA constructs
A ssDNA construct was prepared as described previously (13); additionally, a 7.3 kb construct was prepared analogously starting from a λ-DNA template with primer sequences AACTCAGCTCACCGTCGAACA and 5′ biotinylated GACGCAGGGGACCTGCAG and digested with PspOMI.
Magnetic tweezers assay
A magnetic tweezers setup was used in these experiments as described (34). By using image processing, 5-nm position accuracy of the bead was obtained in all three dimensions (35). To exclude the effect of thermal drift, all positions were measured relative to a 3.2 μm polystyrene bead (Bang Laboratories, Carmel, IN) fixed to the bottom of the flow cell. Polystyrene beads as well as DNA constructs carrying a magnetic bead at one end were anchored to the bottom of a flow cell as described elsewhere (35). The force-extension curve of single DNA molecules was measured. After conformation of the correct contour and persistence lengths, experiments were started by addition of RecA. All measurements were carried out at 25°C.
RecA/DNA reactions
The flow-cell final volume was approximately 100 μl. All reactions were done in 25 mM Tris–HCl (pH 7.5), 5 mM MgCl2 or CaCl2 and 1 mM DTT. RecA was purchased from New England Biolabs. RecA and ATP [final concentrations 1 µM RecA (unless stated otherwise) and 1 mM ATP] were added into the flow cell. Interaction of RecA with the tethered DNA molecule was monitored through measurement of the height of the magnetic bead.
MC simulations
We follow the protocol described in Van der Heijden et al. (13,36).
RESULTS
ATP hydrolysis is essential to form long continuous RecA–ssDNA filaments
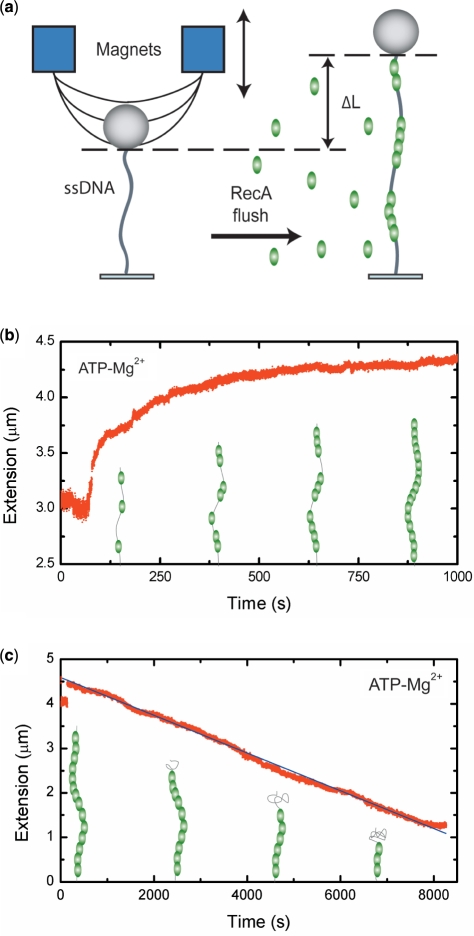
Our magnetic-tweezers single-molecule technique allows monitoring long (∼8 kb) individual DNA molecules. The DNA is bound at one end to a magnetic bead and at the other end to the surface of a flow cell. A force is applied by a pair of external magnets (Figure 1a). Video microscopy is used to monitor the bead position, and thus the DNA end-to-end distance, in real time. The assembly and disassembly of RecA filaments is monitored by measuring changes in the end-to-end distance of the DNA tether. RecA binding increases the tether length as it extends and stiffens ssDNA. Length changes of the tethered DNA can thus be correlated to changes in RecA filament coverage of the ssDNA molecule. Filament assembly was recorded at forces around 6 pN to minimize the influence of secondary structure in the ssDNA on the assembly profile. At 6 pN, the tether length of a ssDNA molecule is approximately equal to the length of a B-form dsDNA molecule with the same number of same number of nucleotides. When a complete RecA filament forms, the tether is expected to extend to 1.5 times the B-form length. Filament disassembly was monitored at forces below 0.5 pN, unless stated otherwise. Below 0.5 pN, the end-to-end distance of ssDNA is negligible compared to the stiff RecA filament. A decrease in the end-to-end distance can therefore be directly correlated to dissociation of the filament.
Figure 1.
RecA filament formation and dissociation on ssDNA as measured by magnetic tweezers. (a) Schematic drawing of the magnetic tweezers setup. (b) A 8.6 kb ssDNA molecule is extended by 48% compared to B-form DNA at a stretching force of 6 pN in the presence of 1 µM RecA, 1 mM ATP and 5 mM Mg2+. (c) Linear disassembly profile of a 8.6 kb RecA–ssDNA filament recorded at 0.5 pN. The black line is a guide to the eye.
Filament assembly was started by introducing 1 μM RecA in the presence of 1 mM ATP and 5 mM Mg2+ into the flow cell. Immediately after buffer flow was stopped, DNA end-to-end distance started to increase, consistent with the formation of DNA-RecA nucleoprotein filament(s) that extend and stiffen the DNA tether (Figure 1b). In contrast to the binding profile seen on dsDNA (Supplementary Figure S1) (7), the profile is strongly non-linear, indicative of a substantially less cooperative assembly. Cooperativity can be defined as the ratio between the rate of filament extension and nucleation. At high ratios (>106), the binding profile displays a linear increase, whereas at low ratios the increase is sigmoidal (Supplementary Figure S2) (36). A low cooperativity will result in incomplete DNA coverage because different nucleation positions can be out of register with each other, leading to gaps of at least 1 or 2 nt considering the 3-nt binding site of RecA. Multimeric binding, where the unit of filament nucleation or extension consists of more than one RecA monomer, will further increase the size of these gaps leading to even less complete coverage. Experimentally we found that the final end-to-end distance of the RecA filaments corresponded to a 48.0 ± 1.4% (N = 19) elongation compared to B-form DNA of the same nt length. The measured elongation is in excellent agreement with electron microscopy and other studies of RecA filaments on ssDNA (5,33). The elongation obtained thus corresponds to a fully covered ssDNA molecule. We note that this is a very surprising result because the shape of the binding profile would suggest multiple nucleation events leading to gaps in the filament as reported for RAD51 (13).
Dissociation measurements also indicated the absence of gaps in the final filament as the obtained profiles were linear, indicative of dissociation from a single filament (Figure 1c). After assembly, dissociation of RecA from ssDNA was initiated by exchanging the buffer in the flow cell for one containing 1 mM ATP and 5 mM Mg2+ but no RecA. This allows continuous ATP hydrolysis and therefore disassembly of the bound RecA filament. The measured disassembly rate of 0.83 ± 0.16 monomers per second (N = 10) compares well to the bulk value of 0.91 monomers per second (37).
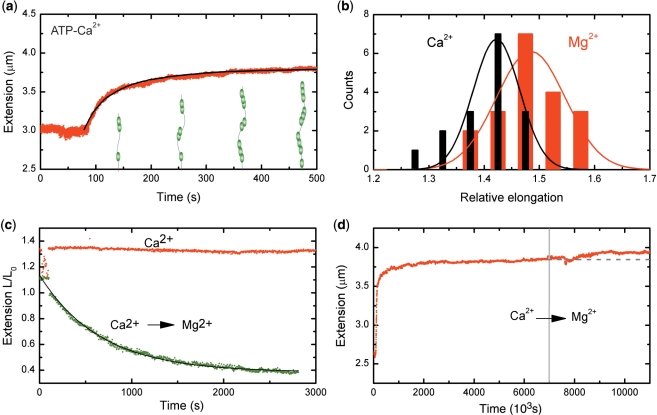
To understand the interplay between ATP hydrolysis driven RecA binding and dissociation during assembly, we directly measured filament assembly under conditions where RecA ATP hydrolysis was suppressed. In the presence of 5 mM Ca2+, RecA binds to ssDNA but ATP hydrolysis is strongly attenuated (4). Dissociation of RecA during filament assembly can thus be inhibited by replacing Mg2+ with Ca2+. In the presence of Ca2+ and ATP, we observed a non-linear growth profile similar to the data for Mg2+ (Figure 2a). The end-to-end distance, however, of the RecA–ssDNA filaments did not reach a full 50% elongation compared to B-form DNA, but instead a significantly lower elongation of 38 ± 2% (N = 16) (Figure 2b). Based on the ratio of measured to expected elongation, this 38 ± 2% (N = 16) corresponds to a fractional coverage of 79 ± 7% of the DNA. The end-to-end distance of the DNA tether did not decrease upon changing the buffer for one containing 1 mM ATP and 5 mM Ca2+ but lacking RecA (Figure 2c). This shows that filament disassembly was indeed inhibited, yielding stable filaments when ATP hydrolysis is suppressed. The exchange of Ca2+ for Mg2+, by changing to a buffer containing ATP and Mg2+, led however to a decrease in the end-to-end distance of the molecule as the filament dissociated (Figure 2c). The disassembly profile was now exponential, indicative of dissociation from the ends of multiple filament patches. The shorter end-to-end distance and presence of gaps in the filament assembled in the presence of Ca2+ could also result from secondary structure in the ssDNA. To test if secondary structure influenced these measurements, we assembled filaments at a force of 20 pN, which effectively removes all secondary structure in the ssDNA (38). The dissociation behavior of these filaments was the same as observed for filaments assembled at low force (Supplementary Figure S3). The marked difference between the RecA–ssDNA filaments formed in the presence of Mg2+ and Ca2+ remained essentially the same (cf. Figures 1c and 2c). This shows that secondary structure does not play a major role in preventing the formation of a continuous RecA filament on ssDNA in the presence of Ca2+.
Figure 2.
RecA–ssDNA filaments in conditions suppressing ATP hydrolysis. (a) Filaments formed on a 8.6 kb ssDNA molecule showed a similar binding behavior at a stretching force of 6 pN in a 5 mM Ca2+ buffer to that in the presence of Mg2+. The black line is a fit from MC simulations, adopting hexamers for nucleation and extension units, used to determine the nucleation and extension rate. (b) Histogram of RecA–ssDNA tether length at a stretching force of 6 pN in the presence of either 5 mM Mg2+ (red) or 5 mM Ca2+ (black). Solid lines are a fit to a normal distribution. (c) RecA disassembly at a stretching force of 0.5 pN from a 8.6 kb ssDNA was blocked in the presence of 5 mM Ca2+ (red points), but readily occurred when Ca2+ was replaced by Mg2+ (green points) The filament dissociation fits well to a single exponential decay (black line). (d) A 7.3 kb RecA–ssDNA filament assembled in the presence of Ca2+ at a stretching force of 6 pN showed a further increase in end-to-end distance when the buffer was exchanged for one containing 1 µM RecA and 5 mM Mg2+. The gray dashed line is a guide to the eye.
The above results indicate that filament formation by random nucleation of RecA on ssDNA creates multiple filament patches that, however, anneal into one continuous filament via ATP-hydrolysis-dependent rearrangements. This implies that a patchy RecA filament, with bare tracts of ssDNA, formed when ATP hydrolysis is suppressed, is able to anneal into one continuous filament when ATP hydrolysis is again allowed. This was indeed observed: A filament that had reached its saturation extension in ATP-hydrolysis suppressing conditions (Ca2+, ATP and RecA) showed a further increase in extension after switching to a buffer that allowed for ATP hydrolysis (Mg2+, ATP and RecA) (Figure 2d). The final extension of the filaments reached 50 ± 3% (N = 7) corresponding to ssDNA fully covered by RecA.
RecA nucleates and extends filaments with multimers
RecA filament assembly involves nucleation followed by filament extension. Nucleation has been reported to occur with 5 or 6 monomers whereas a single monomer was suggested as the functional unit for filament extension (9,10). A monomeric unit for filament extension compared to a multimeric nucleation unit should give rise to a concentration dependence of the cooperativity. This was not supported by our experiments as the shape of the binding profiles is conserved for different concentrations (Supplementary Figure S4), implying that the nucleation and extension unit are the same.
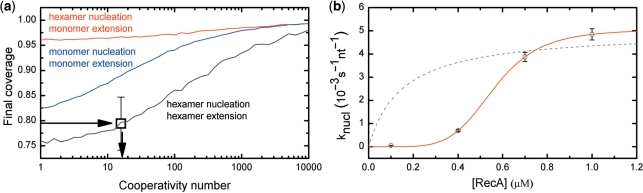
In order to quantitatively understand RecA filament formation, we modeled the nucleoprotein filament formation with MC simulations [for details, see refs (13,36)]. The simulations involve four parameters to describe the RecA filament formation: nucleation rate, cooperativity, nucleation unit (monomers or multimers) and extension unit (again either monomers or multimers). We first simulated filament assembly curves to obtain the final coverage of the molecule for different nucleation and extension units at different cooperativity (Figure 3a). Higher cooperativity resulted in a higher final coverage as fewer gaps were formed. Larger extension units resulted in a lower final coverage as the minimum gap size between filament patches increases. The final occupancy of 79 ± 7% obtained from our measurements in the presence of Ca2+ and ATP corresponds to the regime expected for both nucleation and extension with a multimeric unit and is inconsistent with a monomeric extension unit.
Figure 3.
Final DNA coverage of a RecA filament is determined by the cooperativity as well as the nucleation and extension unit. (a) Monte Carlo simulations of the saturation coverage of RecA on ssDNA in the absence of dissociation. Our measured final coverage of 79% ± 7 (black box) in the presence of Ca2+ is in the range of nucleation and extension by hexamers. (b) Concentration dependent nucleation rates were extracted from multiple growth profiles using the MC simulations. The observed behavior (round circles) differed markedly from Michaelis–Menten kinetics (dotted line) but is accurately fit by the Hill equation with a coefficient of 5.4 ± 0.2. Error bars show SEM.
Because disassembly is inhibited in the presence of ATP and Ca2+, we neglected disassembly to simulate growth profiles obtained in the presence of Ca2+. Nucleation rates were obtained from experimental growth profiles by fitting MC simulations with two free fit parameters (nucleation rate and cooperativity), adopting hexamers for nucleation and extension units. A fit to an experimental growth profile, based on the MC simulations, is shown in Figure 2a. The nucleation rates obtained from experiments at different RecA concentrations show a strong concentration dependency (Figure 3b). A fit of the concentration dependent nucleation rates with the Hill equation yields a Hill coefficient of 5.4 ± 0.2. The large Hill coefficient confirms that nucleation occurs with a multimeric unit, which is expected to be a hexamer as the Hill coefficient sets a lower bound for the number of interacting monomers.
The growth profiles in the presence of ATP and Mg2+ (Figure 1b) can be modeled using the nucleation rate and cooperativity determined for conditions where ATP hydrolysis was suppressed, and the dissociation rate that was independently obtained from the linear disassembly of complete filaments in the presence of ATP and Mg2+. These simulations always resulted in multiple filament patches and 100% coverage was never obtained. Inclusion of the dissociation reduced the gaps due to multimeric nucleation and extension units to 1 or 2 nt, but it did not completely remove these gaps. Thus accounting for assembly and disassembly alone in our simulations was not sufficient to describe the observed formation of one continuous filament in the presence of ATP and Mg2+. An additional element of the filament dynamics is needed to account for the data, which is examined in the ‘Discussion’ section.
RecA dissociation from ssDNA is force dependent
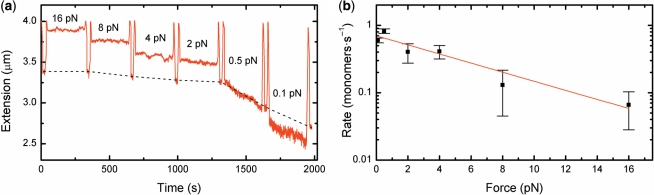
Exerting a force may change the mechanical interactions between RecA and its DNA substrate. Stretching forces applied to a dsDNA molecule enhance binding and slow down dissociation of nucleoprotein filaments (6,19,39). Upon dissociation of RecA in the absence of any force exerted on ssDNA, the end-to-end distance will decrease as the ssDNA behaves as an entropic spring. The free energy related to stretching ssDNA to 1.5 times the B-form DNA length, approximately 3 kBT, could very well contribute to the disassembly process. To test if an applied force influences RecA dissociation from ssDNA, dissociation was initiated as before. Subsequently, the filament was subjected to a constant force (in the range of 16 pN ≥ F ≥ 0.1 pN) during a 270 s interval, while disassembly proceeded. The change in filament length was monitored by periodically reducing the force to 0.5 pN (Figure 4a). RecA–ssDNA filaments subjected to a high force (F ≥ 8 pN) showed almost no change in end-to-end distance. At lower forces, however, the end-to-end distance of the RecA–ssDNA tether decreased considerably, indicating disassembly of RecA from the DNA. The profiles observed during the different force intervals were all linear confirming that disassembly occurs from one single filament.
Figure 4.
Stretching ssDNA strongly affects the disassembly of RecA–ssDNA filaments. (a) Dissociation at constant force intervals between 16 and 0.1 pN. The RecA–ssDNA filament length was determined by temporarily shifting the force to 0.5 pN after each interval. The black dotted line is a guide to the eye showing the disassembly. (b) The RecA dissociation rate is force dependent. Fitting the force dependence with a single exponential k(F) = k0exp(–dF/kBT) (red line) gives a distance to the energy barrier of 0.6 + 0.2 nm. Data in b are means of about five different time traces, error bars show SEM.
A strong decrease in the disassembly rate k is observed with increasing force (Figure 4b). Disassembly rates in monomers per second were calculated as the change in tether length at 0.5 pN divided by 1.5 nm, the length of three bases within a single RecA monomer. The disassembly rate had an exponential dependence on force: k(F) = k0exp(–dF/kBT), where k0 is the disassembly rate at zero force, kBT the thermal energy, F the applied force and d the distance to the energy barrier along the relevant reaction coordinate. Thus, the energy barrier for dissociation is apparently raised by subjecting the RecA-bound DNA to force. The distance to the energy barrier from the force dependency was 0.6 ± 0.2 nm, corresponding to approximately half the length of the three nucleotides held fixed in the RecA–ssDNA filament. This agrees well with RecA dissociation from ssDNA as single monomers.
RecA–ssDNA filaments reversibly switch between extended and collapsed states during ATP hydrolysis
In the RecA–ssDNA filament, a nucleotide cofactor is bound at the important interface between RecA monomers. Changes to this interface are able to influence the structure and conformation of the filament, and modulate the filament's function in the recombination process (11,40,41). To directly determine the influence of nucleotide cofactors on the mechanical state of the filament, we probed the length of the RecA–ssDNA filament, in real time, while exchanging buffers containing different nucleotide cofactors. First, a RecA–ssDNA filament was assembled in the presence of Mg2+ and ATP, as described above. Subsequently, the buffer was exchanged multiple times under continuous flow to induce exchange of the bound nucleotide cofactor. The force during this continuous flow experiment was approximately 3 pN, calibrated by a separate measurement based on the force-extension behavior of ssDNA.
The RecA–ssDNA filament changed length reversibly in response to the presence of a cofactor (Figure 5a). The initial RecA–ssDNA filament, formed in the presence of 5 mM Mg2+ and 1 mM ATP, had an extension corresponding to a fully covered RecA–ssDNA molecule. Upon exchanging the buffer for one without ATP and RecA, the filament converted to a shorter state. In this state, the bound ATP is expected to be hydrolyzed to ADP, and we will henceforth refer to it as the ADP-bound state. Changing the buffer back to one containing 1 mM ATP but no RecA resulted in extension of the filament. The initial length of the filament was not fully recovered, possibly because some disassembly occurred during the conversion to the ADP-bound state. This procedure was then repeated a second and third time on the same molecule showing a similar behavior, indicating that the conversion between the ATP- and ADP-bound states was reversible. Finally, the buffer was exchanged for one containing 1 mM ATPγS, but still lacking RecA. This resulted in an increase in length of the RecA–ssDNA filament that now surpassed that of the ATP-bound state of the previous cycle.
Figure 5.
RecA–ssDNA filament reversibly interconvert between an extended ATP-bound and a compressed ADP-bound state. (a) Real time measurement of extension changes of a RecA–ssDNA filament at a stretching force of 3 pN in response to different nucleotide cofactors. Flushing with a buffer lacking ATP at t = 280 s resulted in a decrease in extension of a RecA–ssDNA molecule (red circles). Reintroduction of ATP (1 mM) resulted in a rapid length increase at t = 480 s. The initial extension was, however, not fully recovered likely due to dissociation of RecA. This procedure was repeated two more times before a buffer containing ATPγS was flushed in at t = 1230 s. This resulted in a fast extension surpassing that of the previous ATP-bound filament state. The calculated response based Michaelis–Menten kinetics accurately predicts the observed behavior (black dotted line) (Figure S5). (b) Force extension behavior of RecA–ssDNA filaments depends on the nucleotide cofactor bound. RecA–ssDNA filaments complexed with ATPγS (green squares) or ATP (blue triangles) are well described by an extensible worm-like chain (solid lines). RecA–ssDNA filaments converted to the ADP-bound state (red circles) are are shorter and more flexible than filaments in the ATP or ATPγS form. The force extension of bare ssDNA (black circles) is shown for comparison. No fit to the WLC model was made for ssDNA as it behaves qualitatively differently.
The transition from the elongated ATP-bound state to the shorter ADP-bound state, upon ATP depletion, was much slower than the fast increase in length observed when ATP was flushed in. This difference in response results from the interplay between the high affinity of RecA for ATP and the time-dependent concentration changes in the flow cell during buffer exchange. The Km of RecA–ssDNA filaments for ATP is in the range of 20–50 μM (42). Only when the concentration of ATP is reduced to the micromolar range will the RecA–ssDNA filament start to accumulate an ADP-bound state. To quantitatively understand this process, we employed a finite-element simulation with a commercial computer program (Comsol Multiphysics) to model the time-dependent concentration in our flow cell (Supplementary Figure S5). This allowed us to predict the time response of the RecA–ssDNA filament under the condition that ATP binding and ADP release are much faster than the changes in ATP concentration. The response was calculated as Θ = [ATP]/(Km + [ATP]), where Θ is the fraction of the RecA–ssDNA filament in the ATP-bound state. This calculated response, using a Km of 40 μM, can be scaled to an extension of the RecA–ssDNA filament. This matches well with the measured data (black dotted line in Figure 5a).
The mechanical properties of the RecA–ssDNA filament in the presence of different nucleotide cofactors were determined by probing its force-extension behavior (Figure 5b). The initial RecA–ssDNA filament, formed in the presence of 5 mM Mg2+ and 1 mM ATP, had a force-extension curve corresponding to a stiff and extended filament. Upon exchanging the buffer for one without ATP and RecA, the filament converted to a shorter and more flexible ADP-bound state. Finally the RecA–ssDNA filament in the presence of 1 mM ATPγS was stiffer and more extended than the ATP-bound state. The mechanical properties of RecA–ssDNA filaments can be described in terms of persistence length and contour length by a worm-like chain (WLC) model. At higher forces, the elastic behavior of the filaments deviates from the WLC and is better described by an extension to the WLC model incorporating enthalpic stretching (43–45). The extensible WLC model including a seventh-order correction term proposed by Bouchiat et al. accurately describes our data for the ATP and ATPγS-bound filaments for forces ranging from 0.02 to 40 pN (Figure 5b) (45). The data for the ADP-bound filaments, however, showed an increase in extensibility for forces above 7 pN and deviated substantially from the extensible WLC model. Force-extension data of ADP-bound filaments were therefore fitted only up to 7 pN. At higher forces, the more extensible behavior of the ADP filament approached that of bare ssDNA. The mechanical properties extracted from the extensible WLC model are summarized in Table 1. RecA–ssDNA filaments in the ADP-bound state had a contour length that is ∼6% shorter than that of the ATP-bound filaments. The ADP-bound filaments were also considerably more flexible and this resulted in even larger differences in end-to-end distance at low forces. Indeed, at 0.01 pN the difference in end-to-end distance between the ADP- and ATP-bound filament was almost 50%. The mechanical differences between the ATP and ADP states of the RecA–ssDNA filament are particularly interesting as these two states continuously interconvert upon ATP hydrolysis in a RecA–ssDNA filament. The rate of buffer exchange was increased for the force-extension measurements to reduce the effects of disassembly of the RecA–ssDNA filament. Under those conditions, complete exchange of the flow cell contents required just tens of seconds. The contribution of disassembly in these force-extension measurements is therefore expected to be very small as ADP-bound filaments showed considerable stiffening and extension surpassing that of the ATP-bound form when a buffer containing ATPγS, but no RecA, was flushed in. More importantly, subsequent addition of a buffer containing both ATPγS and RecA did not further increase the length of the filaments, indicating that there were no bare ssDNA sites available for RecA assembly (Supplementary Figure S6). This confirmed that the change in filament properties was induced solely by the different nucleotide cofactors bound and not due to RecA disassembly.
Table I.
Mechanical properties of RecA-ssDNA filaments
| ssDNA- RecA-ATP | ssDNA- RecA-ADP | ssDNA- RecA-ATPγs | |
|---|---|---|---|
| Rise per base pair, nm | 0.50 ± 0.01 | 0.47 ± 0.01 | 0.53 ± 0.01 |
| Persistence length, μm | 1.2 ± 0.1 | 0.25 ± 0.02 | 2.1 ± 0.1 |
| Stretch modulus, nN | 0.83 ± 0.04 | 0.43 ± 0.03 | 2.5 ± 0.2 |
Errors denote SEM of at least four different experiments.
DISCUSSION
We have shown that the ATPase activity of RecA in the presence of Mg2+ is essential for the formation of a continuous filament over several kbs on ssDNA. RecA polymerizes with a low cooperativity on ssDNA and substantial rearrangement of RecA monomers needs to take place to remove the gaps created by nucleation at multiple sites along the ssDNA. Modeling filament formation with MC simulations involving only assembly and disassembly cannot account for the annealing of these gaps. The gaps in the filament persist even if the cooperativity, assembly or disassembly rates are changed by orders of magnitude (Supplementary Figure S7).
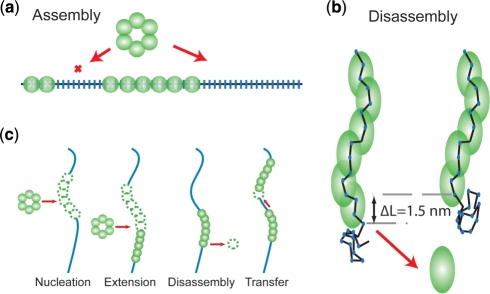
To explain the formation of continuous filaments, we therefore suggest a model where RecA monomers are able to transfer from one filament end to the neighboring filament in a unidirectional fashion (Figure 6c). This fits all our data well for a transfer rate of 2 monomers per second per filament end (Supplementary Figure S7b). Several mechanisms can be envisioned for the transfer of these monomers between the ends of filament patches: monomers could diffuse or hop between the ends. Monomers could also transfer when ends collide and loosely stack end-to-end due to the very flexible ssDNA joints in between (46,47). Upon dissociation of a RecA monomer, the local concentration is very high facilitating rebinding at the neighboring filament patch. A somewhat similar redistribution of proteins was also suggested for single-stranded binding proteins along an ssDNA molecule (48).
Figure 6.
RecA assembly and disassembly mechanisms. (a) Nucleation as well as filament extension on ssDNA by a multimeric binding unit leaves gaps of considerable size in the filament. (b) Dissociation of RecA monomer from ssDNA is aided by the stretching energy stored in the ssDNA. Upon dissociation the total end-to-end distance decreases by ∼1.5 nm, depending on the stretching force applied (c) Mechanisms considered in the Monte Carlo simulations describing RecA interactions with ssDNA.
Filaments can however also appear to be continuous but actually consist of trains of stacked filament patches as observed in electron microscopy (46,47). In this case, any excess of unbound ssDNA could be looped out at the interface between two stacked patches. A stacking mechanism is, however, not able to explain the differences in dissociation profile observed between filaments formed in the presence of Mg2+ and Ca2+. Disassembly curves of filaments formed in the presence of Ca2+ indicate that blocking ATP hydrolysis results in the formation of multiple filament patches containing stretches of bare ssDNA, inconsistent with a strong filament stacking interaction (Figure 2c).
Free RecA can exist in solution predominantly in multimeric form with a preferred stoichiometry between 6 and 12 monomers (49). The RecA assembly state in solution may have an effect on the preferred binding unit during RecA assembly. RecA nucleation rates on ssDNA were best fit by a Hill coefficient of 5.4 ± 0.2. The Hill coefficient sets a lower bound for the number of interacting monomers and we therefore expect a hexameric nucleation unit. Our data indicates that both filament nucleation and filament extension proceeds by RecA multimers, which would leave gaps of up to 17 nt in the filament when ATP hydrolysis is blocked (Figure 6a). This is in apparent contrast to a report by Joo et al. based on FRET measurements that suggested extension occurs by single monomers (9). Several factors may explain these differences. Our assay uses ssDNA of several kilonucleotides in length, while the assay employed by Joo et al. uses very short DNA molecules (20–60 nt) with the reporter probes spaced only 13 nt apart. The length of the employed oligonucleotides has the potential to limit the ability to identify multimeric binding events or influence the RecA interaction with the ssDNA (42). To determine dynamics at the 3′-extending end, Joo et al. exploit their observation that, in their conditions, in the presence of ATPγS, RecA–ssDNA filaments appear unstable. Currently, it is unclear how to interpret this observation, given that numerous other assays found that RecA–ssDNA filaments are stable in the presence of ATPγS over extended periods of time (11,50). Finally, dynamics at the extending end were measured at a very low RecA concentration of 8 nM, which may influence the stoichiometry of free RecA in solution (49).
We note that our conclusion that both filament nucleation and extension occur by multimers is derived from two independent types of data: first, the final extension of the RecA–ssDNA filament under conditions suppressing disassembly indicates the presence of bare ssDNA gaps larger than the 3-nt RecA binding site. Second, the Hill coefficient of 5.4 ± 0.2 indicates that the nucleation unit likely is a hexamer. The shape of the filament assembly profiles is conserved for different RecA concentrations (Supplementary Figure S4), indicating that the cooperativity is independent of RecA concentration and therefore the nucleation and extension unit are of equal size.
In a RecA filament, ssDNA is extended and its conformational freedom is severely restricted. The energy stored in this system is released upon dissociation and we determined the distance to the energy barrier to be 0.6 ± 0.2 nm. This distance is approximately half way between the 1.5 nm to which the ssDNA is extended by a RecA monomer and the negligible end-to-end distance for bare ssDNA. The sub-monomer distance to the energy barrier therefore supports a model where RecA dissociates as a single monomer at a time releasing three bases (Figure 6b). The 0.6 ± 0.2 nm distance to the energy barrier for dissociation from ssDNA is significantly larger than the 0.27 ± 0.04 nm reported for dissociation of RAD51 from dsDNA, or the 0.26 nm reported for polymerization of RecA on dsDNA (6,19). On dsDNA, however, three base pairs, with an end-to-end distance of 1.5 nm in the filament, are released upon disassembly of a recombinase corresponding to a length decrease of 0.5 nm, which is in line with a distance to the energy barrier of around 0.27 nm. The differences in length change of the substrate, 1.5 nm for ssDNA or 0.5 nm for dsDNA, thus readily explain the differences in the distance to the energy barrier. We therefore believe that the larger distance reported here is not recombinase specific but substrate specific and expect the energy barrier for other recombinases on ssDNA to be similar to that determined for RecA here. The larger distance to the energy barrier on ssDNA compared with dsDNA gives rise to a stronger dependence on the dissociation rate on the applied force. On ssDNA, dissociation is already strongly reduced at forces around 10 pN, whereas a force of 48 pN was required to stall dissociation for a RAD51 filament bound to dsDNA (19). These results indicate that the stability of nucleoprotein filaments may be regulated by forces applied to the DNA. During HR, any forces existing between the invading strand and the template strand would thus tend to stabilize the RecA filament on the invading ssDNA. The forces required are on the order of a few pN, indeed the range that can be easily generated by polymerases or helicases.
In the RecA–ssDNA filament, a nucleotide cofactor is bound at the structurally important interface between monomers. Here we show that differences in the bound nucleotide cofactor are able to directly and reversibly change the extension and mechanical properties of the RecA–ssDNA filament. This conversion is a direct response of the RecA–ssDNA filament to ATP concentration and is well explained by Michaelis–Menten kinetics (Figure 5a). Direct conversion between extended and compressed filament states reported here and previously (51), is at odds with studies that found a stoichiometry of 5 nt instead of 3 nt per monomer for the compressed ADP-bound filament (29). We expect that an active filament bound to nucleotide triplets maintains its stoichiometry upon ATP hydrolysis as adjacent RecA-bound nucleotides are unavailable. The reported ADP-bound filament with a stoichiometry of 5 nt may however result from de novo filament formation in the absence of ATP. Maintaining a stoichiometry of 3 nt per monomer during changes in filament extension is consistent with the recent structure of RecA–DNA complexes where changes in the extension of one phosphate bond at the interface of two monomers could account for changes in filament length without rearrangement of the bound nucleotide triplet (5). Force-extension measurements also indicate that interface between monomers in the ADP-bound state is different from the ATP-bound state as the force-extension behavior of the filament becomes more extensible above 7 pN and resembles that of ssDNA (Figure 5b).
The mechanical parameters determined from force-extension data of ATP-bound RecA–ssDNA filaments are in good agreement with values reported in literature (6). However, the persistence length that we determined for the ATPγS filament is much larger: 2.1 ± 0.1 μm compared to the 923 ± 46 nm previously reported (6). We attribute this difference to the different procedure used to assemble the RecA–ssDNA filaments. We converted a fully continuous RecA–ssDNA filament that was preassembled in the presence of ATP-Mg2+ to an ATPγS filament by exchange of the nucleotide cofactor. Hegner et al. assembled filaments directly in the presence of ATPγS. Our measurements show that blocking of ATP hydrolysis will result in the formation of multiple filament patches. Such a patchy filament will have a lower apparent persistence length than the fully continuous RecA–ssDNA filament measured in our experiments.
Changes in the mechanical state of the filament may aid in the recombination process by modifying the affinity of the RecA–ssDNA filament for its homologous target or by inducing forces between the pairing strands. There are a number of processes unique to RecA, such as the bypass of long heterologous regions during strand exchange and the migration of DNA branches that require the generation of torque (30). We suggest that ATP-hydrolysis-driven local contractions and extensions of the RecA–DNA filament likely also result in a change in pitch of the filament and consequently could generate the required torque. These conformational changes in the filament are expected to involve multiple RecA monomers as the ATP hydrolysis cycles of neighboring RecA monomers become coupled when in contact with dsDNA during strand exchange (27,28,52). It will be very interesting to see if the dynamics of the RecA–ssDNA filament can be directly linked to a mechanistic picture of nucleoprotein filaments interactions with its partners in the different steps of HR.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomolecular Physics Program of the Dutch organization for Fundamental Research of Matter (FOM); grants from the Netherlands Organization for Scientific Research (NWO); the Netherlands Genomics Initiative/NWO; European Commission STREP project BioNano-Switch; the Integrated Projects Molecular Imaging and DNA Repair (512113) and a National Cancer Institute-NIH USA program project. Funding for open access charge: The Netherlands Organization for Scientific Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter Veenhuizen, Susanne Hage and Ya-Hui Chien for technical assistance and Iwijn De Vlaminck for critically reading of the manuscript.
Footnotes
Present address: Thijn van der Heijden, Leiden University, Niels Bohrweg 2, 2333 CA Leiden, The Netherlands.
REFERENCES
- 1.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 2.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cromie GA, Connelly JC, Leach DRF. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 4.Menetski JP, Varghese A, Kowalczykowski SC. Properties of the high-affinity single-stranded-DNA binding state of the Escherichia-coli RecA protein. Biochemistry. 1988;27:1205–1212. doi: 10.1021/bi00404a021. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:484–489. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 6.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Heijden T, van Noort J, van Leest H, Kanaar R, Wyman C, Dekker N, Dekker C. Torque-limited RecA polymerization on dsDNA. Nucleic Acids Res. 2005;33:2099–2105. doi: 10.1093/nar/gki512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 9.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 10.De Zutter SK, Knight KL. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- 11.Menetski JP, Kowalczykowski SC. Interaction of Reca protein with single-stranded-DNA–quantitative aspects of binding-affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 12.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJG, Wuite GJL, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 13.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Register JC, Griffith J. The direction of RecA protein assembly onto single-strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 15.Cox MM, Lehman IR. Directionality and polarity in RecA protein-promoted branch migration. Proc. Natl Acad. Sci. USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutreix M, Rao BJ, Radding CM. The effects on strand exchange of 5′ versus 3′ ends of single-stranded-DNA in RecA nucleoprotein filaments. J. Mol. Biol. 1991;219:645–654. doi: 10.1016/0022-2836(91)90661-o. [DOI] [PubMed] [Google Scholar]
- 17.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Wegner A. Head to tail polymerization of actin. J. Mol. Biol. 1976;108:139–150. doi: 10.1016/s0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- 19.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice KP, Eggler AL, Sung P, Cox MM. DNA pairing and strand exchange by the Escherichia coli RecA and yeast Rad51 proteins without ATP hydrolysis–on the importance of not getting stuck. J. Biol. Chem. 2001;276:38570–38581. doi: 10.1074/jbc.M105678200. [DOI] [PubMed] [Google Scholar]
- 21.Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 2005;170:357–366. doi: 10.1083/jcb.200412090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stasiak A, Dicapua E. The helicity of DNA in complexes with RecA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 23.Egelman EH, Stasiak A. Structure of helical RecA-DNA complexes–complexes formed in the presence of ATP-Gamma-S or ATP. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 24.Dicapua E, Schnarr M, Ruigrok RWH, Lindner P, Timmins PA. Complexes of RecA protein in solution–a study by small-angle neutron-scattering. J. Mol. Biol. 1990;214:557–570. doi: 10.1016/0022-2836(90)90198-U. [DOI] [PubMed] [Google Scholar]
- 25.Brenner SL, Mitchell RS, Morrical SW, Neuendorf SK, Schutte BC, Cox MM. RecA protein-promoted ATP hydrolysis occurs throughout RecA nucleoprotein filaments. J. Biol. Chem. 1987;262:4011–4016. [PubMed] [Google Scholar]
- 26.Yu X, Egelman EH. Direct visualization of dynamics and co-operative conformational changes within RecA filaments that appear to be associated with the hydrolysis of adenosine 5′-O-(3-thiotriphosphate) J. Mol. Biol. 1992;225:193–216. doi: 10.1016/0022-2836(92)91036-o. [DOI] [PubMed] [Google Scholar]
- 27.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:231–243. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan Q, Cox MM. RecA protein dynamics in the interior of RecA nucleoprotein filaments. J. Mol. Biol. 1996;257:756–774. doi: 10.1006/jmbi.1996.0200. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 30.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 31.Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic-recombination. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 32.Kowalczykowski SC, Krupp RA. DNA-strand exchange promoted by RecA protein in the absence of ATP–implications for the mechanism of energy transduction in protein-promoted nucleic-acid transactions. Proc. Natl Acad. Sci. USA. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn K, Chrysogelos S, Griffith J. Electron-microscopic visualization of RecA-DNA filaments–evidence for a cyclic extension of duplex DNA. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 34.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Heijden T, Dekker C. Monte Carlo simulations of protein assembly, disassembly, and linear motion on DNA. Biophys. J. 2008;95:4560–4569. doi: 10.1529/biophysj.108.135061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 38.Montanari A, Mezard M. Hairpin formation and elongation of biomolecules. Phys. Rev. Lett. 2001;86:2178–2181. doi: 10.1103/PhysRevLett.86.2178. [DOI] [PubMed] [Google Scholar]
- 39.Bennink ML, Scharer OD, Kanaar R, Sakata-Sogawa K, Schins JM, Kanger JS, de Grooth BG, Greve J. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry. 1999;36:200–208. doi: 10.1002/(sici)1097-0320(19990701)36:3<200::aid-cyto9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 40.Zaitsev EN, Kowalczykowski SC. A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 2000;14:740–749. [PMC free article] [PubMed] [Google Scholar]
- 41.De Zutter JK, Forget AL, Logan KM, Knight KL. Phe217 regulates the transfer of allosteric information across the subunit interface of the RecA protein filament. Structure. 2001;9:47–55. doi: 10.1016/s0969-2126(00)00552-9. [DOI] [PubMed] [Google Scholar]
- 42.Bianco PR, Weinstock GM. Interaction of the RecA protein of Escherichia coli with single-stranded oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:4933–4939. doi: 10.1093/nar/24.24.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odijk T. Stiff chains and filaments under tension. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- 44.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouchiat C, Wang MD, Allemand JF, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Register JC, Griffith J. RecA protein filaments can juxtapose DNA ends–An activity that may reflect a function in DNA-repair. Proc. Natl Acad. Sci. USA. 1986;83:624–628. doi: 10.1073/pnas.83.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiianitsa K, Stasiak A. Helical repeat of DNA in the region of homologous pairing. Proc. Natl Acad. Sci. USA. 1997;94:7837–7840. doi: 10.1073/pnas.94.15.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, Vonhippel PH. On the thermodynamics and kinetics of the cooperative binding of bacteriophage T4-coded Gene-32 (helix destabilizing) protein to nucleic-acid lattices. Biophys. J. 1980;32:403–418. doi: 10.1016/S0006-3495(80)84964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenner SL, Zlotnick A, Stafford WF. RecA protein self-assembly.II. Analytical equilibrium ultracentrifugation studies of the entropy-driven self-association of RecA. J. Mol. Biol. 1990;216:949–964. doi: 10.1016/S0022-2836(99)80013-8. [DOI] [PubMed] [Google Scholar]
- 50.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 51.Nishinaka T, Doi Y, Hara R, Yashima E. Elastic behavior of RecA-DNA helical filaments. J. Mol. Biol. 2007;370:837–845. doi: 10.1016/j.jmb.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 52.VanLoock MS, Yu X, Yang SX, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.