Abstract
Inhibitor of differentiation 2 (Id2) is a natural inhibitor of the basic helix–loop–helix transcription factors. Although Id2 is well known to prevent differentiation and promote cell-cycle progression and tumorigenesis, the molecular events that regulate Id2 activity remain to be investigated. Here, we identified that Four-and-a-half LIM-only protein 2 (FHL2) is a novel functional repressor of Id2. Moreover, we demonstrated that FHL2 can directly interact with all members of the Id family (Id1–4) via an N-terminal loop–helix structure conserved in Id proteins. FHL2 antagonizes the inhibitory effect of Id proteins on basic helix–loop–helix protein E47-mediated transcription, which was abrogated by the deletion mutation of Ids that disrupted their interaction with FHL2. We also showed a competitive nature between FHL2 and E47 for binding Id2, whereby FHL2 prevents the formation of the Id2–E47 heterodimer, thus releasing E47 to DNA and restoring its transcriptional activity. FHL2 expression was remarkably up-regulated during retinoic acid-induced differentiation of neuroblastoma cells, during which the expression of Id2 was opposite to that. Ectopic FHL2 expression in neuroblastoma cells markedly reduces the transcriptional and cell-cycle promoting functions of Id2. Altogether, these results indicate that FHL2 is an important repressor of the oncogenic activity of Id2 in neuroblastoma cells.
INTRODUCTION
Inhibitor of differentiation (Id) proteins are key regulators in a wide range of developmental and cellular processes by regulating transcriptional networks (1,2). Id proteins themselves do not bind to DNA because of lack of a DNA-binding domain, but they preferentially bind to ubiquitously expressed basic helix–loop–helix (bHLH) factor E proteins (E47, E2–2 and HEB) and sequester them from tissue-specific bHLH proteins (3,4). The E transcription factor engaged by Id proteins is no longer able to bind to an E-box DNA target sequence and activate transcription. The factors affecting the balance of the Id/bHLH activity are expected to be critical to the whole cellular equilibrium (5–7).
So far, four individual Id genes, Id1–4, have been identified in mammals. All members of the Id protein family share a similar structure consisting of a highly conserved HLH domain. Among them, Id2 has been extensively studied in several cell types and in animal models and found to be not only an inhibitor of differentiation but also a positive regulator of cell proliferation and oncogenesis (5,6,8). Aberrant elevation of Id2, typically activated by oncoproteins such as Myc and Ews–Fli1, was frequently observed in tumors from the central and peripheral nervous system (9–12). The aberrant accumulation of Id2 contributes to uncontrolled proliferation and neoangiogenesis, two hallmarks of neural cancers (13).
Similar to the regulation of other transcription factors, Id protein activity has been previously reported to be controlled by additional Id partners that are unrelated to transcription factors (8). These studies have largely focused on Id2. Among the partners is the retinoblastoma tumor suppressor protein Rb. Rb binds to and antagonizes Id2 from binding to transcription factors, thus releasing bHLH protein-mediated transcription (10,14,15). Recently, cytoplasmic proteins such as enigma homolog (ENH), polycystin-2 (PC2) and interferon-induced protein p204 were reported to bind to Id2 and sequestrate it into cytoplasm, thus preventing the access of Id2 to its nuclear partners (16–18). Loss of the cytoplasmic adaptor ENH was suggested to boost Id2 activity in aggressive neuroblastomas (16). In view of the oncogenic activity of Id2, proteins that prevent Id2 from binding to bHLH factors may be potential tumor suppressors.
Four-and-a-half-LIM-only protein 2 (FHL2) is the best-studied member of the FHL family, which is expressed in several human adult tissues, including the brain (19,20). FHL2 has no DNA-binding activity but can regulate multiple cell signaling pathways by interacting with various transcription factors known to be involved in development and tumorigenesis (19). The function of FHL2 in cancer is particularly intriguing, because it may act as an oncoprotein or as a tumor suppressor in a tissue-dependent fashion (21). The dual nature of FHL2 is reflected by the finding that it functions as activator or repressor of its interacting transcription factors depending on the cell type. So far, the FHL2 function in neuroblastoma cells is still unclear.
Mediation of the interaction of almost all recognized partners with Id2 is considered to be through its HLH domain. An exception is APC/CCDH1, which interacts with Id2 through the D-box domain of Id2 and mediates ubiquitin-dependent proteasomal degradation of Id2 (22). We supposed that Id2/bHLH activity may also be regulated by proteins interacting with Id2 via the non-HLH region. We used the HLH-deleted and D-box-mutated human Id2 as bait protein in a yeast two-hybrid system to identify novel Id2 interactors. FHL2 was identified to physically interact with Id proteins through an N-terminal conservation region. FHL2, whose expression increased during retinoic acid (RA)-induced differentiation of neuroblastoma cells, competitively prevented Ids from binding to E47 and thus largely restored E47-mediated transcription. Finally, we proposed an anti-proliferation function of FHL2 in neural cells by showing that FHL2 can inactivate the transcriptional and cell-cycle-promoting functions of Id2 in neuroblastoma cells.
MATERIALS AND METHODS
Plasmids and siRNA
The plasmids including pcDNA3–Id2–DBM containing the complete coding sequence of the full-length human Id2 with a D-box mutation (22), 5xE-box-Luc reporter and pcDNA3-E47 were kindly provided by Dr Iavarone (Columbia University Medical Center, New York). The Id2–DBM–δHLH fragment, lacking the entire HLH domain (codons 35–76 aa), was derived from pcDNA3–Id2–DBM by a sequential PCR scheme as described previously. The human full-length Id2 cDNA was obtained from Dr Desprez (California Pacific Medical Center, San Francisco). The human Id4 cDNA was provided by Dr Junjie Wu (Weill Medical College, Cornell University, Ithaca, New York). The full-length Flag-tagged FHL2 and the deletion mutants of FHL2 in pcDNA3 were donated by Dr Qinong Ye as described previously (23). Id1 and Id3 were amplified from a human endometrium cDNA library. The various deletion mutants of Ids and E47 were constructed by PCR and were subsequently subcloned into pcDNA3.1, pGEX-6P1, pBIND or pACT vectors through appropriate digestion sites.
Oligonucleotides for siRNA were chemically synthesized by Shanghai GeneChem Co. (Shanghai). The human FHL2-specific siRNA, 5′-AACTGCTTCTGTGACTTGTAT-3′ (sense strand), was as described previously (24). The unrelated siRNA sequence (sense strand, 5′-AAGACGAACGTGTCACGTATC-3′) was used as a control.
Yeast two-hybrid screening and mammalian two-hybrid assays
The HLH-deleted Id2–DBM (Id2–DBM–δHLH) was inserted in-frame into the Gal4 DNA-binding domain vector pGBKT7. A human MCF-7 cDNA library, prepared by the use of the BD SMART™ kit (Clontech) according to the manufacturer's instructions, with some modifications, was screened according to protocols recommended by the MATCHMAKER two-hybrid system 3 kit.
Mammalian two-hybrid assays were performed in cells as described previously (25).
Cell culture, transfection and colony-forming assay
MCF-7, 293T, SK–N–SH, SH–SY5Y and COS-7 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10–15% FBS (Hyclone). An amount of 1 μM retinoic acid (Sigma, St. Louis, MO) was used to induce differentiation of SK–N–SH and SH–SY5Y cells. DNA transfection involved use of Superfect reagent (Qiagen, Hilden, Germany). Transfection with siRNA duplexes involved use of Lipofectamine 2000 (Invitrogen). For colony-forming assay, transfected cells were selected with G418 (Invitrogen) for 14 days. Colonies were scored in triplicate cultures at least three times.
GST pull-down assay
GST and GST fusion proteins were prepared as described previously (26). 35S-labeled proteins were produced with use of a Promega's TNT-coupled in vitro transcription and translation (IVT) system, with the expression vectors for E47, FHL2 and its derivatives in pcDNA3. GST pull-down assay was performed as described (26).
Coimmunoprecipitation (CoIP) and western blot analysis
Cells were cultured in 10-cm dishes and transfected with or without expression vectors. At 48 h after transfection, cells were harvested and total proteins or nuclear proteins were extracted. The CoIP and western blot assays were performed as described previously (26). The rabbit anti-Flag (Sigma), Myc tag (Abcam, Cambridge, MA), GAL4DBD (Clontech), Id2 (Santa Cruz Biotechnology or Abcam), E47 (Santa Cruz), β-actin (Santa Cruz), FHL2 antibodies (Aviva Systems Biology, Beijing) and the mouse anti-p57Kip2 antibody (Santa Cruz) were used for immunoprecipitation or immunoblotting.
Luciferase assay
The luciferase reporter construct 5xE-box-Luc was cotransfected with pcDNA3-E47, pcDNA3.1-Id1-4 and/or pcDNA3.1-Id-del and/or pcDNA3.1-FHL2, or FHL2-siRNA/control-siRNA into cells. pRL-SV40 was used as an inner control. Forty-two hours later, cells were harvested and the relative luciferase activities were measured as described previously (26).
Northern blot and quantitative RT-PCR (qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) and underwent northern blot analysis as previously described (26). The membranes were probed with a 612-bp fragment of FHL2, a 402-bp fragment of Id2 and a 515-bp β-actin fragment labeled with [32P]dCTP by random priming.
For qPCR experiments, cDNA was prepared by use of Superscript II RNase H− reverse transcriptase (Invitrogen) and 1–2 μg total RNA. The PCR primer sets for p57Kip2, IGF2, H19 and HPRT were as previously reported (27). Reactions were run on a LightCycler (Roche).
G1/S checkpoint assay
SK–N–SH or SH–SY5Y cells were seeded in 3.5-cm culture dishes and transfected with plasmids pcDNA3.1, pcDNA3.1-Id2, pcDNA3.1-Id2-del and/or pcDNA3.1-FHL2 or FHL2-siRNA/control-siRNA. An EGFP-expressing vector was used to identify transfected cells. After 48 h, cells were labeled with 10 μM BrdU for 2 h. Immunostaining involved use of anti-BrdU antibody (Becton Dickinson). The proportion of BrdU and EGFP double-positive cells to EGFP-positive cells was determined by use of an Olympus fluorescence microscope. At least 350 cells from each plate were counted. The mean and SEM were calculated from three separate plates of three independent experiments.
Chromatin immunoprecipitation (ChIP) assay
MCF-7 cells at 80% confluence in 10 cm tissue culture plates were transiently cotransfected with 5 μg of 5xE-box-Luc and 5 μg of siRNA duplexes by use of Lipofectamine 2000 reagent. After 48 h, ChIP assays were performed according to the protocol for the ChIP assay kit (Upstate Biology, NY). Immunoprecipitation involved E47 antibody or non-specific IgG. DNA fragment were purified with use of a QIAquick Spin Kit (Qiagen). The presence of the target DNA sequences in both the input and the recovered DNA immunocomplexes was detected by PCR. The 5xE-box was amplified with its flanking sequence used as the primer set. The forward primer is 5′-AGTGCAAGTGCAGGTGCCAG-3′ and the reverse primer is 5′-GCTCTCCACGGTTCCATCT-3′.
Electrophoretic mobility shift assay (EMSA)
Electrophoretic mobility shift assay (EMSA) involved use of in vitro-translated (IVT) proteins and/or purified proteins from bacteria. Annealed μE5 E2-box oligonucleotide (5′-GATCCCAGAACACCTGCAGCAGGATC-3′) probe was generated by labeling with T4 polynucleotide kinase and [γ-32P]dATP. Purified GST-E47-bHLH (10 ng) and/or GST-Id2 (500 ng), GST-Id3 (500 ng), IVT-FHL2 (1, 3 or 5 μl), or IVT-empty vector (5 μl) were used for each 25 μl reaction mixture in a binding buffer. Proteins were incubated for 20 min at room temperature prior to the addition of 32P-labeled probes (1.75 pmol). A 100-fold excess of unlabeled wild-type or E-box mutant μE5 oligonucleotide (5′-GATCCCAGAACACTTTCAGCAGGATC-3′) was used for competition experiments. An amount of 0.3 μg of anti-E47 (Santa Cruz) or non-specific IgG were used for supershift experiments. The sample was run through a 6% polyacrylamide gel then dried and subjected to autoradiography.
Statistical analysis
Experimental results were expressed as the mean ± standard error of the mean (SEM). Statistical analysis involved use of Statview 5.0 software. Paired Student's t-test or two-way ANOVA followed by the Student–Newman–Keuls test were used where applicable to assess significant differences between groups. P < 0.05 was considered to be statistically significant.
RESULTS
Two-hybrid screening identifies FHL2 as an Id2-interacting factor independent of its HLH domain
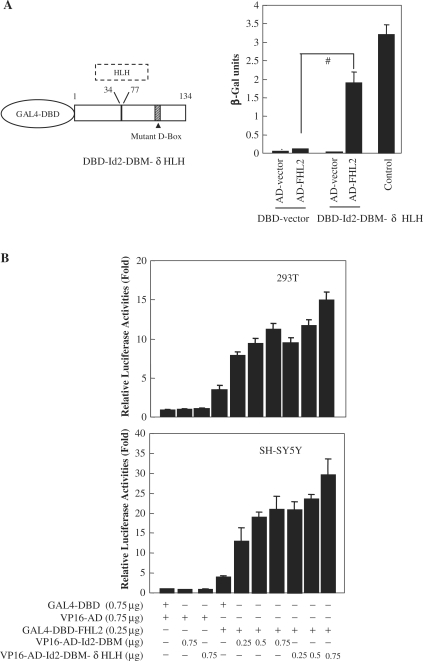
To probe the HLH-independent Id2 interactors, we performed yeast two-hybrid screening using the HLH-deleted and D-box-mutated Id2 (Id2–DBM–δHLH) as a bait in conjunction with a cDNA library of human breast cancer MCF-7 cells. This screening yielded 20 validated cDNA clones corresponding to 19 different proteins. Among them, the cDNA sequence coding for the full-length FHL2 (279 aa) was isolated from two individual clones. Quantification of β-galactosidase activity further showed a specific interaction between FHL2 and Id2–DBM–δHLH but not between either factor and the pairwise control containing only the Gal4 activation domain (AD) or only the Gal4 DNA-binding domain (DBD) (Figure 1A, right panel).
Figure 1.
Two-hybrid screening identifies FHL2 as an Id2-interacting protein. (A) The left panel shows the schematic illustration of DBD–Id2–DBM–δHLH bait protein. The mutant D-box was described in ref. (22). The right bargraph shows the results of β-galactosidase activity assays. Yeast AH109 cells were transformed with the indicated GAL4–DBD and GAL4–AD chimeric constructs, and the β-galactosidase activity was measured by a liquid O-nitrophenyl-β-d-galactoside assay. The experiment was repeated twice, and three different yeast transformants were used for each measurement. Control represents the interaction of p53 with SV40 large T-antigen proteins (#P < 0.01). (B) Mammalian two-hybrid assay was performed with 293T and MCF-7 cells, in which pG5-Luc was cotransfected with the indicated vectors. Approximately 42 h after transfection, the cells were harvested and luciferase activity was measured. The relative luciferase activity levels (normalized) for pACT and pBIND parent vector transfections were arbitrarily assigned a value of 1. All experiments were performed in triplicate and were repeated at least three times, and the results are expressed as mean ± SEM.
To extend these observations from yeast, functional interaction between FHL2 and Id2 was assessed in 293T and SH–SY5Y cells by fusing the full-length coding sequence of FHL2 gene with the GAL4–DBD and fusing Id2–DBM or Id2–DBM–δHLH with the VP16 AD. As shown in Figure 1B, the chimeric GAL4–DBD–FHL2 protein only weakly activated the GAL4-dependent reporter gene, by approximately three- to four-fold in these cells, which is consistent with previous reports (28,29). The luciferase activity was significantly enhanced by cotransfection of GAL4–DBD–FHL2 with the VP16–Id2–DBM or VP16–Id2–DBM–δHLH vector in a dose-dependent manner (Figure 1B).
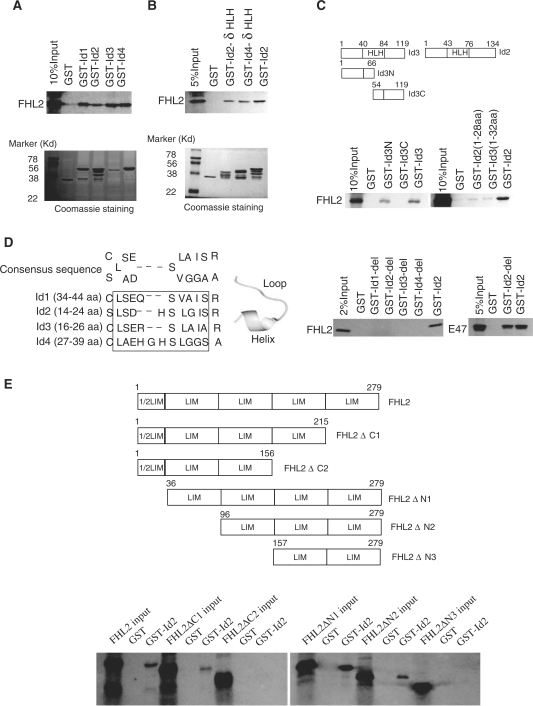
Figure 2.
FHL2 interacts with Id proteins in vitro and in vivo. (A) GST-pulldown assays were performed with in vitro-translated, [35S]-labeled FHL2 in the presence of GST–Id (Id1–4) fusion proteins. GST protein was used as a control. (B) GST-pulldown assays were performed with [35S]-labeled FHL2 and GST alone, GST–Id2–δHLH, GST–Id4–δHLH or GST–Id2. (C) The top panel is the schematic illustration of Id3 and its mutants. GST alone, GST–Id3-N, GST–Id2-C and GST–Id2 were used to pull down the full-length FHL2 (bottom panel). (D) The left panel shows the alignment of the relatively conserved region within the N termini of Id proteins (Id1–4). The sequence in the pane was deleted in Id-del mutants. The middle panel shows the representative loop–helix structure of these conserved amino acids. GST alone, GST–Id-del mutants or GST–Id2 were used to pull down the full-length FHL2 or E47 (right panel). (E) The upper panel is the schematic illustration of FHL2 and its mutants. GST-pulldown assays were performed with various IVT–FHL2 mutants and GST–Id2 or GST alone. (F) COS-7 cells were transfected with Flag-FHL2, Myc-Id2 or their combination. Cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (G) COS-7 cells were transfected with the indicated plasmids. After 48 h transfection, cells were lysed and underwent CoIP experiments using the indicated antibodies. NsIgG, non-specific IgG. NS, non-specific band.
Physical interaction between FHL2 and Id2 in vitro and in vivo
To validate the specificity of the binding between FHL2 and Id2, we used GST fusion proteins and in vitro-translated proteins in pulldown assays. GST–Id2 efficiently bound to the in vitro-translated 35S-labeled full-length FHL2 (Figure 2A). Given the low homology among the regions outside the HLH domain in Id proteins (30), we investigated the possibility of three other Id members (Id1, Id3 and Id4) binding to FHL2 in this assay. As shown in Figure 2A, GST–Id1, GST–Id3 and GST–Id4 bound to the full-length FHL2 as well. The interaction of FHL2 with all Id proteins is not equivalent in this assay, as demonstrated in Figure 2A; the retention of FHL2 by GST–Id3 is substantially greater than that of the others, given the apparent lower level of this GST fusion protein. GST–Id2 and GST–Id4 carrying a deletion of the HLH domain (Id2–δHLH, Id4–δHLH) bound to FHL2 (Figure 2B), which indicates that the HLH domain is not required for FHL2-Id interaction.
To determine which region of Ids was responsible for mediating the interaction with FHL2, the N- and C-terminal deletion mutants of Id3 (Id3-N and Id3-C) were tested for their ability to interact with FHL2 by GST-pulldown assay. The full-length FHL2 was pulled down by GST–Id3-N but not by GST–Id3-C and GST alone (Figure 2C). Next, we further showed that the full-length FHL2 could be pulled downed by the Id2 (1–28 aa) and Id3 (1–32 aa) mutants (Figure 2C), which suggests that the N-terminal region of Ids is sufficient for FHL2 interaction. We noted a relatively conserved region within the N termini of Ids (Figure 2D, left panel) that formed a conserved small loop–helix structure (Figure 2D, middle panel), so we generated mutant Id proteins (Id1–4) with this conserved region deleted (Id1-del to Id4-del), and then performed pulldown assays. Surprisingly, FHL2 could no longer be pulled down by any of these Id mutants; however, the bHLH transcriptional factor E47 could be pulled down by Id2-del (Figure 2D, right panel). These results indicated that the residues in the N-terminal region of Ids are indispensable for FHL2 binding. We postulated that the deletion of residues in the N-terminal region of Ids may cause a gross conformational change of Ids that disrupts the binding of FHL2 but not E47.
To more thoroughly characterize the FHL2–Id interaction, we analyzed a series of 35S-labeled FHL2 domain deletions for interaction with GST–Id2. As shown in Figure 2E, the full-length FHL2, FHL2ΔC1, FHL2ΔN1 and FHL2ΔN2, but not FHL2ΔC2 and FHL2ΔN3, were pulled down by GST–Id2, which demonstrates that both the second and the third LIM domain in FHL2 are required for the FHL2–Id2 interaction.
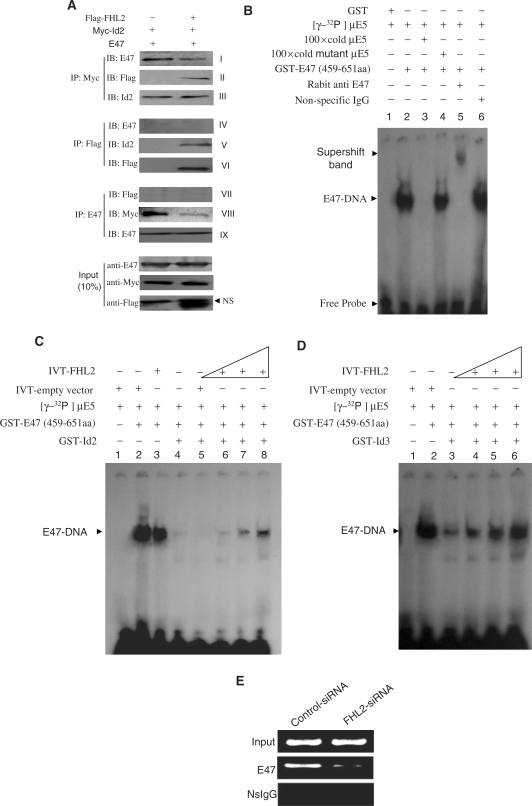
To confirm that FHL2 can interact with Id2 in cells, we transiently transfected COS-7 cells with Flag-FHL2, Myc-Id2, or a combination of Flag-FHL2 and Myc-Id2. Cell lysates were first precipitated with a Flag antibody, then underwent immunoblotting analysis with an anti-Myc tag or Id2 antibody. An intense band corresponding to Myc-Id2 was detected only in immunoprecipitates containing Flag-FHL2 (Figure 2F). The presence of Id2 in the FHL2-containing immune complexes is specific, because an irrelevant GAL4–DBD antibody failed to immunoprecipitate Id2. A reciprocal coimmunoprecipitation experiment with use of an anti-Myc antibody also revealed the presence of Flag-FHL2 in the immune complexes containing Myc-Id2 (Figure 2F). Next, we cotransfected Flag-FHL2 and Myc-Id2-del or Myc-Id2-δHLH into COS-7 cells for CoIP assays. As demonstrated in Figure 2G, Flag-FHL2/Myc-Id2–δHLH but not Flag-FHL2/Myc-Id2-del could be reciprocally coimmunoprecipitated by use of anti-Flag and/or anti-Myc antibodies. These results confirmed that the N-terminal loop–helix structure in Id2 but not the HLH domain is required for the FHL2–Id2 interaction in the cell system.
FHL2 antagonizes the inhibition of E-box-mediated transcription by Ids, but not by Id-dels
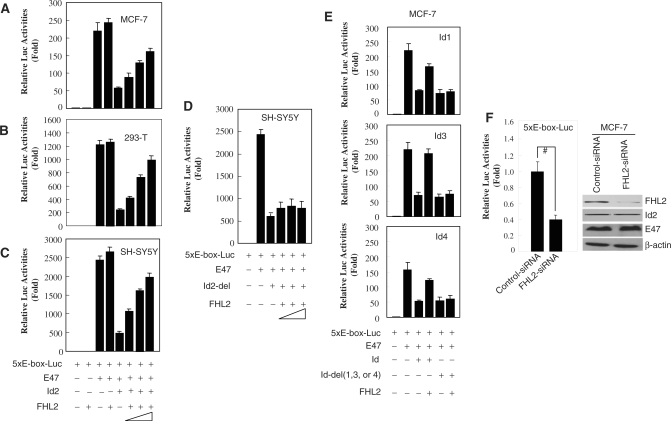
To explore the possible functional significance of the observed FHL2-Id2 interaction, we performed luciferase reporter assays with five multimerized E-boxes driving the expression of luciferase (5xE-box-Luc). Transfection of MCF-7 cells with reporter alone or reporter plus FHL2 expression vector revealed background luciferase activity (Figure 3A, lane 1 and 2). Cotransfection of E-box-Luc plus E47 significantly activated the system in cells (lane 3). Given that E47/Id2 heterodimers lose the ability to bind to the E-box, Id2 cotransfection resulted in a significant repression of the E47-activated luciferase activity (lane 5); however, this repression was markedly antagonized by FHL2 in a dose-dependent manner (lanes 6–8). FHL2 slightly increased E47 activation of the reporter (lane 4), which might be due to FHL2 inhibition of the endogenous Id activities. Similar results were obtained in 293T and SH-SY5Y cells (Figure 3B and C), which suggests that the inhibitory effect of FHL2 on Id2 activity in this assay is independent of other tissue-specific cofactors. To further confirm whether this repression was mediated by the direct interaction of FHL2 and Id2, we used the Id2-del expression vector, a 9-aa deleted (from 15 to 23 aa) Id2 mutant that cannot interact with FHL2 in vitro and in vivo, as shown in Figure 2D and G, for luciferase assays. As expected, Id2-del also significantly inhibited E47-mediated reporter activity as did the wild-type Id2 (Figure 3D, lane 3); however, unlike Id2 repression, Id2-del repression was not antagonized by FHL2. Furthermore, we also observed that FHL2 significantly overcame the inhibition of E47-mediated transcription by Id1, Id3 and Id4 in MCF-7 cells, but this inhibitory effect was abrogated in Id1-del, Id3-del and Id4-del mutants (Figure 3E). Taken together, these data strongly indicate that FHL2 can potently repress Id activities by directly associating with them, thus restoring the transcription programme driven by E47. To further address this, we cotransfected 5xE-box-Luc and FHL2-specific siRNA into MCF-7 cells for luciferase assays. As shown in Figure 3F, knock down of the endogenous FHL2 expression led to a commensurate reduction in endogenous E47 activity.
Figure 3.
Effect of FHL2 on E47-mediated transcription inhibited by Id proteins. (A–E) MCF-7, 293T and SH–SY5Y cells were transiently cotransfected with pRL-SV40 and the indicated vectors. The relative luciferase activity levels were normalized in all cases by 5xE-box-Luc and mock effector transfection and arbitrarily assigned a value of 1. All experiments were performed in triplicate and were repeated at least three times, and the results are expressed as mean ± SEM. (F) MCF-7 cells were cotransfected with 5xE-box-Luc, pRL-SV40 and control-siRNA or FHL2-specific siRNA. The relative luciferase activity levels were normalized by 5xE-box-Luc and control-siRNA transfection and arbitrarily assigned a value of 1. All experiments were performed in triplicate and were repeated at least three times, and the results are expressed as mean ± SEM. (#P < 0.01).
FHL2 antagonizes the binding of Id2 to E47, thus releasing E47 to its cognate DNA
To explore the molecular mechanism by which FHL2 represses Id activities, we analyzed the interactions among E47, Id2 and FHL2 proteins present in the nucleus by performing immunoprecipitation assays with MCF-7 cells cotransfected with E47, Myc-Id2 and/or Flag-FHL2. Id2 present in both Flag and E47 antibody immunoprecipitates (Figure 4A, panels V and VIII) and reciprocal experiment results demonstrated E47 and FHL2 detected in Myc antibody immunoprecipitates (panels I and II). These results further confirmed the interaction of Id2 with FHL2 or E47. However, neither E47 nor FHL2 was detected in their mutual immunoprecipitates (panels IV and VII), which indicates that FHL2 and E47 do not interact and coexist in an Myc–Id2 protein complex in vivo. The lack of evidence of an FHL2–E47 interaction is consistent with another report (31). More importantly, CoIP results showed that the Id2–E47 recognition was markedly blunted in the presence of ectopic FHL2 (panels I and VIII). These results suggest a competitive nature between FHL2 and E47 for binding Id2, whereby FHL2 prevents the formation of an Id2–E47 heterodimer.
Figure 4.
FHL2 competitively blunts the binding of Id2 to E47 and releases E47 to its cognate DNA. (A) FHL2 blunts the E47–Id2 association in MCF-7 cells. Myc-tagged Id2, E47 and/or Flag-tagged FHL2 were transiently cotransfected into MCF-7 cells. After 48 h transfection, cells were harvested and fractionated to a nuclear fraction. Equal amounts of nuclear lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (B–D) FHL2 attenuates Id-inhibited E47–DNA binding in vitro. EMSA was performed as described in Materials and Methods section. (E) Knock down of the endogenous FHL2 in MCF-7 cells sharply attenuated the binding of the endogenous E47 to the exogenous target DNA sequence. ChIP assays were performed as described in Materials and methods section. NS, non-specific band.
On the basis of these results, we further determined whether the FHL2-Id2 association would release E47 to its cognate DNA sequence by performing EMSA with [γ-32P]-labeled μE5, which contains an E-box sequence, a probe that was previously used as a consensus sequence for E47 homodimer binding in vitro (32). We first identified the specific band formed by the E47 and μE5 probe. As shown in Figure 4B, purified GST did not bind to the E5 probe (lane 1). A shifted band representing an E47–E47 homodimer complex bound to DNA was observed in the presence of GST–E47 (459–651 aa) (lane 2), which was confirmed by adding a non-labeled μE5, a mutant probe, or E47-specific or non-specific antibodies (lanes 3–6). The specific E47-DNA band was not affected by adding either an IVT-empty vector or IVT-FHL2 proteins (Figure 4C, lanes 2 and 3). As expected, the E47-DNA band was substantially attenuated when purified GST-Id2 was added to the reaction system (Figure 4C, lane 4). This attenuation was not affected by adding IVT-empty vector proteins (Figure 4C, lane 5) but was reversed by adding IVT-FHL2 in a dose-dependent manner (Figure 4C, lanes 6–8). Similar results were observed with Id3 (Figure 4D). These in vitro data indicated that FHL2 possesses the ability to antagonize the binding of E47 and Id proteins, thereby releasing E47 to its target DNA. To further confirm this in-cell system, we performed ChIP assays after cotransfecting 5xE-box-Luc and FHL2-specific siRNA into MCF-7 cells. As shown in Figure 4E, knockdown of the endogenous FHL2 expression in MCF-7 cells caused a significant reduction of the endogenous E47 binding to the exogenous DNA target.
FHL2 is greatly induced during differentiation of neuroblastoma cells, and ectopic FHL2 overcomes Id2-promoted cell-cycle progression
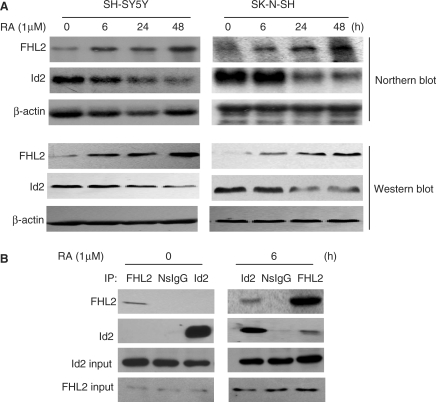
The findings in the previous sections prompted us to investigate whether FHL2 is involved in the biological processes regulated by the Id2–E47 transcriptome. To do so, we used human neuroblastoma SK–N–SH and SH–SY5Y cells, in which the function of the Id2–E47 transcriptome in cell proliferation and differentiation has been well studied (10,11,33,34). In addition, human neuroblastoma cells are frequently used as in vitro models to recapitulate differentiation of the nervous system (35,36). We tested the dynamic expression of FHL2 during retinoic acid (RA)-induced differentiation of these cells. As shown in Figure 5A, FHL2 expression was very weak and even negligible in both SK–N–SH and SH–SY5Y cells. However, treatment with RA (1 μM) induced a progressive and remarkable elevation of both FHL2 mRNA and protein levels. In accordance with previous reports (16,37), RA administration led to a marked inhibition of Id2 gene expression. In RA-untreated SH–SY5Y cells, the endogenous interaction of FHL2 and Id2 could not be detected by CoIP assays but could be detected in SH–SY5Y cells with 6 h RA treatment (Figure 5B). The opposite expression style and interaction of FHL2 and Id2 during cell differentiation induced by RA suggested an antagonistic role of FHL2 for Id2 activity.
Figure 5.
The reverse expression and functional interaction of FHL2 and Id2 was induced by retinoic acid (RA) in human neuroblastoma cells. (A) Northern and western blot analyses of FHL2 and Id2 gene expression in SH–SY5Y and SK–N–SH cells treated with RA (1 μM) for the indicated times. β-actin is used as a loading control. (B) SH–SY5Y cells were treated with RA (1 μM) for the indicated times, lysed and underwent CoIP experiments with the indicated antibodies.
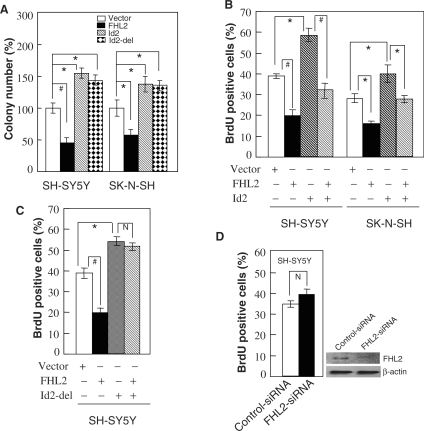
A prominent function of Id2 in neuroblastoma cells is to encourage cell proliferation by promoting the transition from the G1 to S phase of the cell cycle (10,11). Therefore, we investigated whether FHL2 inhibits cell proliferation and opposes Id2-mediated entry into the S phase. FHL2 transfection markedly inhibited the colony formation of both SK–N–SH and SH–SY5Y cells, whereas Id2 or Id2-del transfection promoted colony formation (Figure 6A). These results clearly demonstrate the opposite effect of FHL2 and Id2 on cell proliferation. Next, SK–N–SH and SH–SY5Y cells were cotransfected with FHL2 and Id2 in the presence of a GFP expression plasmid, and the rate of DNA synthesis was measured by incorporation of BrdU in GFP-positive cells. As shown in Figure 6B, ectopic FHL2 strongly inhibited S phase entry and abrogated Id2-stimulated DNA synthesis. To confirm whether this effect resulted from the direct interaction of FHL2 and Id2, we transiently transfected Id2-del or Id2-del plus FHL2 into SH–SY5Y cells and performed BrdU incorporation assays (Figure 6C). Although the significant promotion of S phase entry was observed in both Id2-del and wild-type Id2 cells, the promotional effect in Id2-del cells was not abolished by FHL2 as for wild-type Id2. Next, we performed BrdU incorporation assays in FHL2-siRNA-transfected SH–SY5Y cells. Not unexpectedly, transfection of FHL2-specific siRNA slightly but not significantly promoted the S-phase entry because of the very weak and even negligible expression level of FHL2 in SH–SY5Y cells (Figure 6D). Similarly, FHL2-siRNA transduction did not significantly promote the S-phase entry of SK–N–SH cells (data not shown). Altogether, these data suggest that the ectopic or RA-induced expression of FHL2 can effectively inhibit cell-cycle progression of neuroblastoma cells by antagonizing Id2 activity.
Figure 6.
Ectopic expression of FHL2 significantly inhibited proliferation of human neuroblastoma cells by suppressing Id2-promoted S phase entry. (A) SH–SY5Y and SK–N–SH cells were stably transfected with pcDNA3.1 empty vector, pcDNA3.1–FHL2, pcDNA3.1–Id2 or pcDNA3.1–Id2-del, and colonies were scored after selection in G418. The total number of colonies recovered from the empty vector control transfection was assigned as 100 percent. (B and C) SH–SY5Y or SK–N–SH cells were transiently transfected with the indicated plasmid combination. The pEGFP-N1 plasmid was included to identify transfected cells. Cultures were labeled with BrdU for 2 h and immunostained for BrdU by using a Cy3-conjugated secondary antibody. Cells were assessed for GFP and BrdU, and the percentage of transfected cells positive for BrdU was scored. (D) SH–SY5Y cells were transiently transfected with the indicated siRNA duplexes. After 48 h, cells were assessed for BrdU incorporation as in C. All experiments in A–D were performed in triplicate and were repeated at least three times, and the results are expressed as mean ± SEM (*P < 0.05; #P < 0.01; N, P > 0.05).
FHL2 up-regulates the expression of E47 target genes by inactivating Id2 activity in SK–N–SH cells
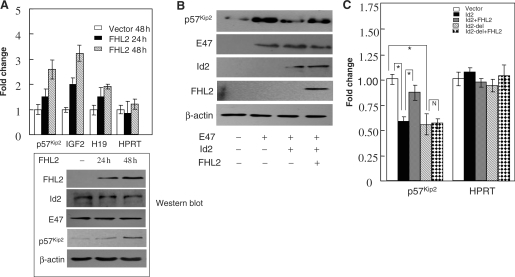
To test whether FHL2 affects the expression of E47–Id2 transcriptome targets, we chose p57Kip2, IGF2 and H19 genes to measure their mRNA expression by performing qPCR. As shown in Figure 7A, FHL2 transfection markedly up-regulated the mRNA expression of p57Kip2, IGF2 and H19. Ectopic FHL2 expression did not change the protein levels of Id2, E47and β-actin but increased the protein level of p57Kip2. To check whether the up-regulation of E47 target genes induced by FHL2 was a consequence of the functional regulation of the E47–Id2 transcriptome by FHL2, we measured the protein level of p57Kip2 in E47, Id2 and/or FHL2-transfected SK–N–SH cells (Figure 7B). Similar to a previous report (27), E47 effectively induced the accumulation of p57Kip2 in cells, and this effect was abolished by Id2. Ectopic expression of FHL2 reversed the repressive effect of Id2 and partially restored E47-induced p57Kip2 up-regulation. Finally, to further validate whether the induction of E47 target genes by FHL2 is directly linked to the FHL2–Id2 association, we performed qPCR for p57Kip2 in SK–N–SH cells (Figure 7C). Id2 induced a significant decrease of p57Kip2 mRNA expression, but this effect was markedly attenuated by FHL2. Similar to wild-type Id2, Id2-del also induced a similar decrease of p57Kip2 mRNA expression; however, this effect was not abrogated by FHL2. Taken together, these data indicate that FHL2 is able to effectively up-regulate the expression of E47 target genes in neuroblastoma cells by directly interacting with and preventing Id2 from binding to E47.
Figure 7.
Downstream targets of E47–Id2 transcriptome are regulated by FHL2 in SK–N–SH neuroblastoma cells. (A) SK–N–SH cells were transiently transfected with the FHL2 expression vector or empty vector and cultured for the indicated times. Expression of the indicated transcript abundance was analyzed by qPCR values. HPRT was used as an internal control. All experiments were repeated at least three times, and the results are expressed as mean ± SEM. FHL2, Id2, E47, p57Kip2 and β-actin protein levels in transfected SK–N–SH cells analyzed by western blot is shown at the bottom. (B) SK–N–SH cells were transiently cotransfected with the indicated combinations of plasmids expressing E47, Id2 and FHL2. Cell lysates were analyzed for the expression of p57Kip2, IGF2, E47, Id2, FHL2 and β-actin. (C) SK–N–SH cells were transiently transfected with the indicated expression vector or different combinations. After 48 h transfection, total RNA was extracted and the abundance of p57Kip2 was analyzed by qPCR. HPRT was used as an internal control. All experiments were repeated at least three times, and the results are expressed as mean ± SEM (*P < 0.05; N, P > 0.05).
DISCUSSION
The biological function of Id proteins is achieved through antagonizing the activity of their associated bHLH transcription factors (1,2). Although the molecular mechanism involved in this process has been well dissected, additional regulators should work in concert with Ids in view of the spatio-temporal variation of Id-controlled gene expression. In this study, the LIM-only protein FHL2, a well-known cofactor or adaptor of several transcription factors (19), was identified as a novel Id2 interactor. In addition, the interaction of FHL2 with three other members of the Id protein family (Id1, Id3 and Id4) was demonstrated in GST-pulldown in vitro assays. We further show that FHL2 blocks Id–E47 coupling by a competitive binding mechanism, which subsequently represses the inhibitory effect of Ids on E47-mediated transcription. Finally, the FHL2 regulation of the E47–Id2 transcriptome was extended to the proliferation of the neuroblastma cells. These findings underscore an important function of FHL2 in regulating the Id2 signaling pathway.
The LIM domain-containing proteins can function as adaptors or scaffolds to support the assembly of multimeric protein complexes and can operate as competitors, autoinhibitors and localizers (38,39). As a member of the LIM protein family, FHL2 has been shown to associate with several transcriptional factors or signaling transducers such as AR, CBP/p300, β-catenin, FOXO1, AP1 and TRAF6, to regulate signal transduction and gene expression (19). In most cases, FHL2 protein appears to participate in functional complexes to modulate the tissue-specific activity of activators or repressors. One possible explanation for the dual role of FHL2 in a transcription or regulating signaling pathway might be that FHL2 acts to stabilize the functional complexes as a type of bridging factor. This explanation was supported by the fact, like that the formation of a ternary complex by FHL2, CBP/p300, and β-catenin could synergistically activate AR-mediated transcription (28), and that FHL2 interacts with titin, a protein that plays a crucial role as organizer of the sarcomere, and functions as an adaptor molecule that links the metabolic enzymes MM-creatine kinase, adenylate cyclase and phosphofructokinase to titin, thereby helping to recruit metabolic enzymes needed for energy provision during muscle contraction (40). In dramatic contrast, we observed that FHL2 acts as a competitor of E47 for Id2 binding and subsequently allows E47 to recruit to its target DNA and execute its transcriptional activity (Figure 3 and 4). Although different epitopes of Id2 are required for FHL2 and E47 binding, the binding site for FHL2 or E47 of Id2 may be masked by a conformational change of Id2 when it is occupied by FHL2 or E47. In addition, Id2 may not provide enough surface space to simultaneously interact with multiple proteins; after all, Id2 is only 18 kDa. Indeed, such competitive binding is also observed for other FHL2-interacting molecules. For example, the cytoplasmic domain of TRAF6, which is formed by few amino acid numbers, can provide a binding surface for only FHL2 or RANK. As a consequence, FHL2 antagonizes the RANK–TRAF6 interaction and blunts the RANK–TRAF6 signaling (41).
Emerging evidence has linked FHL2 to cell-cycle progression. However, the function of FHL2 in cell proliferation is particularly intriguing, because it may exert a positive or negative effect on cell-cycle-regulated processes in a tissue-dependent fashion. This dual nature of FHL2 can be explained by the finding that FHL2 can function as a repressor or activator of transcriptional activity depending on the cell type. For example, FHL2 promoted SKI-induced proliferation of UCD-Mel-N melanoma cells and enhanced proliferation of MDA-MB-231 human breast cancer cells by regulating cell-cycle-dependent p21 expression (42,43). Loss of FHL2 downregulated the expression of cyclin D1 and greatly reduced the proliferative capacity of mouse embryo fibroblasts (44). In contrast, FHL2 antagonized the proliferation of differentiated myoblasts by downregulating cyclin D1 (45) and suppressed liver cancer cell growth through a TGF β-like pathway (24). In this study, we demonstrated an antagonistic effect of FHL2 on Id2-driven proliferation of neuroblastoma cell types. The p57Kip2, IGF2 and H19 genes, belonging to the human chromosome 11p15.5-imprinted cluster, which have crucial functions in differentiation, the cell cycle and oncogenesis (46–48), are within the small group of E47–Id2 targets (27). In accordance with a previous report (27), we also demonstrated that the expression of these genes was strongly induced by E47 and inhibited by Id2. Among them, p57Kip2 is one of the three members of the Cip/Kip family of cyclin-dependent kinase inhibitors and a well-known cell-cycle regulator (49), the induction of which is essential for E47-mediated inhibition of the cell cycle in neuroblastoma cells (27). Ectopic expression of FHL2 in neuroblastoma cells strongly induced the expression of p57Kip2, IGF2 and H19 genes by regulating the Id2–E47 transcriptome (Figure 7). Given the oncogenic activity of Id2 in tumors from the nervous system, our data suggest that FHL2 possesses a tumor suppressor function in these cell types.
Id family proteins were implicated in the control of differentiation in organisms from fly to human (1,6). There is general agreement with the notion that differentiation of a variety of cell types requires elimination of the Id function. Thus, the functional inhibitors of Id proteins might possess the potential to facilitate cell differentiation. Here, we demonstrated a reverse expression relation between Id2 and its functional repressor FHL2 in differentiated neuroblastoma cells induced by RA (Figure 5). In addition, independent evidences suggested a diverse role of FHL2 and Id proteins during the differentiation process of myoblasts and osteoblasts (45,50). These limited data implied that a refined regulation mechanism of Id function by FHL2 is required during the processes of development and cellular differentiation.
In the past decade, numerous reports demonstrated the aberrant accumulation of Id proteins in solid tumors and Ids facilitating tumorigenesis by repressing cell differentiation, stimulating cell proliferation and promoting tumor neoangiogenesis (5–9). As a functional suppressor of Id2, FHL2 is frequently deregulated, which includes being overexpressed and down-regulated in various types of tumors (21). The most common mechanism selected by tumor cells to activate Id2 function is to elevate the expression of Id2 gene. In this study, we demonstrated that the expression of FHL2 is very weak and even negligible in human neuroblastoma cells. Based on our findings, we suggest that tumor cells may target another level in Id biology regulation. The fact that RA induced the reverse expression of Id2 and its functional repressor FHL2 (Figure 5) indicated that FHL2 may be an important stalker against Id2 protein activity in neuroblastoma cells. Loss of FHL2 will release the oncogenic activity of Id2 and may contribute to tumor progression. Testing the ratio of FHL2 to Id2 expression in human neuroblastoma cells might be beneficial to differentiate different clinopathological stages.
In conclusion, these findings provide definite evidence for FHL2–Id2 interaction and its corresponding functional effect. The functional activity of FHL2 acting as an repressor of the oncogenic activity of Id2 in neuroblastoma cells suggests that loss of FHL2 may be involved in tumorigenesis and progression of the nervous system.
FUNDING
National Natural Science Foundation of China (grant 30870507); Ministry of Science and Technology of China (2005CB522603 and 2005CB522400). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Iavarone, Columbia University Medical Center, New York, for providing Id-DBM, E47 and E-box-Luc plasmids; Dr Desprez, California Pacific Medical Center, San Francisco, for providing the Id2 plasmid; and Dr Junjie Wu, Weill Medical College, Cornell University, Ithaca, NY, for providing human Id4 cDNA.
REFERENCES
- 1.Massari ME, Murre C. Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix–loop–helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 3.Atchley WR, Fitch WM. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl Acad. Sci. USA, 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langlands K, Yin X, Anand G, Pronchowink EV. Differential interactions of Id proteins with basic helix–loop–helix transcription factors. J. Biol. Chem. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- 5.Perk J, Iavarone A, Benezra R. Id family of helix–loop–helix proteins in cancer. Nat. Rev. Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 6.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 7.Yokota Y. Id and development. Oncogene. 2002;20:8290–8298. doi: 10.1038/sj.onc.1205090. [DOI] [PubMed] [Google Scholar]
- 8.Iavarone A, Lasorella A. ID proteins as targets in cancer and tools in neurology. Trends Mol. Med. 2006;12:588–594. doi: 10.1016/j.molmed.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Fukuma M, Okita H, Hata J, Umezawa A. Upregulation of Id2, an oncogenic helix–loop–helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene. 2003;22:1–9. doi: 10.1038/sj.onc.1206055. [DOI] [PubMed] [Google Scholar]
- 10.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signaling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 11.Lasorella A, Boldrini R, Dominici C, Donfrancesco A, Yokota Y, Inserra A, Iavarone A. Id2 is critical for cellular proliferation and is the oncogenic effector of N-myc in human neuroblastoma. Cancer Res. 2002;62:301–306. [PubMed] [Google Scholar]
- 12.Nishimori H, Sasaki Y, Yoshida K, Irifune H, Zembutsu H, Tanaka T, Aoyama T, Hosaka T, Kawaguchi S, Wada T, et al. The Id2 gene is a novel target of transcriptional activation by EWS–ETS fusion proteins in Ewing family tumors. Oncogene. 2002;21:8302–8309. doi: 10.1038/sj.onc.1206025. [DOI] [PubMed] [Google Scholar]
- 13.Lasorella A, Rothschild G, Yokota Y, Russell RG, Iavarone A. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol. Cell Biol. 2005;25:3563–3574. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix–loop–helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 15.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 16.Lasorella A, Iavarone A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc. Natl Acad. Sci. USA. 2006;103:4976–4981. doi: 10.1073/pnas.0600168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Luo Y, Starremans PG, McNamara CA, Pei Y, Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix–loop–helix inhibitor Id2. Nature Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 18.Liu CJ, Ding B, Wang H, Lengyel P. The MyoD-inducible p204 protein overcomes the inhibition of myoblast differentiation by Id proteins. Mol. Cell Biol. 2002;22:2893–2905. doi: 10.1128/MCB.22.9.2893-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessena M, Møllera S, Hansena T, Moensa U, van Ghelueb M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol. Life Sci. 2006;63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanahashi H, Tabira T. Alzheimer's disease-associated presenilin 2 interacts with DRAL, an LIM-domain protein. Hum. Mol. Genet. 2000;9:2281–2289. doi: 10.1093/oxfordjournals.hmg.a018919. [DOI] [PubMed] [Google Scholar]
- 21.Kleiber K, Strebhardt K, Martin BT. The biological relevance of FHL2 in tumor cells and its role as a putative cancer target. Anticancer Res. 2007;27:55–62. [PubMed] [Google Scholar]
- 22.Lasorella A, Stegmüller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Zhu J, Zhang H, Lu Q, Huang C, Ye Q. BRCA1 interacts with FHL2 and enhances FHL2 transcription function. FEBS Lett. 2003;553:183–189. doi: 10.1016/s0014-5793(03)00978-5. [DOI] [PubMed] [Google Scholar]
- 24.Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu W, Zhu J, Han J, Zhang H, Lin J, et al. Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-beta-like signaling pathway. J. Clin. Invest. 2009;119:349–361. doi: 10.1172/JCI35930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han WD, Zhao YL, Meng YG, Zang L, Wu ZQ, Li Q, Si YL, Huang K, Ba JM, Morinaga H, et al. Estrogenically regulated ERα target gene LRP16 interacts with ERα and enhances the receptor's transcriptional activity. Endocr-Relat. Cancer. 2007;14:741–753. doi: 10.1677/ERC-06-0082. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Zhao YL, Wu ZQ, Si YL, Meng YG, Fu XB, Mu YM, Han WD. The single-macro domain protein LRP16 is an essential cofactor of androgen receptor. Endocr-Relat. Cancer. 2009;16:139–153. doi: 10.1677/ERC-08-0150. [DOI] [PubMed] [Google Scholar]
- 27.Rothschild G, Zhao X, Iavarone A, Lasorella A. E proteins and Id2 converge on p57KIP2 to regulate cell cycle in neural cells. Mol. Cell Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. Interaction and functional cooperation between the LIM Protein FHL2, CBP/p300 and β-Catenin. Mol. Cell Biol. 2004;24:10689–10702. doi: 10.1128/MCB.24.24.10689-10702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schüle R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wibley J, Deed R, Jasiok M, Douglas K, Norton J. A homology model of the Id-3 helix–loop–helix domain as a basis for structure-function predictions. Biochem. Biophys. Acta. 1996;1294:138–146. doi: 10.1016/0167-4838(96)00008-8. [DOI] [PubMed] [Google Scholar]
- 31.Hill AA, Riley P. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol. Cell Biol. 2004;24:9835–9847. doi: 10.1128/MCB.24.22.9835-9847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen CP, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toma JG, El-Bizri H, Barnabe-Heider F, Aloyz R, Miller FD. Evidence that helix–loop–helix proteins collaborate with retinoblastoma tumor suppressor protein to regulate cortical neurogenesis. J. Neurosci. 2000;20:7648–7656. doi: 10.1523/JNEUROSCI.20-20-07648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix–loop–helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 35.Abemayor E, Sidell N. Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ. Health Perspect. 1989;80:3–15. doi: 10.1289/ehp.89803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidell N, Sarafian T, Kelly M, Tsuchida T, Haussler M. Retinoic acid-induced differentiation of human neuroblastoma: a cell variant system showing two distinct responses. Exp. Cell Biol. 1986;54:287–300. doi: 10.1159/000163368. [DOI] [PubMed] [Google Scholar]
- 37.López-Carballo G, Moreno L, Masiá S, Pérez P, Barettino D. Activation of phosphatidylinositol 3-kinase/AKT signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 38.Bach I. The LIM domain: regulation by association. Mech. Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 39.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 40.Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Elher E. Subcellular targeting of metabolic nzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J. Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- 41.Bai S, Kitaura H, Zhao H, Chen J, Müller JM, Schüle R, Darnay B, Novack DV, Ross FP, Teitelbaum SL. FHL2 inhibits the activated osteoclast in a TRAF6-dependent manner. J. Clin. Invest. 2005;115:2742–2751. doi: 10.1172/JCI24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D, Xu W, Bales E, Colmenares C, Conacci-Sorrell M, Ishii S, Stavnezer E, Campisi J, Fisher DE, Ben-Ze'ev A, et al. SKI activates Wnt/beta-catenin signaling in human melanoma. Cancer Res. 2003;63:6626–6634. [PubMed] [Google Scholar]
- 43.Labalette C, Noüet Y, Sobczak-Thepot J, Armengol C, Levillayer F, Gendron MC, Renard CA, Regnault B, Chen J, Buendia MA, et al. The LIM-only protein FHL2 regulates cyclin D1 expression and cell proliferation. J. Biol. Chem. 2008;283:15201–15208. doi: 10.1074/jbc.M800708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin BT, Kleiber K, Wixler V, Raab M, Zimmer B, Kaufmann M, Strebhardt K. FHL2 regulates cell cycle-dependent and doxorubicin-induced p21Cip1/Waf1 expression in breast cancer cells. Cell Cycle. 2007;6:1779–1788. doi: 10.4161/cc.6.14.4448. [DOI] [PubMed] [Google Scholar]
- 45.Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kühl M, Behrens J, von der Mark K, Starzinski-Powitz A, Wixler V. The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 2002;159:113–122. doi: 10.1083/jcb.200202075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 47.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 48.Chung WY, Yuan L, Feng L, Hensle T, Tycko B. Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms’ tumors. Hum. Mol. Genet. 1996;5:1101–1108. doi: 10.1093/hmg/5.8.1101. [DOI] [PubMed] [Google Scholar]
- 49.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 50.Govoni KE, Amaar YG, Kramer A, Winter E, Baylink DJ, Mohan S. Regulation of insulin-like growth factor binding protein-5, four and a half lim-2, and a disintegrin and metalloprotease-9 expression in osteoblasts. Growth Horm. IGF. Res. 2006;16:49–56. doi: 10.1016/j.ghir.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]