Abstract
The i-motif is a four-stranded structure formed by two intercalated parallel duplexes containing hemiprotonated C•C+ pairs. In order to describe the sequence of reactions by which four C-rich strands associate, we measured the formation and dissociation rates of three [TCn]4 tetramers (n = 3, 4 and 5), their dissociation constant and the reaction order for tetramer formation by NMR. We find that TCn association results in the formation of several tetramers differing by the number of intercalated C•C+ pairs. The formation rates of the fully and partially intercalated species are comparable but their lifetimes increase strongly with the number of intercalated C•C+ pairs, and for this reason the single tetramer detected at equilibrium is that with optimal intercalation. The tetramer half formation times vary as the power −2 of the oligonucleotide concentration indicating that the reaction order for i-motif formation is 3. This observation is inconsistent with a model supposing association of two preformed duplex and suggests that quadruplex formation proceeds via sequential strand association into duplex and triplex intermediate species and that triplex formation is rate limiting.
INTRODUCTION
At slightly acidic pH, the oligonucleotides carrying a cytidine stretch form a tetrameric structure in which two parallel duplexes are associated in a head to tail orientation with their hemi-protonated C•C+ pairs intercalated in a so called i-motif (1). High-resolution structures of i-motif tetramers reported either from NMR or X-ray studies (2,3) show that intercalation and staking of C•C+ pairs generate a rigid core of long-lived base pairs including two narrow and two wide grooves. I-motif structures formed by four identical C-rich oligonucleotides (4), by two hairpins caring two cytidine stretches (5) or by a folded strand caring four cytidine stretches (6,7), have been described but little is known about the sequence of the intermolecular events by which two parallel duplexes can zip together into an interdigitated four-stranded structure.
In favorable circumstances, the formation and dissociation kinetics of a multimeric structure can yield information about its formation mechanism, on the transition states and on the limiting steps. Using this approach, we recently established that the formation of (TGnT)4 tetramolecular G-quadruplexes proceeds via sequential strand association into duplex and triplex intermediate species and that triplex formation is rate limiting in KCl solution (8). Using the same strategy, we investigate in the present work the formation pathway of i-motif tetramers formed by four identical TCn strands. The kinetics studies on i-motif previously reported concern mainly monomolecular (9) and dimeric (10) structures. Using UV absorption spectrophotometry (11), FRET (12) and NMR (10), it was shown that strand association and i-motif dissociation are relatively slow and that the i-motif stability is optimal around the cytidine pK. Nevertheless, these studies could not provide information about the i-motif formation pathway.
Association of CnT (13) and Cn (14) strands into i-motif results at equilibrium in the formation of several species differing by their intercalation topology. For this reason, we selected for the present study the TCn sequences (n = 3, 4 and 5) that adopt at equilibrium a single structure with optimal intercalation and the stacking order: (T1•T1-Cn•Cn-C2•C2 … C2•C2-Cn•Cn-T1•T1) (1,4, J.-L. Leroy, unpublished results).
The formation of an i-motif tetramer cannot result from the synchronized collision of four C-rich strands. Despite of the lack of experimental evidence about the existence of intermediate structures, i-motif formation must result of the association of two preformed parallel hemiprotonated duplexes or of the step by step association of four C-rich strands. Intercalation of two fully hemiprotonated duplexes into an i-motif structure is topologically impossible, but there is no difficulty to imagine intercalation of two incompletely hemiprotonated duplexes. For example, two [TC4]2 duplexes with C2•C2+ and C3•C3+ pairs and non-paired C4 and C5 bases could associate by mutual intercalation of bases C5 at steps T1-C2 and of C4 between pairs C2•C2+ and C3•C3+.
In order to understand the formation pathway of tetrameric i-motif structures, we measured by NMR the effects of temperature, pH and strand concentration on the formation and dissociation rates of the (TCn)4 tetramers. The spectra collected as a function of the time during i-motif formation show that several tetramers differing by the intercalation topology are formed at comparable rate. Nevertheless, the fully intercalated tetramers whose lifetimes are much more longer than those with partial intercalation are the single species detected at equilibrium. We further show that the reaction order for tetramer formation is 3, an observation inconsistent with duplex dimerization suggesting that i-motif formation proceed via duplex and triplex intermediate species.
MATERIALS AND METHODS
Oligonucleotides and NMR samples
The oligonucleotides were synthesized on a 10-μM scale, purified by chromatography on a Diethylaminoethyl (DEAE) column and dialyzed, first against a 0.1 M NaCl solution and then repeatedly against water. The dialysis ended when the equilibrium conductivity in the dialysis bath was <10 μS. At last, the oligonucleotides were dry lyophilized and stock solutions were prepared by dissolving the lyophilized sample in water. The solution pH was adjusted with HCl and NaOH. The strand concentration was determined from absorbance using A260 values of 30 400, 37 500 and 45 000 for TC3, TC4 and TC5, respectively.
The NMR samples, 400 μl in H2O/2H2O (9/1 v/v), were buffered by sodium phosphate of 40 mM. They contained 0.2 mM of sodium acetate and 0.5 mM of dimethyl sulfoxyde. The chemical shift of the methyl protons of acetate, δacetate, provided a marker for pH determination according to: pH = 4.63−log (1.909−δacetate)/(δacetate−2.076) at 0°C. The NMR peak of DMSO was used as an internal marker allowing spectra normalization.
Association and dissociation rate measurements
The protocol of the kinetic experiments has been already described (8). The TCn association rates into i-motif structures were measured using a sample initially melted. To measure the tetramer dissociation rates, a small volume of a concentrated fully tetrameric [TCn]4 solution was diluted into the NMR tube immediately before the beginning of the NMR measurements. The dissociation kinetics experiments were always performed using solutions diluted in such way that the tetramer equilibrium fraction was <10%. In that case, the dissociation rates were found independent of the oligonucleotide concentration and the evolutions of the NMR markers during tetramer dissociation were mono exponential.
All the NMR experiments were performed using a 500 MHz Varian Inova spectrometer using the jump and return sequence for water suppression (15). The dead time of the kinetics measurements was <3 min. The tetramer and monomer fractions were determined from the areas and peak heights of the thymidine methyl and H3 protons. The tetramer fraction was also measured using the cytidine imino proton peaks. Generally, the kinetic experiments were run during a time longer than 10 time constants. In the case of experiments with an extremely slow reaction rate, the NMR tubes were stored at controlled temperature in a thermostated incubator after a first set of measurements and additional kinetics measurements were performed after several days or weeks. The spectra collected versus time were normalized using the DMSO peak as a marker. The area and peak height of the NMR markers selected to determine the monomer and tetramer proportions were measured on each spectra with a home-made software and the time elapsed since the beginning of the kinetics was computed from the parameter file recorded with each spectrum. At last, the data were transferred into the Kaleidagraph software for analysis.
Determination of the reaction order for i-motif formation
The reaction order is a key parameter for the determination of the formation pathway of multimeric structures. The reaction order for the formation of an i-motif tetramer (Q) is the value of the exponent of the monomer concentration (M) in the rate equation:
The half reaction time, τasso, for strand association into a multimeric structure depends on a rate constant, k, on the initial oligonucleotide concentration [M0] and on the reaction order:
| 1 |
The half reaction time for tetramer formation was measured as a function of the oligonucleotide concentration and the reaction order was determined from the slope, −(order−1), of the log–log plot of τasso versus the oligonucleotide concentration.
The tetramer dissociation constant
The tetramer dissociation constant may be expressed as a function of αeq, the monomer equilibrium fraction of an oligonucleotide solution at concentration [M0] by:
| 2 |
We will characterize the tetramer stability by Fi, the free monomer concentration for which αeq = 0.5. Fi that will be designated as the reduced dissociation constant is related to the tetramer dissociation constant by:
| 3 |
It may be noticed that according to this definition, the concentration-dependent association time and the concentration-independent dissociation time are identical in an oligonucleotide solution at concentration M0 = 2 Fi. This property was routinely checked in order to control the consistency of the measured association and dissociation times.
RESULTS
i-motif dissociation kinetics
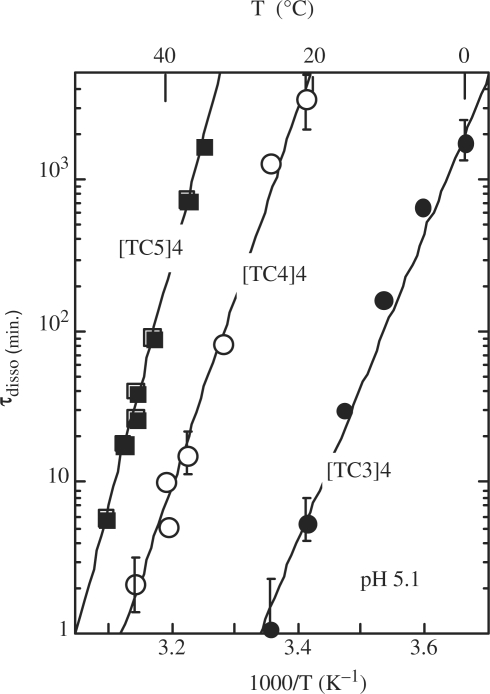
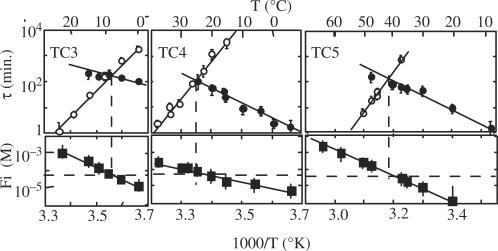
The lifetimes of the [TC3]4, [TC4]4 and [TC5]4 tetramers measured at pH 5.1 versus temperature are displayed in Figure 1. The experiments performed on the tetramer of TC2 show that its lifetime is <3 min at 0°C. The exchange time of the imino proton of the C2•C2+ pair of [TC2]4 is ∼0.3 min at 0°C (4). Hence, considering that the imino proton exchange time is shorter or equal to the tetramer lifetime, we estimate that the lifetime of [TC2]4 is in the range of 0.3–3 min at 0°C.
Figure 1.
Lifetime versus temperature at pH 5.1 of the fully intercalated i-motif tetramers of [TC3]4 (black circles), [TC4]4 (open circles) and [TC5]4 (black squares). The lifetime of [TC2]4 is estimated in the range of 3 s to 3 min at 0°C.
The comparison of the tetramer lifetimes displayed in Figure 1 shows that each additional cytidine increases the [TCn]4 lifetime by one to three magnitude orders. The lifetimes are independent of pH around the cytidine pK (pKN3 = 4.4) and decrease above pH 6 or under pH 4 as shown in Figure 2 for [TC3]4. The half titration pH of C•C+ pairs, 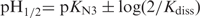 , depends on the cytidine pKN3 and on the base pair dissociation constant Kdiss (16). It is predictable that the outer C•C+ pairs are less stable than the inner one and that they dissociate fist when the pH move away from the pKN3 value. This should result in the progressive disruption of the outer C•C+ pairs by end fraying effect and probably in shorter tetramer lifetime.
, depends on the cytidine pKN3 and on the base pair dissociation constant Kdiss (16). It is predictable that the outer C•C+ pairs are less stable than the inner one and that they dissociate fist when the pH move away from the pKN3 value. This should result in the progressive disruption of the outer C•C+ pairs by end fraying effect and probably in shorter tetramer lifetime.
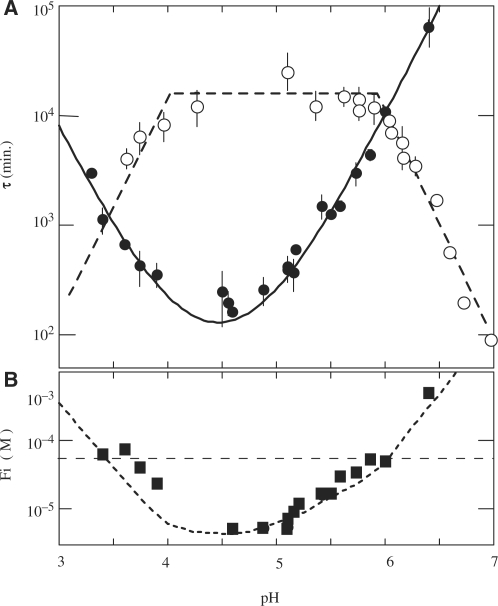
Figure 2.
Effect of pH on the formation and dissociation times and on the stability of the fully intercalated [TC3]4 tetramer. (A) [TC3]4 lifetime (open circles) and half formation time in 10−4 M TC3 solutions (black circles). The half formation time varies with pH as the power −2 of the product of the fraction of neutral cytidine, 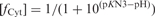 and protonated cytidine:
and protonated cytidine: 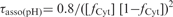 . (B) Reduced dissociation constant versus pH. The observation that Fi is close 10−4/2 (dashed line) at the temperature for which the association time measured in 10−4 M TC3 solution and the dissociation time are equal shows the consistence of the kinetics and equilibrium measurements.
. (B) Reduced dissociation constant versus pH. The observation that Fi is close 10−4/2 (dashed line) at the temperature for which the association time measured in 10−4 M TC3 solution and the dissociation time are equal shows the consistence of the kinetics and equilibrium measurements.
[TC3]4 association kinetics
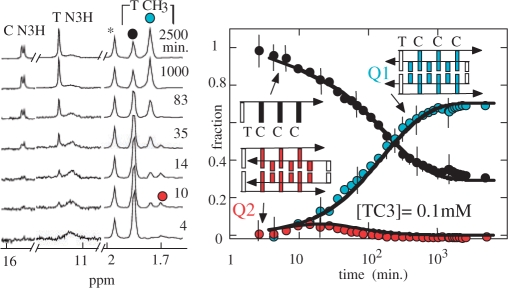
Representative spectra of a TC3 solution initially monomeric collected as the function of the time are displayed in Figure 3. In addition to the fully intercalated tetramer, Q1, the spectra show in the early stage of the kinetics (4–35 min) the accumulation of a small proportion of a multimeric species, Q2, which is tentatively assigned to the i-motif tetramer with an empty site between T1 and C2. The Q2 proportion is maximum ( f(Q2) ≈ 0.07) 20 min after the beginning of the experiment, and decreases towards a value under the detection threshold at equilibrium.
Figure 3.
Association kinetics of TC3 at 0°C pH 5.1 into i-motif structures. The proton spectra of a 10−4 M TC3 solution initially monomeric recorded as a function of the time (left panel) shows the formation of two multimers. The imino protons and the methyl peaks of the fully intercalated tetramer are labeled by blue dots and the methyl peak of the monomer by a black dot. The spectra show the transient accumulations of a short-lived species at the beginning of the kinetics (red dot), which is tentatively identified as the incompletely intercalated structure displayed on the figure. The solid line fitting the time evolution of the monomer and tetramer fractions are computed according to a model involving two parallel reactions (cf. text). The simulation indicates that the formation rate of both tetramers is comparable: 1/60 min and 1/90 min for the red and blue species, respectively, but their lifetimes are extremely different: 5 min and 1900 min for the red and blue species, respectively.
The solid lines accounting for the evolution of the monomer and tetramer fractions displayed in Figure 3 were computed according to a model involving two parallel reactions
| 4 |
The dissociation rate of the fully intercalated species, kof2 = 1/1600 min−1, was obtained from the dissociation time measured on Figure 1. The formation rate of the Q1 and Q2 species, kon1 = 1/90 min−1 and kon2 = 1/60 min−1, respectively, and kof1 = 1/5 min−1, were adjusted to fit the evolution versus time of the monomer, Q1 and Q2 fractions in 0.1 mM TC3 solution. A scheme involving successive monomer/Q1 and Q1/Q2 equilibriums instead of parallel reactions could be considered. However, one may note that this alternative model would involve duplex sliding without tetramer disruption from structure Q1 to Q2 and that such rearrangement is topologically impossible.
Effect of the oligonucleotide concentration on the association kinetics of TC3
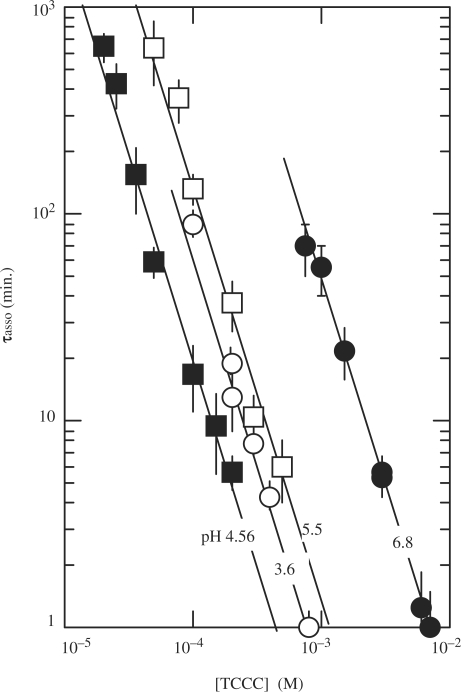
Kinetic experiments performed versus the oligonucleotide concentration (Figure 4) show that the half association time of TC3 varies as the power of −2 of the oligonucleotide concentration and thus establish that according to Equation (1) the order for tetramer formation is 3.
Figure 4.
Half formation time of the fully intercalated [TC3]4 tetramer versus the oligonucleotide concentration at 0°C pH 6.8 (black circles), 5.5 (open squares), 4.56 (black squares) and 3.6 (open circles). The lines drawn through the data points show that the half reaction time increases as power of −2 of the oligonucleotide concentration and therefore establish that the reaction order is 3.
Effect of pH on the association kinetics of TC3
As it may be expected for a reaction involving association of neutral and protonated cytidines into a hemiprotonated structure, the half formation time of [TC3]4 depends on pH. The half association times measured vs. the oligonucleotide concentration show that the reaction order is independent of pH.
The plot of the half association time, τasso(pH), measured in 10−4 M TC3 solution versus pH (Figure 2) shows that 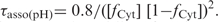 Where
Where 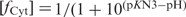 is the neutral cytidine fractions and [1-fCyt] is the protonated cytidine fractions.
is the neutral cytidine fractions and [1-fCyt] is the protonated cytidine fractions.
Effect of the C stretch length on the i-motif formation kinetics
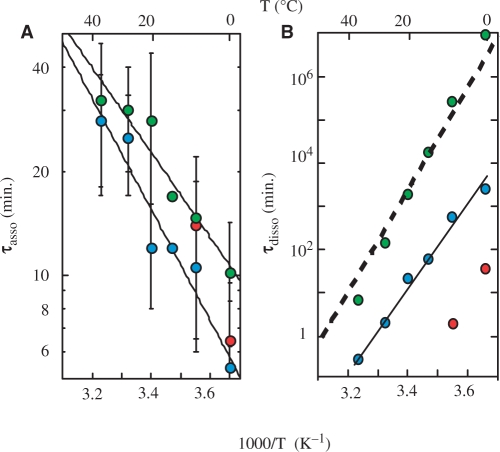
The half association times of the TCn monomers measured in 10−4 M solutions or extrapolated to 10−4 M from measurements performed at higher concentrations (2 × 10−4 to 1.2 × 10−3 M) are displayed in Figure 5 together with the dissociation times and the reduced dissociation constants of the fully intercalated (TCn)4 tetramers. The half association times increase as a function of temperature with negative activation energies ranging from -306 kJ\M for TC5 to -197 kJ\M for TC3. The comparison of the association times of TC3, TC4 and TC5 shows that they decrease by a factor of about 10 for each residue added to the C stretch.
Figure 5.
Effect of temperature on the formation and dissociation times and on the stability of [TC3]4 (left), [TC4]4 (center) and [TC5]4 (right) at pH 5.1. Upper panels: half formation times (open circles) and dissociation times (black circle) of the fully intercalated tetramers. The half formation times were measured in 10−4 M oligonucleotide solutions or extrapolated at 10−4 M from values measured at higher oligonucleotide concentrations. Lower panels: reduced dissociation constants derived from the monomer and tetramer fractions at equilibrium (black squares).
The reduced dissociation constants displayed in the lower panels of Figure 5 were obtained from the monomer and fully intercalated tetramer concentrations measured on the NMR spectra recorded at equilibrium. Fi measurements yield a redundant information that may be independently derived from the association and dissociation times. Nevertheless, the comparison of the Fi values measured at equilibrium with the association and dissociation times provides a good control of the coherence of the equilibrium and kinetics measurements by showing that, according to the Fi definition [Equations (2) and (3)], the reduced dissociation constants measured at equilibrium in 10−4 M [TCn] solutions are equal or close to 10−4/2 at the temperature for which the half association and dissociation times are equal.
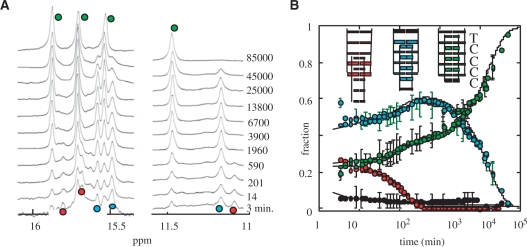
As in the case of TC3 (Figure 3), the spectra recorded during TC4 (Figure 6) and TC5 (Figure S1) association reveals the accumulation of several multimers at the beginning of the kinetics. The evolution of the different species was measured as a function of the time in 0.07, 1.2 and 2 mM oligonucleotide solutions from 0 to 25°C for TC4 and up to 40°C for TC5.
Figure 6.
Association kinetics of TC4 into i-motif structures in 1.2 mM solution at 0°C, pH 5.1. (A) The imino proton region of spectra collected as a function of the time shows the formation of three tetramers. The markers of the fully intercalated tetramer are labeled by green circles. The imino protons peaks labeled by blue circles are tentatively assigned to the species with an empty intercalation site between T1 and C2 and those labeled by red dot to the species whose T1-C2 and C2-C3 intercalation sites are empty. (B) Evolution of the fractions of each species as a function of the time. The monomer fraction (black circle) was determined by the intensity of monomer methyl peak. The full lines fitting the evolution of each species were computed according to a system of parallel reaction between the monomer and each tetramer. The tetramer formation and dissociation times used in the simulation are displayed in Figure 7.
At 0°C, three multimers are observed during TC4 association and at least four in TC5 solution. The gel filtration chromatograms of aliquots took off the NMR tubes during the association kinetics of TC4 and TC5 show that all the multimers have the same elution time than the fully intercalated tetramers and therefore indicate that they correspond to tetramers differing by their intercalation topologies. The signal of the tetramers with partial intercalation reach a maximum value and then slowly decreases toward a value too small to be measured on the last spectra recorded.
We have shown (Figure 1) that the lifetimes of the fully intercalated (TCn)4 tetramers increase strongly with the number of C•C+ pairs. Considering that the lifetimes of the TC4 and TC5 tetramers with incomplete intercalation topology should increase similarly, we tentatively identified the different tetramers by assuming that the number of intercalated C•C+ pairs increase as their lifetimes (cf. Figures 6 and S1).
Numerical simulations of the evolution versus time of the monomer and tetramer fractions were performed according to a model involving parallel reactions between the monomer and each tetramer as described above for [TC3]4 formation [Equation (4)]. The simulations require evaluation of the half formation and dissociation times of each tetramer. The lifetimes of the fully intercalated tetramers were determined from the values displayed in Figure 1. The half formation time of each tetramer and the lifetimes of the non-fully intercalated species were adjusted to get the best fit of the evolution versus time of the monomer and tetramer fractions.
The parameters derived from the numerical simulations of the evolution of the different species observed during [TC4]4 and [TC5]4 formation (Figure 7 and legend of Figure S1) reveal several interesting properties:
Figure 7.
Formation (A) and dissociation times (B) of the fully and partially intercalated tetramer of TC4 derived from the fit of the evolution of the different species in 1.2 mM TC4 solution at 0°C. The color code is the same than in Figure 6. The tetramer formation times are nearly independent of the intercalation topology. The heavy dashed line shows the lifetimes measured (Figure 1) for the fully intercalated TC4 tetramer.
(i) the half formation times of the fully and partially intercalated tetramers are comparable. (ii) In line with the data displayed in Figure 4, the half formation time of the tetramers with full or partial intercalation topologies varies as the power of −2 of the strand concentration and indicates a formation order of three for all the tetramers. (iii) The lifetimes of the partially intercalated tetramers are much more shorter than those of the fully intercalated specie. About 3 years at 0°C, for example, for the fully intercalated (TC4)4 tetramer, 1 day and 50 min for the two other tetramers formed by TC4 (Figure 7B).
Search for a parallel duplex with hemiprotonated C•C+
The i-motif is an extremely stable structure formed by two intercalated duplexes but it is remarkable that the isolated hemiprotonated duplex is never observed. One may imagine that the tetramer is much more stable than the dimer, or that the dimer is in fast exchange with the monomer in the NMR time scale. To examine this point, we synthesized a sequence that can form a hemiprotonated duplex but that cannot be incorporated into an i-motif structure. The (OmC)3C3 sequence contains a C-rich tract of three cytidines with three 2′-O-methy cytidines at the 5′-end. In the i-motif narrow groove, the cytidine H2′ are in close contact at the steps where the intercalated bases are stacked by their faces oriented in the 3′-direction. For this reason, the O-methyl group hinders 2′-OmC intercalation into i-motif structures (17). However, the methyl group at the C2′ position of OmC should not prevent the formation of hemiprotonated OmC•OmC+ pairs. Hence, we expected that intercalation of the C3 tract of (OmC)3C3 would form an i-motif core similar to that of [dC3]4 (14), with two parallel dangling OmC strands at each end and that the OmC, maintained by the i-motif core a in favorable position, would form OmC•OmC+ pairs.
As predicted, the 1D, NOESY and TOCSY NMR spectra of (OmC)3C3 shows the formation of a symmetrical i-motif structure around pH 6 at 0°C. The structure includes three C•C+ pairs whose lifetimes, derived from imino proton exchange measurements, are in the range of 10 ms for the outer C4•C4+ pair to 1 min for the inner C5•C5+ pairs. The amino protons chemical shifts provide a good marker of cytidine protonation (18). Between pH 4 and 6, the H-bonded and exposed amino protons of C4, C5 and C6 are close to 9.5 and 8.5 p.p.m., respectively, as this is always observed for the hemiprotonated pairs of i-motif structures. In contrast, the chemical shifts of the OmC amino protons vary with pH as those of the cytidine monomer (18) indicating that their half titration pH is close to 4.4, the cytidine pK. This, together with the absence of OmC imino protons on the NMR spectrum of (OmC)3C3, shows clearly that the OmC do not form stable OmC•OmC+ pairs. We therefore conclude that it is unrealistic to imagine a stable structure formed by association of two parallel C-rich strand. The origin of the labile character of parallel hemiprotonated C-rich duplexes is not clear. Examination of the balance of hydration, stacking and base-pairing interactions should help to solve this problem.
DISCUSSION
Two salient points revealed by the formation kinetics of i-motif tetramers provide indications about the i-motif formation pathway: (i) in all the pH and temperature conditions tested, the [TCn]4 half formation time varies as the power −2 of the oligonucleotide concentration (Figure 4) indicating that strand association into i-motif tetramer is a third-order reaction. (ii) TCn association results in the parallel formation of several tetramers differing by the number of intercalated C•C+ pairs. It may be noted that with the TCn series, (n − 1) tetrameric species are observed out of n possible intercalation topologies. Considering that the structures with only two mutually intercalated pairs should be hard to detect due to short lifetimes, it is possible that all the intercalation topologies are formed at comparable rate.
The i-motif formation pathway
It may be fist noticed that the observation of a third-order reaction for i-motif tetramer formation is inconsistent with a process involving association of two partially preformed duplexes. If one supposes that duplex formation and duplex association are both rate limiting, tetramer formation should be a fourth-order reaction. If the time required for duplex association is shorter than the lifetime duplex, duplex association should be nonlimiting and the reaction order should be 2. But the reaction order expected for a process involving association of two duplexes cannot be 3.
The reaction order expected for the sequential addition of four TCn monomers (M) via intermediate duplex (D) and triplex species (T) should be 4 if each step is rate limiting. However, the reaction order may be 3 if either triplex or quadruplex formation is not rate limiting.
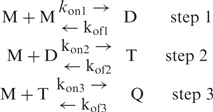 |
5 |
Triplex formation should be nonlimiting if M intercalation into D (Step 2) is fast by comparison with the duplex lifetime. Similarly, quadruplex formation at Step 3 should be nonlimiting if M association to T is faster than T dissociation. Considering that Steps 2 and 3 require both intercalation of a TCn strand into undetectable preformed structures, it is hard to decide which of these steps is nonlimiting. This problem could be investigated by a molecular dynamic analysis of the relative stability of the intermediate D and T species, a method already used to investigate the potential intermediate species in the G-quadruplex formation pathway (19). However, the absence of spectroscopic indications about the presence of a duplex species and the study of (OmC)3C3 (see above) suggests that D is in fast pre-equilibrium with the monomer at the first step of the association pathway and that this is more likely the association of M to T (Step 3) that is the nonlimiting step.
The effect of pH on the TCn association times reflects the fact that i-motif formation requires association of neutral and protonated cytidines. For this reason, the relevant variable in the expression of the tetramer formation rate is not the oligonucleotide concentration but the product of the oligonucleotide concentration by [(fcyt) (1 − fcyt)] the neutral and protonated cytidine fractions (Figure 2).
The factors that shift the equilibrium of Step 1 toward D should enhance the i-motif formation rate. This may account for the acceleration of the i-motif formation rate at low temperature. In addition, the increase of the TCn association rates with the cytidine number may be related to the enhancement of the duplex stability with the length of the C tract.
The parameters derived from the fit of the evolution of the fully and partially intercalated tetramers show that their formation rates are comparable. According to the formation pathway described just above, this indicates that the intercalation rates of M into the D and T intermediate species (Steps 2 and 3) are dependent weakly on the number of intercalated cytidines. This observation suggests that strand intercalation is controlled by a nucleation step, for instance intercalation of a single cytidine in the preformed duplex.
The similarity of the formation rates of tetramers with full or partial intercalation shows that the equilibrium fraction of each species is proportional to their lifetimes and therefore that the predominance of the fully intercalated species at equilibrium is related to its long lifetime. This observation cannot be generalized since many examples have been reported in the literature showing the coexistence of two intercalation topologies at equilibrium (13,20,21).
Comparison with the formation pathway of G-quaduplexes
The stand arrangement in G- and C-rich DNA tetrameric structures are completely different, nevertheless the formation pathways of both structures exhibit obvious similarities.
As shown in a recent NMR study, G-quadruplex formation is a third-order reaction in conditions favorable to the stability of G-rich multi-stranded structures (i.e. high K+ or Na+ concentration and low temperature), but the reaction order is 4 at low salt concentration, in the presence of Li+ and at high temperature (8). On the basis of these observations, it has been assumed that [TGnT]4 formation results of the step by step association of four TGnT strands according to a reaction scheme similar to that used above to describe i-motif formation [Equation (5)]. To explain that G-quadruplex formation may be either a third- or fourth-order reaction, it was conjectured that Step 3 is nonlimiting at high NaCl concentration or at low temperature in experimental conditions showing a third-reaction order and that the shorter triplex lifetime at low NaCl concentration or high temperature results in fourth-reaction order.
The i-motif formation kinetics shows the parallel formation of incompletely and fully intercalated tetramers. Similarly, parallel reactions to that leading to the formation of the thermodynamically stable [TGnT]4 quadruplexes result in the formation of mismatched quadruplexes that are kinetically trapped. Due to the extraordinary long G-quadruplex lifetimes, these species may be trapped during a time so long that formation of the canonical tetramer is practically inaccessible.
The half formation times of i-motif tetramers and of the G-quadruplexes decrease similarly with temperature and the reduction of the [TGnT]4 half formation times with the G number (14,22) is comparable to that observed for the i-motif half formation times with the C number (Figure 5). In all the cases, we assume that the acceleration of the association rate at low temperature and with the number of residue is related to the stabilization of the D and T intermediate species.
i-motif and i-motif nanowires formation pathways
The thymidine at the 5′-end of the TCn oligonucleotides orients certainly duplex formation at the first and third steps of the pathway described above [Equation (5)] by avoiding unfavorable T-C mismatches and slipped strands. Nevertheless, we observe that several intercalation topologies of the TCn oligonucleotides are formed. It is very likely that association of Cn homopolymers should result in the formation of a larger number of species, in particular in tetramer with a core of C•C+ pairs with dangling strands of nonpaired C residues either at the 3′- or the 5′-end of the i-motif core.
A recent study shows that oligonucleotides such as C7 can associate into long linear nanowire structures and suggests that nanowire formation results of the association of incompletely intercalated [dC7]4 structures (23). It is clear that the thymidine at the 5′-end of the sequences used in our kinetic study prevents formation of such structures, but the observation that fully and incompletely intercalated tetramers are formed at comparable rate provides arguments in favor of the mechanism postulated for nanowire formation. Considering the results of our kinetics study, it seems reasonable to assume that short lived Cn i-motif tetramers with incomplete intercalation could associate into long-lived i-motif structures by mutual intercalation of their dangling non-intercalated cytidines. A good knowledge of the formation kinetics of the i-motif structure is certainly a prerequisite condition to the understanding of the formation pathway of i-motif nanowire.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gehring K, Leroy J-L, Guéron M. A tetrameric DNA structure with protonated cytidine-cytidine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 2.Patel D, Bouaziz S, Kettani A, Wang Y. Structure of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centomeric and triplet repeat disease DNA sequences. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. Oxford University Press; 1999. pp. 389–454. [Google Scholar]
- 3.Cai L, Chen L, Raghavan S, Ratliff R, Moyzis R, Rich A. Intercalated cytosine motif and novel adenine clusters in the crystal structure of the tetrahymena telomere. Nucleic Acids Res. 1998;26:4696–4705. doi: 10.1093/nar/26.20.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leroy JL, Guéron M. Solution structure of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. doi: 10.1016/s0969-2126(01)00138-1. [DOI] [PubMed] [Google Scholar]
- 5.Nonin S, Phan AT, Leroy J-L. Solution structure and base-pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a non canonical structure with possible role in chromosome stability. Structure. 1997;5:1231–1246. doi: 10.1016/s0969-2126(97)00273-6. [DOI] [PubMed] [Google Scholar]
- 6.Han X, Leroy J-L, Guéron M. An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCTTTCCTTTACCTTTCC) J. Mol. Biol. 1998;278:949–965. doi: 10.1006/jmbi.1998.1740. [DOI] [PubMed] [Google Scholar]
- 7.Phan AT, Leroy J-L, Guéron M. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomer. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 8.Bardin C, Leroy J-L. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008;36:477–488. doi: 10.1093/nar/gkm1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Zeng Z-X, Kan Z-Y, Hao Y-H, Tan Z. The folding and unfolding kinetics of the i-motif structure formed by the C-rich strand of human telomere DNA. Chembiochem. 2005;6:1957–1960. doi: 10.1002/cbic.200500175. [DOI] [PubMed] [Google Scholar]
- 10.Canalia M, Leroy J-L. Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC) Nucleic Acids Res. 2005;33:5471–5481. doi: 10.1093/nar/gki843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mergny JL, Lacroix L. Kinetics and thermodynamics of i-DNA formation: phosphodiester versus modified oligodeoxynucleotide. Nucleic Acids Res. 1998;26:4797–4803. doi: 10.1093/nar/26.21.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mergny JL. Fluorescence energy transfer as a probe for tetraplex formation: the i-motif. Biochemistry. 1999;38:1573–1581. doi: 10.1021/bi982208r. [DOI] [PubMed] [Google Scholar]
- 13.Kanaori K, Maeda A, Kanehara H, Tajima K, Makino K. 1H nuclear magnetic resonance study on equilibrium between two four-stranded solution conformations of short d(CnT) Biochemistry. 1998;37:12979–12986. doi: 10.1021/bi980492g. [DOI] [PubMed] [Google Scholar]
- 14.Leroy JL, Snoussi K, Guéron M. Investigation of the CH-O hydrogen bonds in the DNA i-motif via the equilibrium between alternative topologies. Magn. Reson. Chem. 2001;39:S171–S176. [Google Scholar]
- 15.Plateau P, Guéron M. Solvent signal suppression in NMR. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 16.Leroy J-L, Nonin S, Han X, Phan AT, Guéron M. Switching and looping in i-motif structures. In: Sarma RH, Sarma MH, editors. Structure and Motion and Expression of Biological Macromolecules. Albany, NY: Adenine Press; 1998. pp. 49–62. [Google Scholar]
- 17.Collin D, Gehring K. Stability of chimeric DNA/RNA cytosine tetrads: implications for i-motif formation by RNA. J. Am. Chem. Soc. 1998;120:4069–4072. [Google Scholar]
- 18.Raska M. Mononucleotides in aqueous solution. Proton magnetic resonance studies of amino groups. Biochemistry. 1974;13:4616–4622. doi: 10.1021/bi00719a023. [DOI] [PubMed] [Google Scholar]
- 19.Stefl R, Cheatham TE, Spackova N, Fadrna E, Berger I, Koca J, Sponer J. Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates. Biophys. J. 2003;85:1787–1804. doi: 10.1016/S0006-3495(03)74608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanaori K, Shibayama N, Gohda K, Tajima K, Makino K. Multiple four-stranded conformations of human telomere sequence d(CCCTAA) in solution. Nucleic Acids Res. 2001;29:831–840. doi: 10.1093/nar/29.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esmaili N, Leroy J-L. i-motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats. Nucleic Acids Res. 2005;33:213–224. doi: 10.1093/nar/gki160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saccà B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplexes-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghodke HB, Krishnan R, Vignesh K, Kumar P, Narayana C, Krishnan Y. The I-tetraplex building block: rational design and controlled fabrication of robust 1D DNA Scaffolds through non-Watson-Crick interactions. Angew. Chem. Int. Ed. 2007;46:2646–2649. doi: 10.1002/anie.200604461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.