Abstract
Androgen receptor (AR) is a ligand-controlled transcription factor frequently deregulated in prostate carcinomas. Since there is scarce information on the action of AR on the chromatin level, we have elucidated the molecular mechanisms underlying the androgen-dependent regulation of immunophilin FKBP51 in prostate cancer cells. In comparison to the canonical AR target PSA, FKBP51 is more rapidly and strongly induced by androgen, with the regulation occurring merely at the transcriptional level. FKBP51 locus harbors 13 in silico-predicted androgen response elements (AREs), with most of them located downstream from transcription start site (TSS) and capable of binding AR in vitro. Chromatin immunoprecipitation assays in VCaP and LNCaP prostate cancer cells indicate that activation of the locus by the AR relies on four major intronic sites, with the compound ARE-containing sites ≥90 kb downstream from the TSS playing critical roles. Binding of agonist-loaded AR onto these sites in vivo was accompanied with significant recruitment of RNA polymerase II and BRM-containing chromatin remodeling complexes to the FKBP51 locus, which resulted in changes in the histone density of the locus. Our results indicate that very distal AREs act as genuine and robust enhancers, highlighting the importance of long-range regulation of transcription by the AR.
INTRODUCTION
Androgen receptor (AR) is a hormone-inducible transcription factor that belongs to the nuclear receptor superfamily (1). Many of the genes regulated by androgens and AR program the development and maintenance of the male phenotype and sexual characteristics. AR also plays a critical role in controlling the malignant growth of prostate tumors, and androgen ablation is a standard therapy for prostate cancer. However, the therapy usually eventually fails and cancer turns into a lethal hormone-refractory state that overexpresses AR and is thereby sensitized to low androgen levels (2,3).
Transcription is regulated at many levels. DNA sequence forms platforms for binding of transcription factors that can either activate or attenuate transcription of a given gene. Androgen receptor's binding sites in DNA, androgen response elements (AREs), are essential for AR-mediated transcriptional activation, but not necessary for transcriptional repression by the receptor (4,5). However, not all AREs that are capable of binding AR in vitro necessarily function in vivo, since epigenetic regulation of chromatin structure and packaging can prevent their accessibility. Major epigenetic mechanisms that regulate chromatin structure include modifications of nucleosomes and DNA methylation (6,7).
Activation of genes by the AR requires, in addition to basal transcription machinery and RNA polymerase II, transcriptional coregulator proteins. Many coregulatory proteins harbor or recruit enzymatic activities that covalently modify histones (8). Agonist-bound nuclear receptors can interact with coactivator proteins linked to histone acetyltransferase activity, which acetylates specific lysine (K) residues in nucleosomal histones. Also histone methylation has recently been implicated in nuclear receptor signaling (9). In contrast to histone acetylation that is connected to active transcription, methylation of histones by K- or arginine (R)-specific activities have been linked to both activation and repression of transcription. The effect of these histone methylations on gene activity depends on the identity of the amino acid residue in a specific histone modified. For example, methylation of histone H3 at K9 and K27 are repressive modifications (transcriptional ‘off’ state), whereas methylation of H3 at K4 and K36 are needed for active transcription (transcriptional ‘on’ state) (6,10).
Nuclear receptors also attract chromatin remodeling complexes which utilize the energy of ATP to alter the histone-DNA association. Consequences of remodeling by SWI/SNF complexes include nucleosome sliding, alterations in nucleosome structure and histone octamer transfer (11–13), which can influence the accessibility of transcription factor binding sites. SWI/SNF complexes contain either BRM (SMARCA2) or BRG1 (SMARCA4) as a central ATPase subunit (14). It is not totally clear whether these ATPases are fully replaceable with each other or if there is some tissue or gene specificity in their function (15).
Perturbations in the transcriptional programs of AR are important, but poorly understood, events in prostate cancer. Knowledge of the AR action under genuine chromatin milieu is therefore essential for better understanding of the molecular mechanisms underlying prostate cancer development. However, regulation of only few AR target loci has thus far been characterized in a detailed fashion at the chromatin level. Most studies have been focusing on the prostate specific antigen (PSA) (16–20) of which protein product's serum concentration is used in prostate cancer diagnostics (21). Even though PSA has been studied as an example of ‘canonical’ AR target gene in prostate cells, it does not appear to be very sensitive to androgen levels in vivo, as PSA mRNA levels were not significantly suppressed in prostatectomy patients in response to androgen deprivation therapy (22). Compared to PSA mRNA, for example FKBP51 (FKBP5) mRNA much better reflects the androgen concentration and activity in human prostate (22).
In the present study, we have focused on the androgen regulation of FKBP51 in VCaP cells derived from a hormone-refractory prostate cancer (23). Our results show that in comparison to PSA, FKBP51 mRNA is more robustly induced by androgens, with the regulation occurring merely at the level of transcription, as it is cycloheximide-independent but actinomycin D sensitive. Quantitative chromatin immunoprecipitation scans and enhancer activity analyses indicate that the androgen activation of the FKBP51 locus occurs via several distal intronic enhancers, with the compound AREs located at ∼90 and ∼100 kb from the transcription start site playing the major role. Binding of AR to these distal enhancers is accompanied with chromatin remodeling throughout the locus. In LNCaP cells, that contain less AR than VCaP cells, AR occupancy and induction of the locus in response to androgen were less robust than in VCaP cells, but the pattern of the receptor loading was similar and the same intronic enhancers were important in both prostate cancer cell types. A model for long-range regulation by AR transcription complexes on the FKBP51 locus in vivo is presented.
MATERIALS AND METHODS
Cell culture
VCaP and LNCaP cells were from American Type Culture Collection (ATCC). VCaP cells were maintained in DMEM containing 10% US defined FBS (HyClone, Logan, Utah, USA), 25 U/ml penicillin and 25 µg/ml streptomycin in a 5% CO2 atmosphere at 37°C. LNCaP cells were grown in RPMI 1640 supplemented according to instructions from ATTC and FBS as above. COS-1 cells were maintained in DMEM containing 10% FBS, penicillin (6.25 U/ml) and streptomycin (6.25 µg/ml).
DNA constructs
Selected regions of FKBP51 were PCR cloned from human genomic DNA using Phusion DNA polymerase (Finnzymes, Espoo, Finland) and subcloned into the TATA box-containing pGL3-basic (Promega, Madison, WI, USA). Primers used were the same as listed for ChIP except for region 12 the forward primer was 5′-TCTTGCCTCCAACACTGCTG-3′ (Supplementary Table S1). All sequences were verified by DNA sequencing using the ALFexpress system. pCMV encoding β-galactosidase was from Clontech (Mountain View, CA, USA).
Antibodies
Primary antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA): AR (sc-816), BRG1 (sc-10768), TRAP220 (sc-8998), FKBP51 (sc-13983), GADPH (sc-25778), and normal rabbit IgG (sc-2027); from Abcam (Cambridge, UK): H3 (ab1791), H3K4me3 (ab8580) and H3K36me3 (ab9050); from Covance (Emeryville, CA, USA): PolII (MMS-126R); from UpState (Lake Placid, NY, USA): H3K9,K14ac (06-599); and from BD Biosciences (San Jose, CA, USA): BRM (610389).
Isolation of RNA and quantitative real-time RT–PCR (qRT-PCR)
VCaP and LNCaP cells were seeded onto 6-well plates (8 × 105 and 3.3 × 105 cells/well, respectively) and grown 36 h in transfection medium [DMEM containing 2.5% charcoal-stripped FBS (CCS-FBS)] devoid of steroids. Subsequently, cells were treated with or without synthetic androgen agonist R1881 (Perkin Elmer Inc., Waltham, MA, USA), 5α-dihydrotestosterone (Steraloids Inc., Newport, RI, USA) or bicalutamide (Zeneca Pharmaceuticals, Macclesfield, UK) as indicated. Total RNA was extracted using TRIzol® Reagent (Invitrogen Lifetechnologies, Carlsbad, CA, USA) and converted to cDNA using Transcriptor First Strand cDNA synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany) following manufacturer's instructions. cDNA was used as a template in qRT-PCR, which was carried out using MX3000P real-Time PCR System (Stratagene, La Jolla, CA, USA), FastStart SYBR Green Master (Roche) and specific primers for FKBP51, PSA, TMPRSS2 and GAPDH (Supplementary Table S1). Analyzed GAPDH mRNA levels were used to normalize the amounts of total RNA between the samples. Fold changes were calculated using the formula 2−(ΔΔCt), where ΔΔCt is ΔCt(R1881)–ΔCt(EtOH), ΔCt is Ct(gene X)–Ct(GAPDH) and Ct is the cycle at which the threshold is crossed.
Electrophoretic mobility shift assay (EMSA)
For production of AR for EMSAs, COS-1 cells were transfected either with empty pSG5 or pSG5-hAR and cell extracts were prepared as described (24). Twelve micrograms of cell extract was used for 20 µl EMSA reaction. The mock extract was treated with ethanol and AR extract with 10 nM R1881. 32P-labeled ds-oligomer probe was added and protein–DNA complexes were allowed to form for 1 h (25). The intronic ARE 5′-AGTACGtgaTGTTCT-3′ (core) of rat prostatein peptide C3(1) was used as a positive control. Probes contained the 27 bp native sequences (Supplementary Table S1). The complexes were separated on 4% non-denaturing PAGE. The gels were dried and detected using phosphoimager (FLA3000, Fuji, Japan).
Reporter gene assay (RGA)
VCaP and LNCaP cells were seeded onto 12-well plates (3.2 × 105 and 1.4 × 105 cells/well, respectively) and grown 4 h in transfection medium. The cells were transfected with reporter gene constructs (1.8 µg pLUC and 0.2 µg pCMVβ/well) using jetPEI™ (Polyplus-transfection SA, Illkirch, France) transfection reagent. COS-1 cells were seeded onto 12-well plates (1.4 × 105 cells/well) and grown 4 h in transfection medium. The cells were transfected with reporter gene constructs (0.46 µg pLUC, 0.02 µg pCMVβ/well and 0.02 µg pSG5-hAR) using TransIT–LT1 (Mirus Bio Corporation, Madison, WI, USA) transfection reagent. One day after transfection, the cells were grown for 16 h with either vehicle (ethanol) or 1 nM R1881 and lysed in Reporter Lysis Buffer (Promega, Madison, WI, USA) and LUC activity was measured with Luciferase Assay System (Promega) using Luminoskan Ascent® (Thermo Electron, Helsinki, Finland) luminometer and β-galactosidase activity as described (25).
Chromatin immunoprecipitation assay (ChIP)
VCaP and LNCaP cells were seeded at ∼70% confluence onto 75 cm2 bottles and allowed to grow in steroid-depleted transfection medium for 48 h prior to ChIP. The experiments were performed essentially as previously described (26), except for short-term assays (≤2 h), the cells were treated with 2.5 µM α-amanitin (Sigma-Aldrich, St Louis, MO, USA) for 2 h to further synchronize the cell population (27) prior to addition of androgen. α-Amanitin was removed by washing twice with PBS, and R1881 was added to the medium for indicated times. Specific primers for different regions are listed in Supplementary Table S1. Quantitative PCR analyses were carried out with FastStart SYBR Green Master and MX3000P Real-Time PCR System. Results were calculated using the formula E−(ΔCt) × 10, where E (efficiency of target amplification) is a coefficient of DNA amplification by one PCR cycle for a particular primer pair and ΔCt is Ct(ChIP-template)–Ct(Input). In Figures 5 and 6, results were presented as fold over the value of normal rabbit IgG-precipitated samples.
Figure 5.
Androgen-induced recruitment of BRM-containing complexes onto the FKBP51 locus. VCaP cells were treated with R1881 or vehicle as in Figure 4 and ChIP assays were performed using antibodies against BRM (A) or BRG1 (B) followed by quantitative PCR analysis. Results are shown as fold over the normal rabbit IgG-precipitated samples and represent the mean ± SD of three experiments.
Figure 6.
Effect of androgen and antiandrogen on FKBP51 transcription and AR loading in VCaP and LNCaP cells. (A) Accumulation of FKBP51 mRNA in response to exposure of VCaP and LNCaP cell to R1881 (1 nM) or bicalutamide (BIC, 10 μM) alone or their combination for 12 h. Messenger RNAs were analyzed by qRT-PCR analysis as described in Figure 1. (B) Immunoblotting analysis of AR protein levels in VCaP and LNCaP cells in the presence and absence of BIC for 2 or 12 h as indicated. (C) Loading of AR onto the FKBP51 locus in response to androgen or antiandrogen exposure for 2 h in VCaP cells and LNCaP cells. Cells were treated with vehicle, R1881 or BIC (10 μM) as in Figure 4 and ChIP assays were performed using antibody against AR followed by quantitative PCR analysis. Results are shown as percentages of the input samples and represent the mean ± SD of three experiments.
Immunoblotting
Cell monolayers were washed with ice-cold TBS and harvested in TBS. Cell pellets were suspended in SDS-PAGE sample buffer and lysed by sonication and lysed by sonication 2 × 10 s. Samples were heated for 5 min at 95°C and separated on 7.5% SDS-PAGE gels. Proteins were transferred onto nitrocellulose membranes and visualized by indicated antibody and horseradish peroxidase-conjugated anti-rabbit antibody using the enhanced chemiluminescence detection reagents according to the manufacturer's instructions (Pierce). For protein quantification, the proteins were visualized by goat anti-rabbit IgG DyLight™ 680 conjugated secondary antibody. Quantification of the immunoblots was performed by scanning with Li-COR Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA) at 700 nm channel according to manufacturer's instructions. AR and FKBP51 protein amounts were normalized with GADPH amounts (assessed with a specific antibody) in each sample.
RESULTS
Rapid and robust induction of FKBP51 transcription by androgen
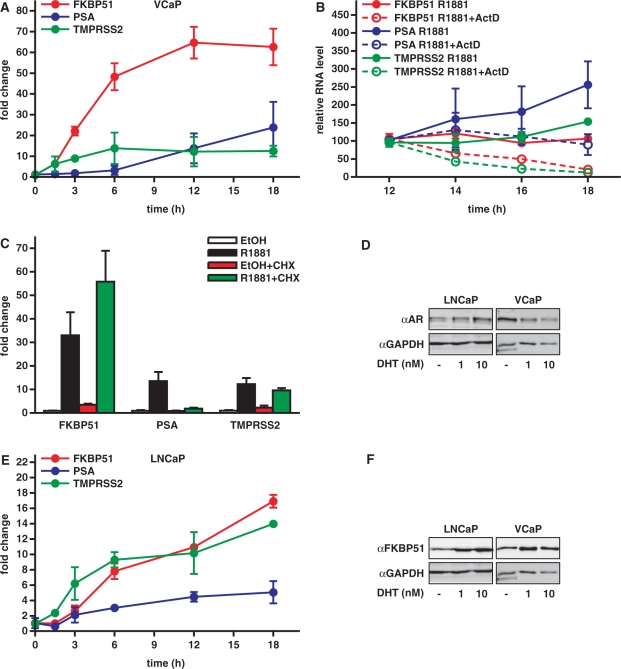
To study the androgen regulation of FKBP51 of human prostate VCaP cancer cells, we isolated RNA at different periods after addition of androgen (R1881; 0, 1.5, 3, 6, 12 and 18 h) and performed qRT-PCR analysis. As shown in Figure 1A, FKBP51 mRNA is rapidly and strongly induced after androgen exposure, with the induction being >20-fold at 3 h and peaking (≥60-fold) at 12 h. Compared to the FKBP51, the induction of PSA was much slower and less impressive. Another AR target gene TMPRSS2 showed similar induction kinetics as FKBP51, but its overall induction remained modest (∼10-fold) in comparison to FKBP51. Since the FKBP51 mRNA persisted at a high level after 12 h, we checked whether that was mainly due to new mRNA synthesis or by mRNA stabilization. To that end, we terminated mRNA synthesis by adding actinomycin D to the cells at 12 h after the androgen exposure and monitored the amount of FKBP51 mRNA at further time points. Decline of the FKBP51 mRNA level after addition of actinomycin D indicates that the AR-mediated transcriptional response continues for prolonged times, at least until 36 h after androgen exposure (Figure 1B, data not shown). To confirm that the gene is a direct transcriptional target for AR, the cells were treated with or without translation inhibitor cycloheximide (CHX) and qRT-PCR analyses were performed. CHX did not inhibit the androgen induction of FKBP51 (but actually enhanced it; p < 0.001) or TMPRSS2 mRNA (p > 0.05), whereas it nearly abolished the androgen induction of PSA mRNA (p < 0.05) (Figure 1C). These results indicate that, in contrast to PSA, the androgen regulation of FKBP51 and that of TMPRSS2 does not require de novo protein synthesis. The effect of CHX on the PSA mRNA is in line with previous findings in other prostate cancer cell types (26,28).
Figure 1.
Rapid and robust augmentation of FKBP51 transcription in response to androgen exposure in prostate cancer cells. (A) Accumulation of FKBP51, TMPRSS2 and PSA mRNA in response to androgen treatment in VCaP cells. Cells were treated with 1 nM synthetic androgen R1881 for indicated times and mRNAs were analyzed by qRT-PCR analysis. (B) To test whether the FKBP51 mRNA accumulation after 12 h androgen treatment was caused by new mRNA synthesis or mRNA stabilization, cells were further grown in the presence or absence of actinomycin D (1 μg/ml) for indicated times and qRT-PCR was performed. (C) VCaP cells were treated with vehicle (ethanol, EtOH) or R1881 for 24 h in the presence and absence of translation inhibitor cycloheximide (CHX, 10 μg/ml) and qRT-PCR analyses were performed. Total RNA levels between samples were normalized using mRNA levels of GAPDH. Fold changes were calculated as described in Materials and Methods. Columns represent the mean ± SD of three independent experiments. (D) Immunoblotting analysis of AR protein levels in VCaP and LNCaP cells grown for 18 h with vehicle (EtOH) or androgen (1 or 10 nM 5α-dihydrotestosterone, DHT) as indicated. AR was detected using anti-AR antibody as described in Materials and Methods. (E) Time course of AR target gene mRNA accumulation in LNCaP cells was carried out as in panel A. (F) Immunoblotting analysis of FKBP51 protein levels in VCaP and LNCaP cells cultured in the presence and absence of androgen as in panel D.
We next tested whether the androgen activation of FKBP51 in VCaP cells differs from that in LNCaP cells which are often used as a model of prostate cancer cells. We first compared the AR protein levels in the two cell lines by immunoblotting with anti-AR antibody. In keeping with the amplification of AR in VCaP cells (23), but not in LNCaP cells, quantification of the AR bands showed that VCaP cells contain ∼3–7-fold more AR protein than LNCaP cells, with the difference being dependent on the presence of androgen in the culture medium (Figure 1D). Interestingly, androgen exposure had a reciprocal effect on AR levels in the two cell lines; it repeatedly decreased the amount of the receptor in VCaP cells and increased it in LNCaP cells.
In comparison to VCaP cells, the fold-induction of FKBP51 mRNA by androgen was weaker and it peaked later in LNCaP cells (Figure 1E). Silencing of AR expression in VCaP cells was paralleled by a marked reduction in the androgen-induced accumulation of FKBP51 mRNA (Supplementary Figure S1), supporting the notion that the weaker induction of FKBP51 in LNCaP cells is mainly due to the lower amount of AR in those cells. Moreover, the curve of FKBP51 mRNA accumulation in response to increasing androgen concentrations was shifted to the right compared to the situation in VCaP cells (Supplementary Figure S2). In both prostate cancer cell lines, induction of FKBP51 by androgen was also manifested on the FKBP51 protein level (Figure 1F).
FKBP51 locus harbors several potential AR-binding sites
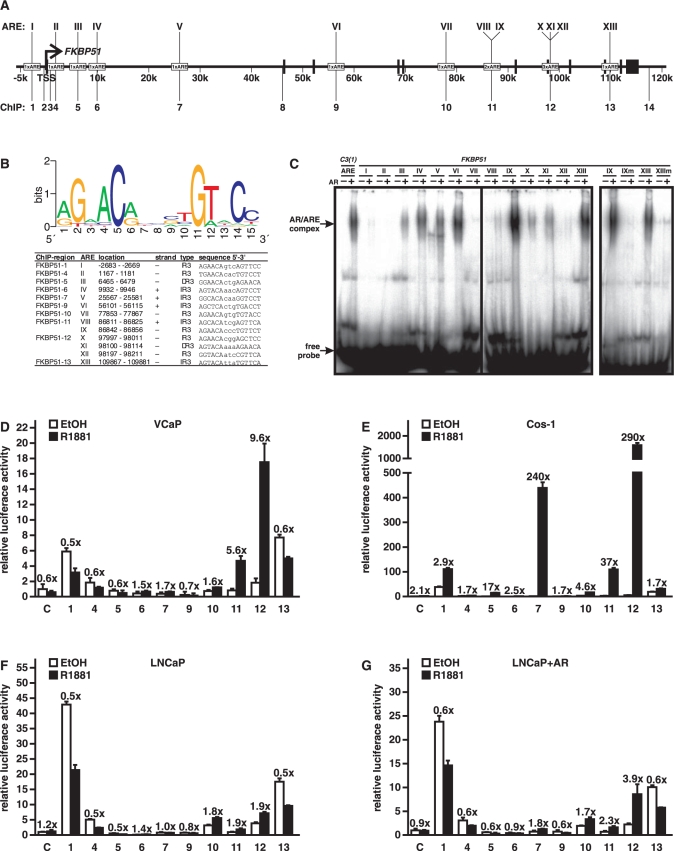
The above data strongly suggest that FKBP51 is directly targeted by AR and mainly at the transcriptional level. Moreover, the robust androgen induction of FKBP51 implies that there may be more than one ARE-containing region within the gene. To identify potential AREs in the FKBP51, we performed in silico analysis from –20 kb upstream of its TSS to 20 kb downstream from its last exon (exon 11) spanning ∼155 kb in chromosome 6 using the CONSITE program (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite/). The score cutoff was adjusted to 80% and the position weight matrix provided by the program was used. After program-based screening, putative AREs were further manually sifted using G at position 2 and C at position 5 in the half-site as the criterion (Figure 2B). This process resulted in identification of 13 putative AREs (AREI–AREXIII, Figure 2B) of which all, excluding AREI, were located within the introns of FKBP51 (Figure 2A). Only two of the AREs, AREIII and AREXI, show characteristics of selective AREs consisting of two hexameric half-sites arranged as a so-called androgen direct repeat separated by a 3-nt spacer (DR3), whereas the other 11 elements resemble 3-nt-spaced inverted repeats of (IR3) of a 5′-TGTTCT-3′-like half-site (29). Interestingly, AREVIII and AREIX in the fifth intron (at ∼87 kb from the TSS) are separated from each other only by 16 nt. Similarly, AREX, AREXI and AREXII are clustered within 215 nt in the 7th intron (at ∼98 kb from the TSS).
Figure 2.
Binding of AR to putative FKBP51 AREs in vitro and their function as enhancers. (A) Schematic locations of the AREs and regions analyzed in reporter and ChIP assays. Bars depict the positions of exons and an arrow indicates the transcription start site. Roman numerals depict the AREs, and Arabic numerals indicate the regions amplified for enhancer analyzes and ChIP assays. (B) Sequences of the potential AREs in FKBP51 gene. (Top) Position weight matrix used for identification of putative AREs. The frequency of nucleotides at each of the 15 positions in the ARE in the matrix is shown. (Lower) List of the in silico-identified AREs, their localization in the FKBP51 gene, sequences and type of ARE. (C) Electrophoretic mobility shift assays were performed in the presence of AR-containing COS-1 cell extracts (AR + ) or extracts from empty vector-transfected cells (AR-), and 32P-labeled dsAREs or mutated (m) versions of AREIX and AREXIII. Protein–DNA complexes were resolved on non-denaturing 4% polyacrylamide gels. The intronic ARE of prostatein peptide C3(1) was used as a positive control. (C, D) Transcriptional activity of the ARE-containing 300-bp FKBP51 fragments was assessed by reporter gene assays. VCaP (D), COS-1 (E), LNCaP (F and G) cells were transfected with TATA-LUC constructs driven by indicated regions of FKBP51 as described in Materials and Methods. For COS-1 (E) and LNCaP + AR (G) analyses, pSG5-hAR (0.02 or 0.2 µg/well, respectively) was cotransfected with the reporter constructs. Cotransfection of pCMVβ and β-galactosidase activity was used for normalization of transfection efficiency. The cells were treated with vehicle (EtOH) or 1 nM R1881 for 16 h before harvesting the cells for reporter analyses. Results are shown as relative LUC activity and fold inductions of androgen-treated samples in the relation to the activity of ethanol-treated samples are shown above the columns. Columns represent the mean ± SD of three independent experiments.
To test the capability of the putative 13 AREs to bind AR in vitro, we performed electrophoretic mobility shift assays with recombinant human AR from COS-1 cells. AR was incubated with dsDNA oligomers containing the 15 bp core element of a given putative ARE surrounded by 6 bp native flanking sequences. The intronic ARE of the rat prostatein peptide C3(1) that binds efficiently AR in vitro was used as a positive control (30). In EMSAs, nine AREs, AREIII–VI, AREVIII–XI, and AREXIII, showed AR–DNA complex formation that was >25% of that of the positive control ARE (Figure 2C and Supplementary Figure S3). EMSAs with mutated oligomers containing A instead of G or C in the 2nd and 5th position of one half-site of AREIX (IXm) and AREXIII (XIIIm) confirmed the specificity of AR–ARE interaction and the importance of both half-sites for the interaction (Figure 3C, on the right). The high portion of the predicted AREs capable of efficiently interacting with the receptor indicates the biochemical relevance of our in silico screen.
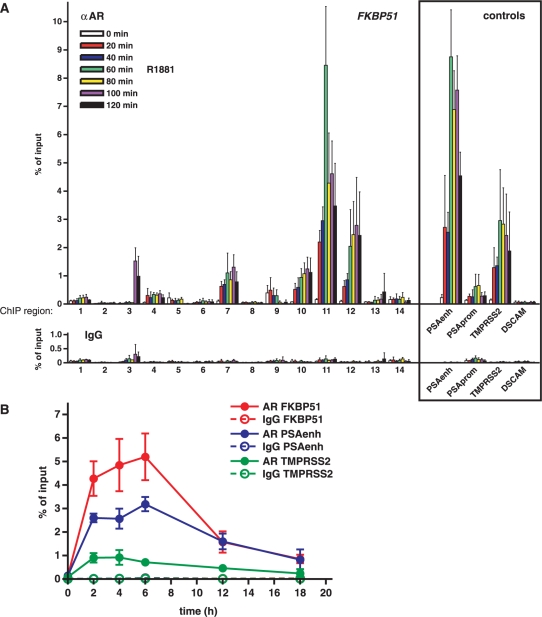
Figure 3.
Spatial and temporal loading of AR onto the FKBP51 locus in response to androgen exposure in VCaP cells. (A) Cells were treated with 1 nM R1881 for indicated times and chromatin immunoprecipitations with anti-AR antibody followed by real-time PCR were carried out as described in Materials and Methods section to investigate the occupancy of AR within 14 different regions of FKBP51 gene. Enhancer and promoter regions of PSA and distal enhancer region of TMPRSS2 served as known positive references, and a region in the middle of DSCAM was as a negative control. ChIP assays with normal IgG monitored nonspecific binding. (B) Unsynchronized VCaP cells were treated with R1881 for indicated times and ChIP samples were analyzed by ChIP with anti-AR or nonspecific IgG. The precipitated DNAs were used as templates for qPCR. Results are shown as percentages of the input samples. Columns represent the mean ± SD of three experiments.
Function of the FKBP51 AREs as enhancers
The above EMSA results suggest that there are several potential AREs within the FKBP51 locus. We next tested their function as androgen-regulated enhancers in prostate cancer cells. Approximately 300-nt regions covering the AREs (cf. Figure 2A) were cloned in front of a TATA-box containing luciferase plasmid. VCaP cells were transfected with the plasmids and treated with or without androgen. Empty TATA-box containing luciferase plasmid served as a negative control. As shown in Figure 2D, region 11 harboring the AREVIII and AREIX and region 12 harboring the AREX, AREXI and AREXII were the only regions conferring significant androgen induction in VCaP cells, with the region 12 showing the strongest activity and androgen induction (∼10-fold). These two FKBP51 regions displayed strong androgen induction also in COS-1 cells cotransfected with an AR expression vector. In addition, region 7 that showed only a modest induction (1.7-fold) in VCaP cells conferred in COS-1 cells a strong androgen induction (Figure 2E). Androgen induction of the regions 11 and 12 was weaker in LNCaP than in VCaP cells (cf. Figure 2D and F). Ectopic expression of AR to some extent strengthened the induction through the region 12 in LNCaP cells (Figure 2G), suggesting that the difference in the effect of androgen between the two prostate cell lines reflects the amount of AR in these cell lines. However, the region 12 harboring three AREs conferred the strongest androgen induction in all studied cell types. Taken together, these results suggest that there are two major distally located enhancers (≥87 kb from the TTS) that function in the androgen regulation of the FKBP51 in prostate cancer cells.
Loading of AR onto FKBP51 chromatin in response to androgen
We next analyzed the interaction of AR with the FKBP51 locus in vivo by performing chromatin immunoprecipitations (ChIP) in VCaP cells with anti-AR antibody followed by real-time PCR analysis. Altogether 14 regions of FKBP51 were scanned. In addition to the regions harboring the in silico-predicted AREs, these assays included four regions without predicted AREs, two on both sides of the TSS (regions 2 and 3), one within first intron (region 8) and one downstream (region 14) the gene (Figure 2A). PSA upstream (∼4 kb) enhancer and promoter, and TMPRSS2 upstream (∼13.5 kb) enhancer regions served as positive controls (20,26), and a region located in the middle of non-androgen responsive DSCAM gene (in chromosome 22) served as a negative control. VCaP cells were exposed to androgen for various time points (0–120 min at 20 min intervals) prior to ChIPs. Loading of AR was clearly detectable onto several regions within the FKBP51 locus at the 20 min time point (Figure 3A). Regions 11 and 12 were the two FKBP51 regions showing the most abundant occupancy of holo-AR, but androgen induced marked enrichment of the receptor (≥10-fold) also onto regions 7 and 10. Binding of holo-AR peaked at the 60 min (region 11) or 80 min time point (regions 7, 10 and 12). Similar kinetics of AR loading was observed with the PSA and TMPRSS2 enhancers. Thus, AR can in vivo interact with several separated sites that are very distal and downstream from the FKBP51 TSS. However, the occupancy level of holo-AR on those sites in chromatin did not directly correlate with their isolated enhancer activity in VCaP cells, in that the region 11 showed less activity than the region 12 as an enhancer and the interaction of AR with regions 7 and 10 in the chromatin was more prominent than expected on the basis of their very weak function as isolated androgen-regulated enhancers. Interestingly, there was also a delayed loading of holo-AR at the 100 min time point at region 3 that does not contain any potential ARE.
Since actinomycin D experiments (Figure 1B) indicate that the FKBP51 is still actively transcribed after 12 h of the androgen addition, we also monitored the loading of AR onto the region 11 at later time points. ChIP assays showed that the amount of AR bound to the FKBP51 decline after 6 h, i.e. earlier than the observed peak of the FKBP51 mRNA accumulation (cf. Figures 3B and 1A). These results suggest that after the initial peaking of AR binding to the FKBP51 and activation of transcription, declining amounts of AR are still sufficient to maintain the maximal androgen-activated transcription of the locus.
Histone marks and density reflect active chromatin structure in FKBP51 locus
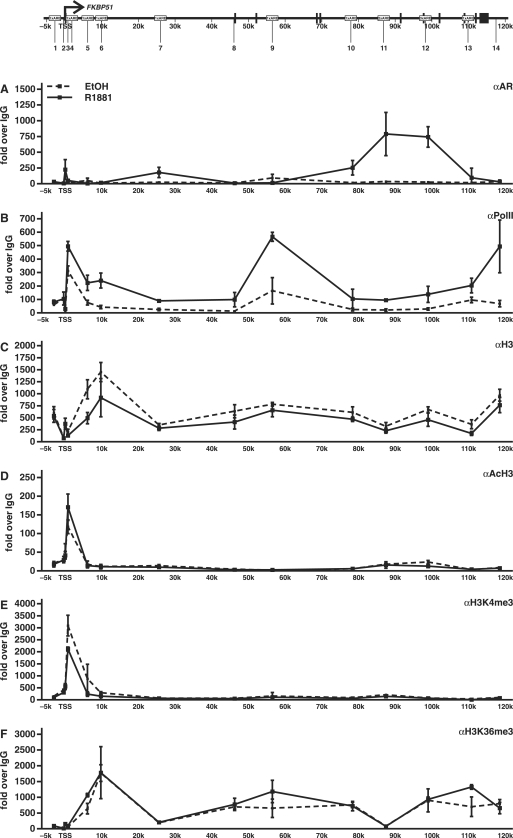
Since the above ChIP assays indicate that most of the AR binding to the FKBP51 locus occurs to sites that are very distal to the TSS, it was of interest to monitor the occupancy and recruitment of RNA polymerase II (PolII) within the locus. In keeping with the augmenting effect of androgen on the FKBP51 transcription, addition of androgen was accompanied with a clear increase in the occupancy of PolII throughout the locus as assessed by ChIP using an antibody against the largest subunit of the PolII in VCaP cells (Figure 4B). The relative occupancy of PolII was higher just downstream from the TSS, in the middle of the locus, and at the 3′ end of the gene than other gene regions. Thus, PolII occupancy does not mirror the loading of AR to the locus, suggesting that chromatin loop formation is involved in the recruitment of PolII by the holo-AR.
Figure 4.
Occupancy of RNA Polymerase II and occurrence of H3 modifications within the FKBP51 locus in VCaP cells. The cells were treated with R1881 or vehicle (EtOH) for 2 h and ChIP assays were performed using antibodies against AR (A), PolII (B), H3 (C), AcH3 (H3K9, K14ac) (D), H3K4me3 (E) or H3K36me3 (F). ChIP samples were used as templates in quantitative PCR. Results are shown as fold over the normal rabbit IgG-precipitated samples and represent the mean ± SD of three experiments.
We next investigated possible changes in chromatin structure of the FKBP51 locus upon androgen induction. Covalent modifications of core histones reflect the chromatin activity, i.e. whether it is in ‘closed’ or ‘open’ conformation. For example, acetylation (ac) of histone 3 K9 and K14 and trimethylation (me3) of both H3K4 and H3K36 are linked to active chromatin conformation (31–34). We scanned the occurrence of the above modifications by using H3 modification specific antibodies in ChIP analyses. As shown in Figure 4D and E, both H3K9,14ac and H3K4me3 levels clearly peaked around and immediately downstream the TSS, whereas H3K36me3 marks were elevated more downstream from the TSS and remained elevated throughout the reading frame of the gene. Since the androgen-induced changes in these histone marks at most sites were modest, we next studied whether androgen induction also brings about changes in histone density. Even though the total amount of H3 in the absence of androgen was similar at most sites in the locus, the occupancy of H3 around the TSS was lower than in other parts of the gene. Interestingly, androgen induction resulted in a decrease in the H3 occupancy at many sites in the locus, with the greatest drop around regions 5 and 6 (Figure 4C). Similar changes were seen in H2B levels, strongly suggesting that the changes in the H3 mirror nucleosome density in general (data not shown).
Androgen-induced recruitment of BRM-containing chromatin-remodeling complexes onto FKBP51 locus
The above changes in the histone density suggest that chromatin remodeling complexes may be involved in the androgen activation of the FKBP51 locus. Since SWI/SNF complexes containing BRM or BRG1 as the major ATPase subunit can catalyze nucleosome sliding and histone transfer (12,13), we investigated whether androgen induction is linked to any changes in the occupancy of BRM- or BRG1-containing SWI/SNF complexes in the locus. ChIP assays with antibodies against BRM showed that androgen treatment of VCaP cells brings about very robust recruitment of BRM onto regions 5 and 6, whereas the occupancy of BRG1 in the locus remained very low after the androgen exposure (Figure 5). Interestingly, the regions around ∼10 kb from the TSS occupying most BRM were the same ones showing the most prominent decrease in H3 density in response to androgen exposure. These results suggest that AR influences to the chromatin structure of FKBP51 via recruitment of BRM-containing complexes to the locus.
Effect of antiandrogen on FKBP51 transcription and AR loading
Since increased AR protein levels have been reported to convert antiandrogens, such as bicalutamide, to agonists (35), it was of interest to investigate whether the difference in the amount of AR between VCaP and LNCaP cells is manifested in a differential response to the pure antagonist bicalutamide (BIC). However, BIC alone displayed no or minimal agonistic activity on FKBP51 mRNA accumulation in LNCaP and VCaP cells (Figure 6A). BIC was also capable of inhibiting R1881-induced accumulation of FKBP51 mRNA in both cell lines, albeit the pure antagonist was somewhat more potent in LNCaP cells than in VCaP cells (≥90% versus ∼80% inhibition). As in the case of pure agonists, the pure antagonist down-regulated the AR protein in VCaP cells but not in LNCaP cells (Figure 6B). We next compared the loading of AR to the FKBP51 chromatin in response to R1881 and BIC in LNCaP and VCaP cells by ChIP and qPCR analysis. As shown in Figure 6B, the overall pattern of agonist-induced loading of AR to the locus was very similar in both cell lines, but the level of AR occupancy was lower in LNCaP cells than in VCaP cells. The latter finding very likely reflects the difference in the amount of AR between the cell lines. Interestingly, BIC alone was capable of promoting binding of AR to the FKBP51 locus in VCaP cells but not in LNCaP cells (Figure 6C). However, in agreement with the minimal mRNA induction of FKBP51 upon BIC exposure in VCaP cells, the BIC treatment did not bring about significant changes in the occupancy of PolII in the locus (Supplementary Figure S4).
DISCUSSION
Knowledge of the molecular mechanisms of AR-dependent transcription is essential for better understanding of its role in prostate cancer development. On chromatin and target locus level, however, detailed analyses of AR-targeted loci have been thus far limited to the PSA gene (16–20). Moreover, only one model cell line, LNCaP, has been employed in most of these studies. Here, we have gathered molecular insight into the AR action by investigating the regulation of FKBP51 transcription using VCaP and LNCaP cells as model systems. The VCaP cells are derived from a bone metastasis and express amplified levels of wild-type AR, thus perhaps better resembling the situation in hormone-refractory prostate cancer than the LNCaP cells that correspond to a lymph node metastasis with a ligand-binding domain T877A-mutated version of the receptor (23). The rationale for choosing the FKBP51 as a model gene is based on the recent finding that the FKBP51 expression is more sensitive to depletion of intraprostatic androgens than any other AR target gene (22). Response of FKBP51 mRNA to androgens has previously been observed also in LNCaP cell and CWR22-R xenograft models (36–38). FKBP51 expression is also upregulated by glucocorticoids and progestins in lymphocytes and breast cancer cells, respectively (39,40). However, the molecular mechanisms of the activation of FKBP51 chromatin by AR or glucocorticoid receptor (GR) have not been addressed in detail.
FKBP51 (FKBP5) encodes a 51 kDa FK506-binding protein that belongs to the immunophilin protein family (41). The exact role of FKBP51 is not known, but it appears to play a role in protein folding and trafficking (41). FKBP51 was discovered as a human homolog for avian FKBP54 (42) that in turn was originally found in progesterone receptor (PR) complexes (43). AR can interact with FKBP51 and an increase in the immunophilin level may augment the activity of AR (37). Interestingly, FKBP51 protein levels are elevated in prostate cancer, which may sensitize AR signaling (37,38). Also our statistical analysis of the microarray data retrieved from the Oncomine website (www.oncomine.org) indicates significantly increased expression of FKBP51 in prostate cancer in comparison to non-malignant prostate (Supplementary Figure S5).
The reason for the fast and robust induction of FKBP51 by AR, in comparison to PSA, appears to be twofold. Firstly, the FKBP51 is regulated purely at the level of transcription, whereas a regulatory component requiring new protein synthesis is involved in the activation of PSA in VCaP cells and LNCaP cells (26). Secondly, several AR-binding sites appear to synergize in the activation of the FKBP51. Our in vitro testing of the potential AREs together with quantitative ChIP scans revealed the presence of at least seven functional AREs within four distinct enhancer regions located in the introns of the gene. Interestingly, the two major enhancer regions harboring multiple AREs are located very distal from the TSS, at ∼87 kb in the 5th intron (two AREs) and ∼98 kb in the 7th intron (three AREs), with the farthermost acting as the most active AR-regulated enhancer region of FKBP51 in isolation. Even though the single ARE-containing regions at ∼25 kb (1st intron) and ∼77 kb (5th intron) possessed in isolation only weak enhancer activity, they showed significant androgen-induced loading of AR both in VCaP and LNCaP cells and are thus likely to cooperate with the compound AREs in vivo.
Interestingly, the AREVIII and the AREIX in the 5th intron of FKBP51 are conserved in the corresponding intron of mouse gene (at ∼65 kb from TSS), and the region was recently shown to occupy AR in mouse prostate but not GR in mouse thymus (44). The lack of GR occupancy in the 5th intron of Fkbp51 in the GR’s target tissue thymus in vivo is somewhat unexpected, since both the GR and the AR (and progesterone receptor, PR) recognize in vitro with a similar affinity the non-selective regulatory elements that are composed of 3-nt-spaced inverted repeats of a 5′-TGTTCT-3′-type half-site (29,45). The AREs in the 5th intron as most of the other elements of the FKBP51 correspond to the latter, non-selective- type response elements, which is in keeping with the regulation of FKBP51 by several steroid hormones. Two of the three AREs in the 7th intron of human FKBP51 that were found and validated in this study are conserved in the corresponding intron of the mouse gene, but their function as AREs or GREs has not been addressed in the previous studies. In contrast to the role of distal elements, ChIP assays have recently revealed that the regulation of Fkbp51 by progestins occurs in breast cancer cells via an intronic enhancer that resides relatively close (∼1.2 kb) to the TSS (46). In transfection-based reporter gene assays however, the 5th intronic region of the human gene has been reported to confer progesterone response in breast cancer cells (39). These results thus emphasize the importance of validation of the putative steroid response elements under chromatin environment by ChIP assays. Thus, even though AR, GR and PR all recognize similar DNA elements in vitro and often regulate same genes, such as FKBP51, these steroid receptors seem to employ alternative binding sites in chromatin to activate transcription in vivo. Since the above analyses were carried out in different cell types, it is likely that differences in chromatin milieu significantly contributed to the observed differences in vivo. The role of other ‘pioneering’ transcription factors binding close to steroid receptor-occupied sites may also be important (20,47), as GATA2 was shown to cooperate with the PR in the regulation of Fkbp51 (46). However, according to our unpublished data, GATA2 or FoxA1 do not play a major role in the androgen regulation of FKBP51 in VCaP cells (Makkonen,H., unpublished results).
Scanning of the chromatin structure of FKBP51 in VCaP cells by ChIP showed a considerable enrichment of acetylated H3 as well as trimethylated H3K4 nearby the TSS, and K36 trimethylated H3 was prevalent throughout the whole coding region of the gene. These marks along their locations are typical to genes in active chromatin (6,10). However, the above H3 modifications were only modestly influenced by androgens, suggesting that the FKBP51 locus existed in an active conformation already prior to AR binding and the receptor was targeting pre-opened chromatin in prostate cancer cells. Similar results have been reported for the PSA locus in LNCaP cells (16). In keeping with the key role of AR in the activation and elongation of FKBP51 transcription, androgen induction brought about a clear increase in the occupancy of PolII throughout the gene, whereas antiandrogen bicalutamide-bound receptor failed to do so, even though it was in VCaP cells capable of promoting AR binding to the locus. Androgen induction also resulted in a decrease in histone density within the locus, which is likely to be at least in part caused by recruitment of BRM-containing chromatin remodeling complexes. The temporal and spatial correlation between AR loading and recruitment of BRM is in agreement with the previous reports that AR predominantly employs a subfamily of SWI/SNF complexes that depend on the BRM, but not on the BRG1, ATPase (48,49).
The very distal enhancer elements are thought to communicate with the general transcription machinery at the TSS thought chromatin looping. However, the challenges associated with testing the existence of such loops and the coordination of interactions of several AREs spanning ∼100 kb in FKBP51 are daunting. In our schematic model (Figure 7), we propose that intricate cooperation between multiple AREs orchestrate recruitment of coregulatory complexes and PolII to the FKBP51 locus, which leads to enhanced transcriptional rate of the PolII. Mediator proteins, such as TRAP220, may potentially bridge AR to PolII (50,51). However, our ChIP assays with anti-TRAP220 antibody do not support a prominent role for the mediator subunit as a bridging factor, which may not be surprising, since there appears to be a lot of functional redundancy among the AR coregulators (19).
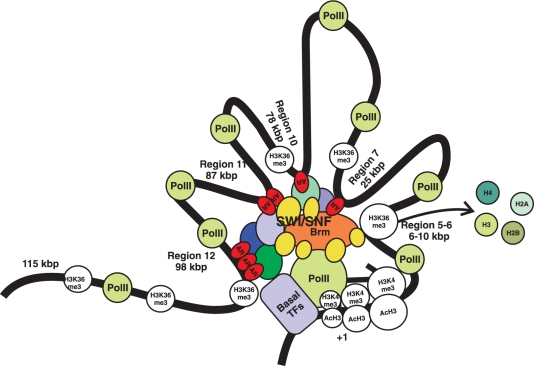
Figure 7.
A spatial model of loop formation in the long-range regulation of the FKBP51 transcription by the AR transcription complex. ARs are first primarily loaded onto two very distal regions at ∼98 and ∼87 kb downstream from the TSS. These two compound-ARE-containing enhancers interact via coregulatory proteins, including BRM-containing SWI/SNF complexes, and cooperate with AR-occupying single-ARE regions at ∼78 and ∼25 kb, which brings about further decrease in nucleosome density and recruitment of PolII tracking through the locus.
Classic models of steroid receptor action view steroid receptor-binding sites as regulatory elements in gene promoters or their proximity as upstream enhancer elements. However, recent wider screens for their binding sites imply that steroid-regulated promoters in most cases lack proximal response elements. For example, genome-wide screening of estrogen receptor binding sites by ChIP combined with tiled microarray analysis indicated that only a minority of estrogen receptor-binding sites localize within 1 kb promoter-proximal regions (52). The same appears to hold true for AR-regulated genes, as most of the AR-binding sites in chromosomes 21 and 22 as well as among a selected sample of target genes seem to reside >10 kb from TSSs, often downstream of them (20,53). Our current validated binding and functional data of the FKBP51 locus also highlight the emerging biological importance of very long-range regulation via distal enhancers by AR.
The nuclear organization of genome, i.e. how transcription factors, their cognate binding sites and chromatin-modifying complexes are organized with respect to each other within the nucleus, is critical for the proper function of our genome (54). Notably prostate cancers show striking genomic aberrations (55). The capability of AR to regulate promoters from a long distance in chromatin should thus be considered when evaluating the role of AR in targeting novel genes and gene programs, especially in malignant cells. Moreover, the issue concerning the insulating mechanisms that should prevent distally bound steroid receptors from promiscuously activating other than their genuine target genes warrants emphasis in further studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland, Finnish Cancer Foundation, Sigrid Jusélius Foundation, and Kuopio University Foundation. Funding for open access charge: Academy of Finland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Merja Räsänen for skillful technical assistance.
REFERENCES
- 1.Gao W, Bohl CE, Dalton JT. Chemistry and structural biology of androgen receptor. Chem. Rev. 2005;105:3352–3370. doi: 10.1021/cr020456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Culig Z, Klocker H, Bartsch G, Hobisch A. Androgen receptors in prostate cancer. Endocr. Relat. Cancer. 2002;9:155–170. doi: 10.1677/erc.0.0090155. [DOI] [PubMed] [Google Scholar]
- 3.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr. Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 4.Kallio PJ, Poukka H, Moilanen A, Jänne OA, Palvimo JJ. Androgen receptor-mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol. Endocrinol. 1995;9:1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- 5.Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, Jänne OA. Mutual transcriptional interference between RelA and androgen receptor. J. Biol. Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 9.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Farrants AK. Chromatin remodelling and actin organisation. FEBS Lett. 2008;582:2041–2050. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 12.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol. Cell Endocrinol. 2007;265–266:162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Jia L, Shen HC, Wantroba M, Khalid O, Liang G, Wang Q, Gentzschein E, Pinski JK, Stanczyk FZ, Jones PA, et al. Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol. Cell Biol. 2006;26:7331–7341. doi: 10.1128/MCB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang Z, Jänne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 2004;18:2633–2648. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 18.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. J. Clin. Oncol. 2003;21:383–391. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 22.Mostaghel EA, Page ST, Lin DW, Fazli L, Coleman IM, True LD, Knudsen B, Hess DL, Nelson CC, Matsumoto AM, et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007;67:5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 23.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 24.Thompson J, Saatcioglu F, Jänne OA, Palvimo JJ. Disrupted amino- and carboxyl-terminal interactions of the androgen receptor are linked to androgen insensitivity. Mol. Endocrinol. 2001;15:923–935. doi: 10.1210/mend.15.6.0647. [DOI] [PubMed] [Google Scholar]
- 25.Palvimo JJ, Kallio PJ, Ikonen T, Mehto M, Jänne OA. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol. Endocrinol. 1993;7:1399–1407. doi: 10.1210/mend.7.11.8114755. [DOI] [PubMed] [Google Scholar]
- 26.Makkonen H, Jaaskelainen T, Pitkanen-Arsiola T, Rytinki M, Waltering KK, Matto M, Visakorpi T, Palvimo JJ. Identification of ETS-like transcription factor 4 as a novel androgen receptor target in prostate cancer cells. Oncogene. 2008;27:4865–4876. doi: 10.1038/onc.2008.125. [DOI] [PubMed] [Google Scholar]
- 27.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 28.Prescott J, Jariwala U, Jia L, Cogan JP, Barski A, Pregizer S, Shen HC, Arasheben A, Neilson JJ, Frenkel B, et al. Androgen receptor-mediated repression of novel target genes. Prostate. 2007;67:1371–1383. doi: 10.1002/pros.20623. [DOI] [PubMed] [Google Scholar]
- 29.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl Acad. Sci. USA. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallio PJ, Palvimo JJ, Mehto M, Jänne OA. Analysis of androgen receptor-DNA interactions with receptor proteins produced in insect cells. J. Biol. Chem. 1994;269:11514–11522. [PubMed] [Google Scholar]
- 31.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 32.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 33.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 34.Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct. Genomic Proteomic. 2006;5:209–221. doi: 10.1093/bfgp/ell028. [DOI] [PubMed] [Google Scholar]
- 35.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 36.Amler LC, Agus DB, LeDuc C, Sapinoso ML, Fox WD, Kern S, Lee D, Wang V, Leysens M, Higgins B, et al. Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22-R1. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- 37.Febbo PG, Lowenberg M, Thorner AR, Brown M, Loda M, Golub TR. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J. Urol. 2005;173:1772–1777. doi: 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- 38.Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, Greenberger LM, Frost P, Bai W, Zhang Y. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology. 2004;145:3913–3924. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- 39.Hubler TR, Scammell JG. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones. 2004;9:243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: A potential marker for glucocorticoid sensitivity, potency, and bioavailability. J. Clin. Endocrinol. Metab. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 41.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 42.Nair SC, Rimerman RA, Toran EJ, Chen S, Prapapanich V, Butts RN, Smith DF. Molecular cloning of human FKBP51 and comparisons of immunophilin interactions with Hsp90 and progesterone receptor. Mol. Cell Biol. 1997;17:594–603. doi: 10.1128/mcb.17.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DF, Faber LE, Toft DO. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J. Biol. Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 44.Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 45.Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 46.Magklara A, Smith CL. A composite intronic element directs dynamic binding of the progesterone receptor and GATA-2. Mol. Endocrinol. 2009;23:61–73. doi: 10.1210/me.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Link KA, Balasubramaniam S, Sharma A, Comstock CE, Godoy-Tundidor S, Powers N, Cao KH, Haelens A, Claessens F, Revelo MP, et al. Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity. Cancer Res. 2008;68:4551–4558. doi: 10.1158/0008-5472.CAN-07-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE. Differential requirement of SWI/SNF for androgen receptor activity. J. Biol. Chem. 2003;278:30605–30613. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 51.Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034–4041. doi: 10.1158/0008-5472.CAN-06-3039. [DOI] [PubMed] [Google Scholar]
- 52.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 53.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 55.Morris DS, Tomlins SA, Montie JE, Chinnaiyan AM. The discovery and application of gene fusions in prostate cancer. BJU Int. 2008;102:276–282. doi: 10.1111/j.1464-410X.2008.07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.