Abstract
RNA-interference (RNAi) is a potent tool for specific gene silencing. In this study, we developed an adenovirus for conditional replication in p53-dysfunctional tumor cells that uses p53-selective expression of a microRNA-network directed against essential adenoviral genes. Compared to a control virus that expressed a scrambled microRNA-network, antiviral RNAi selectively attenuated viral replication in cells with transcriptionally active p53, but not in p53-dysfunctional tumor cells where both viruses replicated equivalently. Since these results were confirmed by an in vivo comparison of both viruses after infection of p53-knockout and normal mice, we could demonstrate that attenuated replication was indeed a result of p53-selective exhibition of antiviral RNAi. Addressing the therapeutic applicability, we could show that the application of RNAi-controlled virus efficiently lysed p53-dysfunctional tumors in vitro and in vivo but resulted in drastically reduced load of virus-DNA in the liver of treated mice. We have generated a broadly applicable adenovirus for selective destruction of p53-dysfunctional tumors and thereby demonstrate that virus-encoded RNAi-networks represent an efficient and versatile tool to modify viral functions. RNAi-networks can be applied to all transcriptionally regulated DNA-viruses to remodulate viral tropism and thus provide means to generate specifically replicating vectors for clinical applications.
INTRODUCTION
RNA-interference (RNAi) has been established as versatile tool for targeted gene repression during recent years. The potent and specific gene silencing effect of RNAi promises enormous potential for the treatment of disorders with defined genetic etiology. RNAi has been successfully applied to revert oncogenesis by downregulation of K-rasV12 or Bcr-Abl (1,2), or to prevent acute liver failure by silencing Fas or Caspase-8 (3,4).
Exogenous delivery of RNAi, directed against viral RNA, has been shown to reduce viral titers of HIV, poliovirus, HCV and Adenovirus (5–8), indicating that RNAi can provide eukaryotic cells with a specific antiviral defence mechanism. An endogenous human microRNA (miR-32) has been discovered that negatively interferes with replication of primate foamy virus-1 (9) and it has been suggested that cell type-specific patterns of microRNAs contribute to the cellular tropism of certain viruses (10). Recently, virus tropism has been successfully remodulated by incorporating target sites for natural, tissue-specific microRNAs into the 3′-UTR of essential virus genes (11–13). These studies indicate the potential of RNAi-based mechanisms for the generation of tropism-modified viruses.
Some DNA-viruses encode for self-reactive miRNAs, that have been considered to play a pivotal role in the correct timing of the lytic life cycle (14,15). The use of microRNAs as a regulative tool to manipulate the viral tropism seems to be ideal since miRNAs are non-antigenic structures of short length in contrast to the high immunogenicity of viral proteins and with respect to the size constraints in viral genomes. However, it has not yet been investigated whether tissue-specific expression of engineered, antiviral miRNAs can be used to manipulate the viral tropism.
Tissue-specific control of replication is an important tool to remodulate the tropism of adenoviruses for the treatment of solid tumors and several concepts have successfully addressed prominent pan-cancer associated molecular targets such as telomerase, Rb or p53 (16).
In the majority of human cancers p53-dependent transcriptional activity is either suppressed or missing due to genetic loss or inactivating mutations, but also due to epigenetic inhibition. Loss of transcriptional functions of p53 (further referred to as p53-dysfunction) is a fundamental step towards malignant cell transformation and represents an excellent broad range target for molecular interventions. In this study we have redesigned an adenovirus for conditional replication in p53-dysfunctional tumors by p53-selective expression of an antiviral RNAi-network. We could demonstrate that p53-dependent expression of such a network is able to selectively control viral replication due to efficient knockdown of essential viral genes and allows for efficient replication and lysis in p53-dysfunctional tumors in vitro and in vivo.
MATERIALS AND METHODS
Cell lines and culturing
Hep3B, H1299, HepG2, Huh7, A549, 293, HCT-116 and primary human IMR90-fibroblasts were obtained from ATCC. HCT116 and HCT116-p53(–/–) were kindly provided by B. Vogelstein. All cells were maintained in DMEM + Glutamax (Life Technologies Inc.) supplemented with 10% heat-inactivated FBS (Life Technologies Inc.), 100 units/ml penicillin and 100 μg/ml Streptomycin (Seromed) at 37°C in 5% CO2. For culturing of HCT116-p53(–/–) the medium was additionally supplemented with 0.4 mg/ml G 418-Sulfate (Calbiochem) and 0.1 mg/ml Hygromycin (Boehringer Mannheim).
RNAi target sequences
The following target sequences were chosen for construction of antiviral RNAi-networks. Firefly luciferase: GAUUUCGAGUCGUCUUAAUGU [shRfLuc-774, as described previously (17)]; E1A: GGAGGCGGTTTCGCAGATTTT (shRE1A-736), GAAGGGATTGACTTACTCACTTTT (shRE1A-785); E4: GACAGGAAACCGTGTGGAATA (shRE4-33056), CCATGTTTAATCAGAGGTTTATA (shRE4-33623); E1B: GGAAGCATTGTGAGCTCATAT (shRE1B-3630); pTp: CGGAGTACCTACACAACAATT (shRpTp-9651); Adpol: GCGAATACGTGCAGCTAAACAT (shRAdPol-6361). Further sequence details of used oligonucleotides can be provided upon request.
Genetic construction
Expression vectors for miR30-shRNA-motifs were constructed using pBluescript provided with synthetic polylinker, a SV40-polyadenylation signal derived from AdApt (Introgene), and a promoter, such as the wildtype CMV-promoter (from pBK-CMV, Stratagene), U6-promoter (pSuperRetro, Addgene), or the p53-responsive promotor prMinRGC from prMinRGC-Luc (18). For the generation of miR30-shRNA-motifs, two partially overlapping oligonucleotides were annealed and overhangs were then filled-in using Herculase (Stratagene). Restriction sites were introduced by a second PCR using TTCTCGAGAAGGTATATTGCTGTTGACAGTGAGCG and AAAACTAGTGAATTCCGAGGCAGTAGGCA as primers. The obtained fragments were inserted into the aforementioned expression vectors. All miRNA-sequences for miR30-shRNAs were designed using a publicly available algorithm (http://katahdin.cshl.org:9331/siRNA/RNAi.cgi?type=shRNA). For experimental evaluation of candidate RNAi sequences, the psiCHECK-2 reporter system (Promega) was used. DNA fragments corresponding to viral target messengers [spanning following regions: 541–926 (E1A), 3608–4070 (E1B), 33 704–32 881 (E4), 10 214–9365 (pTp), 6809–6106 (Adpol)] were generated by PCR using adenoviral DNA as template and cloned into the 3′-UTR of the Renilla reporter gene.
Shuttle plasmids for the generation of recombinant adenovirus were constructed on the basis of the plasmid pHM3. pHM3–E1 was constructed by insertion of ΔN22–E1A under control of the wildtype E1A-promotor. The vector also contained the E1B region up to the translational stop of E1B–55k (pos. 3509) followed by the adenoviral sequence according to pos. from 3614–3661 to obtain a further target site within the 3′UTR of E1B–55k. To further enhance RNAi efficacy, pHM3–E1–3′-UTR was generated by additional insertion of a fragment harboring the target sequences of shRE1A-785, shRE1A-736 and shRE1B-3630 into the Hpa I site of the E1A–3′-UTR in pHM3-E1. p53-dependent expression cassettes for multiple miR30-shRNAs were inserted downstream of the E1 regions of pHM3–E1 and pHM3–E1–3′-UTR to generate shuttle vectors for subsequent transfer into pAdHM4.
Full-length E1A was PCR-amplified from adenoviral DNA using the primers GGGACTAGTAAAATGAGACATATTATCTGCCACGG and AAGCGGCCGCTTATGGCCTGGGGCGTTTACA and placed in an expression vector under control of an EF1α-promoter (obtained from pLVTHM, kindly provided by D. Trono).
Further details concerning cloning procedures or used oligonucleotides can be provided upon request.
Recombinant adenovirus generation and preparation
Recombinant adenoviruses were constructed according to a previously described method (19). The viral vectors Ad-iREP-C1, Ad-iREP-1, Ad-iREP-C2 and Ad-iREP-2 were generated by ligating PI-Sce I/I-Ceu I fragments of shuttle pHM3-vectors described above into pAdHM4. For the production of infectious particles the resulting plasmids were Pac I-digested and transfected into 293 cells until a cytopathic effect became visible. Infectious particles were further amplified in 293 cells and then purified using Adpack20-Kit (Vivascience). Infectious titers were determined using the Rapid-Titer-Kit (Takara/Clontech).
Luciferase assay
Using the calcium phosphate precipitation method or LipofectAMINE™ (Life Technologies), cells were cotransfected with 1 μg of pCMV-fLuc as reporter, 1 μg shRNA expression plasmids and 0.5 μg SV40-LacZ for normalization. Total DNA-amount was adjusted to 5 μg per 60 mm. Cellular extracts were prepared 24–48 h post transfection and analyzed for luciferase activity in a Berthold Lumat LB9501 and normalized by β-galactosidase activity measurements.
For evaluation of shRNA-sequences, psiCHECK™-2 derived vectors were cotransfected as reporter and the Dual-Luciferase® Reporter Assay System (Promega) was applied.
Infection of IMR-90-fibroblasts
To allow efficient infection of IMR-90-fibroblasts, adenoviral vectors were coated with the bifunctional adapter protein CARex-Tat as described previously (20). For this purpose, viruses were mixed with purified recombinant CARex-Tat protein (100 ng/104 ifu) and incubated for 15 min. Total 5 × 105 IMR-90-fibroblasts were then infected with pretreated viral particles at MOI 0.1. Using the QIAmp-DNA-Mini Kit (Qiagen) total DNA was isolated and subjected to qPCR to determine the viral DNA-content.
Viral DNA-quantification
Quantitative PCR (qPCR™ Mastermix Plus, Eurogentec) was performed with 100 ng of DNA using adenoviral hexon-specific primer probes as described before (21). As internal controls, the Ubiquitin-C-Control-Kit (Eurogentec) was used for human DNA, and the 18S-Genomic-Control-Kit (Eurogentec) was applied for murine DNA.
Reverse transcription qPCR
Total 1.5 × 106 A549 and Huh7 cells were seeded in 60-mm-dishes and infected (MOI 0.05) with adenovirus as indicated in the figure legends. Total RNA was isolated using peqGOLD-RNAPure™ (peQLab), digested with DNase, and further purified using the RNeasy-Mini-Kit (Qiagen). One hundred nanograms of RNA was subjected to reverse transcription using random hexamer primer and TaqMan™ Reverse Transcription Reagents (Applied Biosystems). cDNA was then used for quantification using SYBR-Green PCR Master Mix (Applied Biosystems) and 30 cycles with 15 s at 95°C and 60 s at 60°C after an initial step for 10 min at 95°C. The fluorescence signal was acquired at 60°C. Amplicon signals were normalized by GAPDH as internal control. Primer sequences were GCCGGAGCAGAGAGCCTT (E1A-fw); CGTCGTCACTGGGTGGAAAG (E1A-rev); GGCCCCAATTTTAGCGGTAC (E1B-fw): GTCCAGGCTTCCACACAGGTA (E1B-rev); AACTGGTCCGTTATGGCCAA (pTp-fw); TGCACCTGCGTGAGGGTAG (pTp-rev); TTTGTGGACCACACCAGCTC (Fiber-fw); CAAGTATTTGACTGCCACATTTTGT (Fiber-rev); TGGGCTACACTGAGCACCAG (GAPDH-fw); GGGTGTCGCTGTTGAAGTCA (GAPDH-rev).
Detection of mature microRNAs/shRNAs
Mature microRNAs/shRNAs were detected and quantified according to the miR-Q PCR method as described previously (22). For this purpose 100 ng of isolated total RNA of infected cells were used for reverse transcription of mature miRNA with specific reverse primer: TTTTGTCAGGCACCGTATTCACCGTGAGTGGTACGGAGGCG (QRT736) and TTTTGTCAGGCACGTTTCACCGTGAGTGGTACGACAGGA (QRT33 056). TaqMan™ Reverse Transcription Reagents (Applied Biosystems) were applied according to the manufacturer's protocol. Generated miR-cDNA was then applied for quantification using SYBR-Green PCR Master Mix (Applied Biosystems). The reaction was performed using 100 nM QRev736: CCCCGTCAGATGTCCGAGTAGAGGGGGAACGGCGACAAAATCTGCGAAACCGC or QRev33056: CCCCGTCAGATGTCCGAGTAGAGGGGGAACGGCGACTATTCCACACGGTTTCC and 300 nM of reverse and forward primers: TGTCAGGCAACCGTATTCACC (Q-fw) and CGTCAGATGTCCGAGTAGAGG (Q-rev). The amplification was carried out by a first step at 95°C for 10 min, followed by 30 cycles with 15 s at 95°C and 60 s at 62°C. The fluorescence signal was acquired at 62°C. Amplification of GAPDH was used as internal control.
Western blot analysis
For western blot analysis cell extracts were prepared by treating the cell-layer with a lysis buffer containing 25 mM Tris-phosphate, 2 mM EDTA, 2 mM DTT, 10% glycerol and 1% Triton X-100. Ten micrograms of protein were separated by SDS–PAGE and transferred onto a PVDF-membrane (Immobilon®, Millipore). E1A-proteins were detected using a polyclonal rabbit anti-E1A-antibody (Santa Cruz) and visualized with Western-Lightning™ chemiluminescent-reagent Plus (Perkin-Elmer).
Cytolysis assays
Target cells were seeded in 24-well plates at a density of 4 × 104 cells/well and allowed to adhere overnight. Cells were then infected with adenoviruses (MOI of 5, 0.5, 0.05 and 0.005) and incubated for 8–10 days. Crystal-violet staining was then used to visualize the destruction of the cell layer. After a gentle rinse with PBS to remove dead cells, 10% formalin in PBS was added for fixation (30 min). The fixation solution was replaced and cells were then stained for 30 min using an aqueous solution of 0.1% crystal-violet in 10% ethanol. Finally, plates were rinsed with water and air dried.
Animal experiments
Six-week-old, homozygous p53-knockout mice (inbred from strain B6.129S2-Trp53tm1Tyi/J, Jackson Laboratory, Maine, USA) were provided by the Central Animal Facility at the Helmholtz Centre of Infection Research (HZI, Braunschweig, Germany). Six weeks old C57BL/6 and NMRI nu/nu mice were obtained from the Animal Research Institute of the Hannover Medical School. All experiments were performed according to the German legal requirements (TSchG). Tumor xenografts were established by subcutaneous injection of 1 × 107 Hep3B or H1299 cells into the left flank of mice. Tumor nodules were grown to a size of approximately 250 mm3 before initial treatment. Infection of tumors was performed by intratumoral injection of 1 × 109 ifu in 150 µl and was repeated every five days. Tumor volume was calculated as follows: V(tumor) = (length × (width2)/2. For each group six animals were assessed.
For the analysis of intrahepatic viral DNA load, Hep3B cells were infected at MOI 25 to achieve almost complete infection. Twenty-four hours later, infected cells were collected and 5 × 105 cells were injected intravenously. DNA from liver tissue was purified using the QIAmp-DNA-Mini Kit (Qiagen) and viral DNA was quantified by qPCR as described above.
Statistics
For the comparison of two groups, the Student's t-test was used to determine statistical significance. P-values < 0.05 were considered as statistically significant.
RESULTS
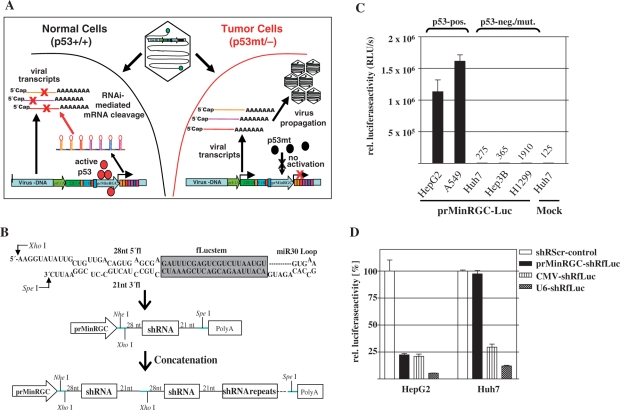
RNA-polymerase II (pol II)-dependent expression of shRNA or microRNA has already been successfully established, suggesting that such RNAi mediators can be expressed by virtually any promoter and that RNAi can therefore be brought about in a tissue-specific manner (23,24). To investigate in a therapeutically relevant setting whether cell-type specific expression of virus-encoded antiviral shRNA restricts virus propagation to defined tissues, we aimed to generate a conditionally replicating, RNAi-controlled adenovirus for targeted therapy of p53-dysfunctional tumors. To redirect adenoviral replication to p53-dysfunctional tumor cells, our concept (Figure 1A) envisioned the expression of a multiple hairpin transcript (referred to as antiviral RNAi-network) that efficiently silences a set of essential viral genes under transcriptional control of a p53-responsive promoter. This strategy allows selective onset of antiviral RNAi and subsequent inhibition of viral replication in response to activated p53 in normal cells. In contrast, the antiviral RNAi-network is not expressed in p53-dysfunctional tumor cells thus enabling normal adenoviral replication.
Figure 1.
Concept of p53-dependent, RNAi-mediated modification of the adenoviral tropism. (A) Scheme of tumor selectively replicating adenovirus that expresses a self-inhibitory, antiviral RNAi-network in response to p53. (B) Used miR30-shRNA-motifs and concatenation to generate RNAi-networks (miR30-shRfLuc as example). (C) Activation of the p53-selective promoter prMinRGC in cell lines with different p53-status. Cells were cotransfected with prMinRGC-fLuc and CMV-LacZ. Luciferase activity was determined 48 h post transfection (mean ± SD). The prMinRGC-promoter is highly activated in p53-positive cell lines whereas activation is absent in p53-negative or mutated cells. (D) HepG2 and Huh7 cells were cotransfected with CMV-fLuc, SV40-LacZ and shRNA-expression plasmids under control of a U6-, CMV- or prMinRGC-promoter (shRfLuc relative to the corresponding shRScr-control, mean ± SD). The results show that RNAi can be expressed in a p53-selective manner.
p53-dependent gene silencing by a multiple shRNA-transcript consisting of concatenated, minimalized miR30-derived hairpins
The genetic information for shRNAs can be encoded by DNA of relatively short length. The concatenation of different shRNAs to construct an ‘RNAi-network’ should further allow synchronous silencing of different genes involved in viral replication. As single modules for such an RNAi-network, we considered a minimalized miR30-derived hairpin (termed miR30-shR, Figure 1B) according to designed hairpins described before (24).
To allow for p53-selective expression of shRNA we chose the artificial, p53-responsive promoter prMinRGC that we have described previously (18). As shown in Figure 1C, this promoter is highly active in p53-normal cells but silent in p53-dysfunctional cells. The prMinRGC-promoter therefore displays an outstanding signal/noise ratio with regard to the p53-status of the target cell. To investigate whether shRNA can be expressed in a p53-selective manner, the experiment was carried out in p53-positive HepG2 and p53-mutant Huh7 cells (Figure 1D). Gene silencing efficacy was then determined after shRNA-expression under control of the pol III-dependent U6-promoter and pol II-dependent CMV- and prMinRGC-promoters. We could observe efficient RNAi in HepG2 cells when shRNAs were expressed under control of the prMinRGC-promoter, whereas RNAi was absent under p53-dysfunctional conditions. Following CMV-promoter dependent expression of shRNA the observed extent of RNAi was almost equivalent in both cell lines. Though pol II-dependent RNAi did not reach the extent of pol III-dependent RNAi, prMinRGC-promoter-dependent expression of shRNA led to a similar extent of RNAi in response to p53 compared to the strong CMV-promoter. This promoter has already been successfully used for efficient exhibition of RNAi (23). The comparison of RNAi caused by prMinRGC-promoter-dependent expression of shRNA in cells with different p53-status indicates that RNAi was indeed p53-dependent. As these results were confirmed in other p53-active or p53-inactive cells (data not shown), efficient RNAi can be exhibited in a p53-selective manner.
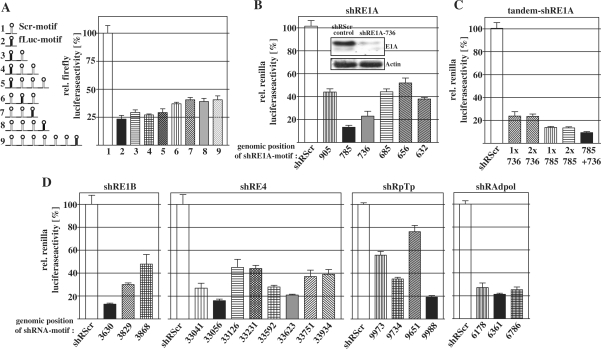
Next, we were interested how gene silencing efficacy is influenced when miR30-shRs are concatenated to generate complex RNAi-networks (concatenation is illustrated in Figure 1B). For this purpose we tested several arrangements that either varied in length of the network or the position of a single miR30-shRfLuc-motif within the network (Figure 2A). Since expression of all arrangements led to an almost equivalent repression of the cotransfected luciferase-reporter we could show that neither the length of the RNAi-network nor the position of an active hairpin significantly influence gene silencing efficacy. Together, these data suggest that p53-dependent expression of multiple hairpin-transcripts as RNAi-networks can be applied to achieve effective and synchronous gene silencing and demonstrate the versatility of the RNAi-network concept.
Figure 2.
Optimization of an antiviral RNAi-network targeting essential adenoviral genes. (A) p53-dependent expression plasmids for complex RNAi-networks with variable hairpin numbers and variable position of a single, embedded miR30-shRfLuc-motif were cotransfected with CMV-fLuc and SV40-LacZ in HepG2 cells and investigated as described above (mean ± SD). (B) The psiCHECK-2-derived plasmids containing target sequences of adenoviral E1A-mRNA within the 3′-UTR of the renilla luciferase were used to evaluate effective miR30-shRNAs against E1A. HepG2 cells were cotransfected with miR30-shRNA-expression constructs under control of the prMinRGC-promoter. Forty-eight hours later, cells were lysed and luciferase activity was determined. Application of miR30-shRE1A-785 and miR30-shRE1A-736 led to effective RNAi (mean ± SD). RNAi efficacy was confirmed when the full-length messenger E1A was addressed. For this purpose, an expression construct for the complete E1A-mRNA was cotransfected with a miR30-shRE1A-736-construct and E1A-level was analyzed by western blot (inset). (C) A psiCHECK-2-plasmid containing an E1A-target fragment was cotransfected in HepG2 cells with plasmids for expression miR30-shRE1A tandem-arrangements as indicated and luciferase activity was determined (mean ± SD). In a multiple miR30-shRNA transcript, RNAi is more efficiently exhibited by different motives compared to motif-repeats. (D) RNAi-efficacy by diverse miR30-shRNA-motifs against psiCHECK-2-derived reporters of E4, E1B, pTp and AdPol was determined. The black and dark grey bars indicate the most effective miR30-shRNA-motifs that were selected for the antiviral RNAi-network.
Antiviral RNAi-network silences essential viral genes following viral infection
We considered the mRNAs of the adenoviral E1A, E1B, E4, polymerase (AdPol) and terminal protein (pTp) as preferable targets for an antiviral RNAi-network. E1A and E4 proteins are most essential for replication and E1B-55k is decisively involved in the shutdown of the cellular p53-response. Terminal protein is bound to the viral DNA–termini and attracts AdPol for the initiation of replication. First, miR30–shR-derivatives of several potential hairpin-motifs per target were screened for their gene silencing efficacy. Complete test results are summarized in Figure 2B–D). To point out, we found two efficient hairpins against E1A that were also able to successfully silence full-length E1A as confirmed by western blot analysis (Figure 2B). Interestingly, we could observe that a tandem combination of the same miR30–shR-motifs against E1A did not increase knockdown efficacy whereas the combination of different motives resulted in additional gene silencing (Figure 2C).
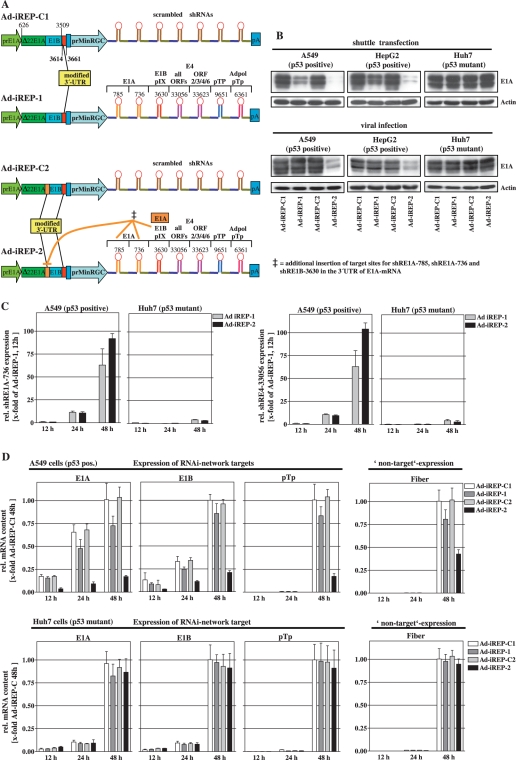
Most effective hairpins were then concatenated for an antiviral RNAi-network that was subsequently introduced into an adenoviral backbone under the control of the prMinRGC-promoter (Ad-iREP-1 and Ad-iREP-2, Figure 3A). To confer improved accessibility for RNAi mechanisms, we positioned additional RNAi target-sites in the 3′-UTRs of E1B (both viruses) and E1A (only Ad-iREP-2). In all vectors we used an N-terminally deleted E1A that reduces negative interference between E1A and p53/p300 and is known to allow wildtype-like viral replication (25). We additionally generated genetically identical control viruses (Ad-iREP-C1 for Ad-iREP-1, and Ad-iREP-C2 for Ad-iREP-2, respectively, Figure 3A) for expression of a nonfunctional shRNA-network containing scrambled hairpins, instead of the antiviral RNAi-network present in Ad-iREP-1/-2. Since we have characterized the p53-status of the used cell lines with regard to the on/off-response of the prMinRGC-promoter (see Figure 1C), the comparison of Ad-iREP-1/-2 and their corresponding control viruses can be directly used to determine the influence of the antiviral RNAi-network.
Figure 3.
p53-selective expression of the antiviral RNAi-networks inhibits expression of adenoviral target genes in vitro. (A) Design of p53-selective, RNAi-controlled viruses containing either an inhibitory RNAi-network (Ad-iREP-1 and Ad-iREP-2) or a noninhibitory RNAi-network (consisting of scrambled hairpins, Ad-iREP-C1 and Ad-iREP-C2) under control of the prMinRGC-promoter. All viruses expressed an N-terminally deleted version of E1A under control of the E1A-wt promoter (prE1A). Ad-iREP-C2 and Ad-iREP-2 were provided with additional target sites by insertion into in the 3'-UTR of E1A-mRNA. (B) Shuttle plasmids were cotransfected with the luciferase reporter pGL2-Prom as internal standard into cells with different p53-status. E1A-silencing was investigated after 24 h posttransfection by western blot analyses. Transfection efficacy was controlled by luciferase measurements (upper panel). Similar analyses were performed 24 h after cell infection with corresponding adenoviral particles at MOI 0.05 (lower panel). (C) A549 and Huh7 cells were infected with Ad-iREP-1 and Ad-iREP-2 at MOI of 0.05. Total RNA was isolated and mature miRNAs of miR30-shRE1A-736 (left) and miR30-shRE4-33056 (right) were quantified using the miR-Q-RT-PCR method (mean ± SD) 12, 24 and 48 h later. The data confirm that the RNAi-network is efficiently expressed in p53-positive cells but not in p53-dysfunctional cells. (D) Adenoviral expression of different RNAi-targets (E1A, E1B and pTp) and fiber as nontarget was investigated in cells with different p53-status following infection with Ad-iREP-1, Ad-iREP-2 and corresponding controls (A549: upper panel; Huh7: lower panel). At time-points indicated mRNA levels were determined by RT-qPCR (mean ± SD). The data show that p53-selective expression of the antiviral RNAi-network by Ad-iREP-2 successfully silenced viral target genes.
First, we analyzed E1A protein levels after transfection of the adenoviral shuttle plasmids into cells with different p53-status. As shown in the upper panel of Figure 3B, transfection of both Ad-iREP-1 and Ad-iREP-2 shuttle plasmids, but not the control vectors, led to down regulation of E1A under p53-positive conditions. In contrast, no difference in E1A-levels was observed in p53-dysfunctional cells suggesting that effective repression of E1A could be dependent of p53-selective activation of RNAi. Important to note that E1A-silencing after transfection of Ad-iREP-2 was more effective than Ad-iREP-1 suggesting that the additional target-site within the E1A-3′-UTR conferred an improved accessibility for RNAi. It has been shown that transient transfection of cells is associated with significant cell stress followed by a strong accumulation of p53 (26). It has to be considered that viral infections, in particular after low-dose application, may induce p53 to a lower extent compared to the transient transfection situation which could lead to an overestimation of transfection results as described above. Consequently, a less potent p53-response upon viral infection could compromise effective induction of antiviral RNAi. We therefore compared E1A-expression after cell infection with the corresponding viruses. Consistent with the transfection results, E1A-levels were significantly decreased in p53-positive cells after infection with Ad-iREP-2 (Figure 3B, lower panel). E1A was not silenced after infection with the control virus Ad-iREP-C2. Similar E1A-levels after infection of p53–dysfunctional Huh7 cells with both viruses confirmed p53-selectivity of the E1A repression observed in the p53-positive cell lines. Interestingly, E1A-silencing could not be observed after infection with Ad-iREP-1. In the particular context of viral infection, Ad-iREP-1-dependent RNAi insufficiently suppressed E1A thereby confirming the beneficial influence of the manipulations performed in the 3′–UTR of E1A. This finding also suggests that a low E1A threshold is sufficient to allow the onset of replication. This observation also underlines the importance of stringent regulative mechanisms for efficient suppression of E1A to allow targeted regulation of viral replication.
Next, we directly determined the expression of the antiviral RNAi-network by detection of mature miRNA using the PCR-based miR-Q method as described previously (22). Two hairpins against E1A and E4 were selected to exemplify successful RNAi-network processing. We could show a strong and equivalent increase of both investigated hairpins after infection of p53-positive cells with Ad-iREP-1 and Ad-iREP-2 (Figure 3C). Since we could not detect significant amounts of hairpins in p53-mutant Huh7 cells at early stages, our data clearly demonstrate that the RNAi-network was expressed in a p53-selective manner. In Huh7 cells only a small background could be detected at 48 h, a time-point that includes a full replication cycle. Interestingly, Ad-iREP-1 expressed comparable amounts of the RNAi-network as Ad-iREP-2 at early stages though this virus failed to repress E1A efficiently. Again, this observation confirms the relevance of the E1A-3′UTR manipulations to achieve better accessibility of the target messenger. We then monitored expression of several targeted genes (E1A, E1B and pTp) following infection of p53-positive A549 cells to investigate the activity of other RNAi-network components. As shown in Figure 3D (upper panel), the expression of target genes was effectively reduced. In contrast, this was not the case following infection of p53-mutant cells (Figure 3D, lower panel). However, it has to be considered in these experiments that levels of the corresponding mRNAs are influenced by the number of viral genomes if replication is indeed allowed or compromised. We therefore determined the expression of the viral fiber-mRNA as a non-target of the antiviral RNAi-network. We could measure a significant reduction of fiber suggesting that replication of viral DNA was actually alleviated. But, the non-target fiber was less reduced than the true targets E1A, E1B and pTp suggesting that the hairpins directed against E1B and pTp actually exhibited RNAi. However, it is impossible to exactly calculate the contribution of the other network components since these genes are more or less downstream functions of genome amplification in the course of replication and thus of E1A expression.
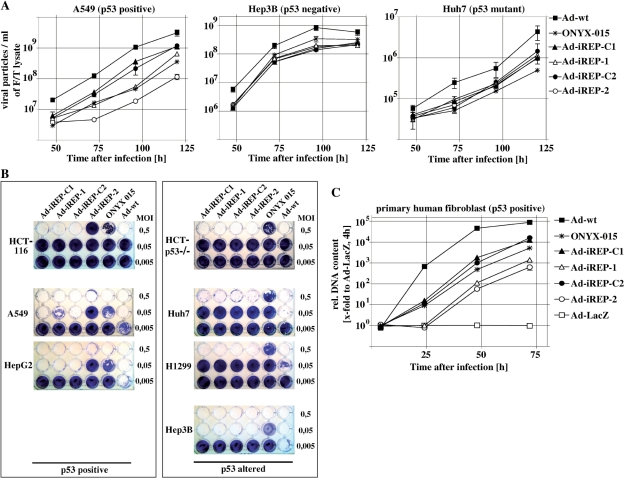
Ad-iREP-2 mediates efficient lysis of p53-dysfunctional cells but is selectively attenuated in normal cells
Next, we investigated whether the achieved knockdown of adenoviral genes was sufficient to inhibit the generation of infectious progeny. We therefore monitored time-dependent progression of adenoviral titers with regard to the cellular p53-status (Figure 4A). For comparative purposes we also included the adenovirus wildtype and ONYX-015, an adenovirus mutant that has been initially described to replicate preferentially in p53-dysfunctional cells and has already been tested in clinical trials. In p53-positive, A549 cells efficient attenuation of viral replication could be observed only after infection with Ad-iREP-2, when compared to the non-regulated Ad-iREP-C2 as standard. In line with the lower efficacy of E1A-silencing described above, attenuation of Ad-iREP-1 was modest. Since all RNAi-controlled viruses replicated to a similar extent compared to their control viruses in p53-dysfunctional Huh-7 and Hep3B cells we confirmed that the inhibition of viral replication was due to antiviral RNAi-activity in response to active p53. These results could be further confirmed in other cell lines such as p53-positive HepG2 and p53-deleted H1299 cells (data not shown). p53-selectivity of Ad-iREP-2 was improved compared to ONYX-015, since ONYX-015 replicated like Ad-iREP-1 in A549 cells. In contrast, ONYX-015 showed weaker replication than RNAi-controlled viruses in all p53-altered cell lines except Hep3B. However, Hep3B cells are known to intrinsically support adenoviral replication by expression of HBx (27). Most likely due to extensive genetic manipulations and the used N-terminally deletion mutant of E1A, all RNAi-viruses showed slightly reduced replication when compared to the wildtype adenovirus.
Figure 4.
Ad-iREP-2 dependent replication and cell lysis are selectively inhibited in the presence of active p53. (A) Cells were infected at MOI 0.01, harvested at indicated time-points and lysed by repeated freezing/thawing to determine viral titers. In A549, but not in Hep3B cells or Huh7 cells, replication of Ad-iREP-2 is significantly inhibited compared to the control Ad-iREP-C2 [mean ± SD, statistical analyses for A549 at 120 h: Ad-iREP-1 versus Ad-iREP-C1 (P = 0.044); Ad-iREP-2 versus Ad-iREP-C2 (P = 0.003); Ad-iREP-2 versus Ad-iREP-1 (P = 0.01); Ad-iREP-2 versus ONYX-015 (P = 0.030); Ad-iREP-1 versus ONYX-015 (not significant); Ad-iREP-C1 versus Ad-iREP-C2 (not significant)]. (B) For cytolysis assays cells were infected at different MOIs as indicated. As demonstrated by crystalviolet stainings, Ad-iREP-2 shows improved lytic efficacy and p53-selectivity compared to ONYX-015. (C) Primary human IMR-90 fibroblasts were infected at MOI 0.1. To mediate efficient transduction viral particles were treated with CARex-Tat as described in the methods section. At indicated time-points, total DNA was isolated and the viral DNA-content was determined. The results indicate that DNA-replication of both Ad-iREP-1 and Ad-iREP-2 was significantly attenuated in primary human fibroblasts [mean ± SD, statistical analyses for A549 at 72 h: Ad-iREP-1 versus Ad-iREP-C1 (P = 0.008); Ad-iREP-2 versus Ad-iREP-C2 (P = 0.003); Ad-iREP-2 versus Ad-iREP-1 (P = 0.041); Ad-iREP-2 versus ONYX-015 (P = 0.012); Ad-iREP-1 versus ONYX-015 (P = 0.017); Ad-iREP-C1 versus Ad-iREP-C2 (not significant)].
Effective oncolysis is a central requirement for conditionally replicating adenoviruses as extensive genetic manipulations can also affect lytic efficacy. Comparing both p53-selectivity and lytic efficacy of RNAi-viruses, ONYX-015 and Ad-wt in vitro, we found that only Ad-iREP-2-mediated oncolysis was significantly attenuated in p53-positive cells, since Ad-iREP-1/-2 and the non-regulated Ad-iREP-C1/-C2 lysed p53-dysfunctional Huh7 and H1299 cell to the same extent (Figure 4B). Furthermore, we could observe that Ad-iREP-2 oncolysis was attenuated in p53-positive HCT-116 cells but not in their isogenic counterpart HCT-116 p53–/–. Interestingly, Ad-iREP–1-mediated lysis was comparable to Ad-iREP-C1 showing that the observed differences in viral titers were insufficient to result in p53-selective cell lysis. With exception of Hep3B, wherein only small differences in oncolysis could be observed, and consistent with the titration results described above, Ad-iREP-2 was able to lyse p53-dysfunctional cells significantly more efficient than ONYX-015. Though RNAi-viruses did not reach the lytic efficacy of the wildtype, our data demonstrate that Ad–iREP–2 combined higher selectivity for p53 and improved lytic efficacy compared with ONYX-015.
Transformed cells provide a convenient microenvironment for viral replication and may not reflect the spectrum of a natural p53-response in primary cells. We therefore investigated viral replication in primary adult human fibroblasts (Figure 4C). However, fibroblasts are highly resistant against adenoviral transduction. For efficient transduction at low MOI, viral particles were pretreated with the bispecific CARex-Tat as described before (18). This allows for efficient cell entry and subsequent monitoring of replication for at least one cycle. Due to these constraints, the more sensitive determination of viral DNA amplification as indicator of replication was chosen. Our results demonstrate that DNA replication of both RNAi-controlled viruses Ad-iREP-1 and Ad-iREP-2 was significantly inhibited compared to the control viruses thus demonstrating the p53-selective effect of antiviral RNAi in primary human cells. In these cells both Ad-iREP-1 and Ad-iREP-2 showed improved selectivity compared to ONYX-015.
RNAi-controlled adenoviral replication allows for efficient oncolysis in mouse xenograft models and leads to reduced intrahepatic load of viral DNA
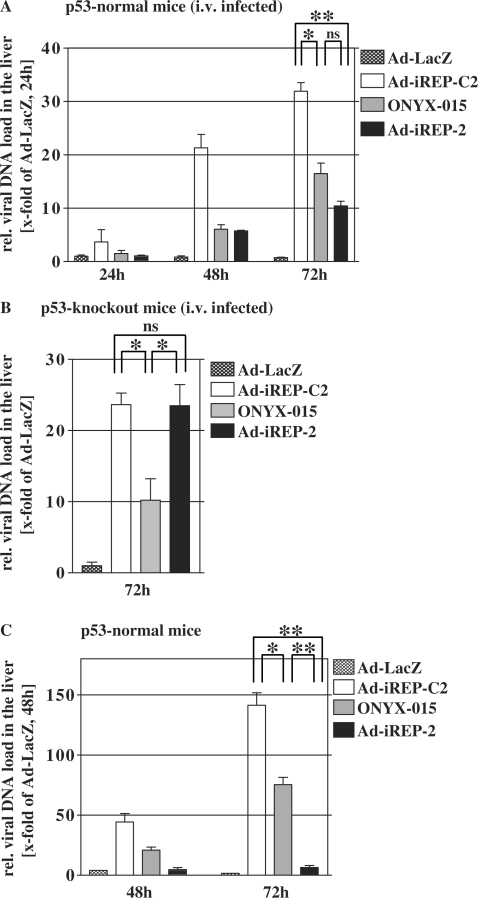
In the course of a tumor treatment with conditionally replicating adenoviruses, viral progeny is permanently released by infected malignant tissue. Due to the dominant hepatotropism of adenoviral particles in vivo, adenoviral hepatitis poses a considerable threat to the liver. Therefore, tight control of conditional replication is decisive to reduce the liver burden. We addressed this question by experiments in mice though we were aware that adenovirus does not produce infectious progeny in murine cells. However, murine cells almost normally amplify the adenoviral genomes which allows for an estimation of a possible liver threat. In a first experiment, we tested intrahepatic load of adenoviral DNA after systemic injection of virus in p53-normal mice (Figure 5A), and p53-knockout mice respectively (Figure 5B). We could observe that viral DNA-levels after application of replication-competent virus types increased in time. This was a result of DNA-replication since application of a replication-incompetent Ad-LacZ-virus did not lead to increasing levels of intrahepatic adenoviral DNA. Compared to the control Ad-iREP-C2, the amount of Ad–iREP–2-DNA was substantially reduced in the liver of normal mice but not of p53-knockout mice suggesting that p53-dependent expression of the antiviral RNAi-network inhibited viral replication in vivo. In contrast, Ad-iREP-2 and the control-virus replicated to a similar extent in p53-knockout mice demonstrating that inhibited replication in the livers of normal mice can be clearly attributed to the p53-dependent function of the RNAi-network.
Figure 5.
Ad-iREP-2 administration in vivo leads to reduced intrahepatic viral DNA-replication and viral DNA-load in the livers of treated mice. p53-normal (A) or p53-knockout mice (B) were injected intravenously with 1 × 107 infectious particles. At indicated time-points, DNA was prepared from liver tissue and the content of viral DNA was determined by qPCR. In normal mice, Ad-iREP-2 replication was inhibited compared to the control Ad-iREP-C2 whereas both viruses replicated comparably in p53 knockout mice thus proving the inhibitory activity of the RNAi-network in vivo (mean ± SD; *P < 0.05; **P < 0.01; ns = not significant). (C) Viral replication in the liver was investigated in a model that simulates a virotherapeutic setting. 5 × 105 Preinfected Hep3B cells/mouse () were injected with into the tail vein and viral DNA load was investigated in liver tissue as described above. The data suggest that Ad-iREP-2 replication is almost completely inhibited and selectivity is significantly improved compared to the control Ad-iREP-C2 (mean ± SD; *P < 0.05; **P < 0.01).
In this experimental setup in normal mice, Ad-iREP-2 replication was only slightly lower than ONYX-015. However, intravenous injections do not sufficiently reflect the constant release of adenoviral progeny that can be expected during virotherapy of tumors. The pulsed entry of high particle doses results in overinfection of peripheral regions in the portal fields of the liver and it has been shown that high multiplicities of infection can result in diminished selectivity of conditionally replicating adenoviruses (28). We therefore investigated adenoviral liver load in an improved murine model of viral oncolysis that simulates a more constant release of viral progeny into the blood flow. To this end we injected preinfected Hep3B cells into the tail vein of nude mice. Hep3B cells provide a comparable production and release of the investigated viruses (see Figure 4A and B). As shown in Figure 5C, we observed a strong DNA-replication of the control virus and to a slightly lesser extent of ONYX-015 in the livers of infected mice. In contrast, replication of Ad-iREP-2 was almost undetectable, confirming the tight control of viral replication due to p53-transcriptionally regulated RNAi-network expression in vivo.
To investigate the efficacy of Ad-iREP-2-virotherapy in vivo, subcutaneously grown H1299 and Hep3B tumors were treated with repeated intratumoral adenovirus injections (Figure 6A and B). Subsequent monitoring of tumor growth revealed that both Ad-iREP-2 and ONYX-015 comparably retarded the growth of Hep3B-tumors. In contrast, Ad-iREP-2 lysed H1299-tumors more effectively compared to the rather ineffective ONYX-015. However, intratumoral application of Ad-iREP-2 resulted in a strong inhibition of tumor growth in this aggressively growing tumor model. On the other hand, systemic delivery of oncolytic adenoviruses following a similar administration scheme did not result in a significant therapeutic benefit due to relatively poor tumor transduction (data not shown).
Figure 6.
Ad-iREP-2 virotherapy efficiently inhibits growth of p53-inactive tumors in vivo. For the investigation of oncolytic efficacy in vivo, subcutaneously grown Hep3B (A) or H1299 (B) tumors on nude mice were treated by repeated intratumoral injection of 1 × 109 pfu/injection. Virus administrations are marked by arrows. Tumor growth was monitored until the trial was aborted due to the tumor burden of controls. Ad-REP-2 application resulted in efficient tumor growth inhibition in both xenograft models and was improved compared to ONYX-015 in H1299 [mean ± SD, statistical analyses for Hep3B tumor at Day 20: Ad-iREP-2 versus w/o virus (P = 0.035); Ad-iREP-2 versus ONYX-015 (not significant); ONYX-015 versus w/o virus (P = 0.047); for H1299 tumor at Day 20: Ad-iREP-2 versus w/o virus (P = 0.004); Ad-iREP-2 versus ONYX-015 (P = 0.005); ONYX-015 versus w/o virus (not significant)].
Together, our results indicate that Ad-iREP-2 regulates its own replication in response to p53-dependent onset of antiviral RNAi. Furthermore, Ad-iREP-2 is an effective oncolytic adenovirus for the oncolytic therapy of p53-altered tumors.
DISCUSSION
The special attractiveness of tumorselective viral replication is the idea that oncolytic viruses reproduce themselves at the therapeutic locus functioning thereby like molecular surgery. Inactivation of the tumor suppressor p53 represents an excellent molecular target for the development of broadly applicable anti-neoplastic agents such as oncolytic viruses. In our study, we could demonstrate that the engineered oncolytic adenovirus Ad-iREP-2 selectively lyses cells with missing transcriptional activity of p53, a dysfunction that encompasses most tumor-associated p53 alterations including gene loss, inactivating mutations and expression of proteins that negatively interfere with p53-function and stability. Interdependence between Ad-iREP-2-replication and p53-transcriptional deficiency was achieved by p53-dependent onset of vector-encoded antiviral RNAi that suppresses essential adenoviral genes such as E1A, E1B, E4, pTp and AdPol. Since we could prove effective replication and potent lytic properties in p53-dysfunctional cells and effective inhibition of tumor growth in vivo, Ad-iREP-2 represents an oncolytic vector for the majority of human cancers. Regarding p53-selectivity and lytic potency, Ad–iREP–2 was significantly improved compared to ONYX-015, an E1B-55k-deleted adenovirus that has been initially proposed to selectively replicate in p53-deficient cells. Though ONYX-015 revealed favorable toxicity and therapeutic efficacy in clinical studies, at least in combination with chemotherapy, it has been reported that ONYX-015 replication does not correlate with the p53-status (29,30) but with other molecular characteristics of the target cell (31,32). These findings can explain the significant attenuation of ONYX-015 in many cancer cells and suggest that ONYX-015-replication is only effective in a limited subgroup of tumors that can intrinsically compensate for the E1B-55k-deficiency. Consistently, in our assays ONYX-015 replicated less efficiently than Ad-iREP-2 in most tumor cells except in Hep3B that intrinsically support adenoviral replication by expression of the HBV-derived X-protein (27). Therefore deletion of crucial viral genes such as E1B-55k can result in a severe loss of oncolytic potency. To ensure potent replication in all cancer cells Ad-iREP-2 provides expression of wildtype E1B-55k that exhibits several essential functions such as support of viral mRNA-export, host protein shut-off and enhancement of cyclin E-expression (33,34).
On the other hand, E1B-55k is a potent inactivator of p53-dependent transcriptional activity, either alone or in concert with E4orf6. Additionally, p53-dependent transcription appears to be generally inhibited by adenoviral replication, even in the presence of mutated E1B-55k and accumulated p53, suggesting that cellular p53-dependent transcription and adenoviral replication are mutually exclusive (35). Therefore, with regard to the presence of wild-type E1B-55k, stringent regulative tools are needed to prevent that the balance turns towards the onset of replication. E1A is expressed first after adenoviral DNA has entered the nucleus and is essential for cell cycle entry, feed-forward activation of viral promoters and viral replication. We could demonstrate that the p53-dependent expression of an antiviral RNAi-network is able to efficiently inhibit E1A-expression resulting in selective attenuation of viral replication in both primary human cells and transformed human cells with active p53.
In another study, it has been demonstrated that the adenovirus 01/PEME, targeting both p53-dysfunction and E2F-activation in tumors, was attenuated in primary human cells (25). However, the p53-dependent expression of an E2F-antagonist, used by 01/PEME to achieve conditional replication, was not sufficient to inhibit replication in A549 and HepG2 cells, despite transcriptionally active p53. As Ad-iREP-2-replication was significantly inhibited in these cell lines, we could prove the regulative stringency of the antiviral RNAi-network.
Finally, the p53-dependence of RNAi-controlled viral replication was also confirmed in vivo. Assessment of viral DNA-load in the livers of intravenously infected mice revealed a significant inhibition of Ad-iREP-2 replication compared to the control virus Ad-iREP-C2 that express a scrambled network in response to p53. This inhibition could not be observed after infection of p53-knockout mice confirming the p53-selective effect of the antiviral RNAi-network. However, both intravenous and intratumoral injection of oncolytic viruses result in a short term systemic viral burst constraining a reasonable evaluation of the specificity of viral replication. In an improved in vivo model that simulates constant, systemic release of viral progeny during oncolytic therapy, we could demonstrate an almost complete inhibition of Ad-iREP-2-replication, confirming the high specificity of RNAi-controlled viral replication in vivo.
Natural microRNAs frequently occur as long clustered transcripts that are expressed by RNA-polymerase II, and polycistronic plasmid vectors for expression of RNAi-network have already been established (36) suggesting that multiple short-hairpin or microRNA-transcripts can be used as convenient tool for differential gene regulation (37). By application of a vector-encoded antiviral RNAi-network to restrict adenoviral replication to tumor cells we were able to demonstrate that RNAi is powerful enough to interfere with viral replication and provides a mean to manipulate the virus tropism. We achieved a decisive improvement of the selective regulation by positioning additional miRNA target sites within the 3′-UTR of E1A to render it more susceptible to RNAi. It can therefore be assumed that the antiviral RNAi-network concept leaves enough space for further optimization as merely this single manipulation led to a visible advancement. RNAi-networks offer an outstanding versatility regarding the number of genes that can be synchronously targeted. As they need only little space compared to conventional protein-based gene regulation systems, they can be easily incorporated in viral vectors with limited capacity for the uptake of heterologous DNA.
In conclusion we have shown that virus-encoded antiviral RNAi can be used for the self-control of viral vector replication. The tumor selectivity of our virus was achieved by an RNAi-network directed against essential viral genes that is solely expressed in response to transcriptionally active p53. RNAi-controlled viral replication showed an improved specificity and antitumoral efficacy in vitro and in vivo compared to an isogenic control virus that expresses a nonfunctional RNAi-network. As our concept is principally applicable to other transcriptionally regulated DNA viruses, the strategy of antiviral RNAi-networks provides a general method for the generation of a broad spectrum of selective viruses.
FUNDING
EU-Commission [Contract No. 12 948 ‘Netsensor’], the German Research Council [KFO119/1-1] and the Mildred-Scheel-Foundation. Funding for open access charge: EU-Commission and the German Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 2.Wilda M, Fuchs U, Wossmann W, Borkhardt A. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi) Oncogene. 2002;21:5716–5724. doi: 10.1038/sj.onc.1205653. [DOI] [PubMed] [Google Scholar]
- 3.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 4.Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, Waltemathe M, Gosling T, Flemming P, Malek NP, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc. Natl Acad. Sci. USA. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 7.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl Acad. Sci. USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung YS, Kim MK, Lee WJ, Kang C. Silencing E1A mRNA by RNA interference inhibits adenovirus replication. Archiv. Virol. 2007;152:1305–1314. doi: 10.1007/s00705-007-0951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 10.Cullen BR. Viruses and microRNAs. Nat. Genet. 2006;38(Suppl.):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 11.Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol. Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- 12.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 13.Ylosmaki E, Hakkarainen T, Hemminki A, Visakorpi T, Andino R, Saksela K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J. Virol. 2008;82:11. doi: 10.1128/JVI.01608-08. 009–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl Acad. Sci. USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 16.Jounaidi Y, Doloff JC, Waxman DJ. Conditionally replicating adenoviruses for cancer treatment. Curr. Cancer Drug Targets. 2007;7:285–301. doi: 10.2174/156800907780618301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhnel F, Zender L, Wirth T, Schulte B, Trautwein C, Manns M, Kubicka S. Tumor-specific adenoviral gene therapy: transcriptional repression of gene expression by utilizing p53-signal transduction pathways. Cancer Gene Ther. 2004;11:28–40. doi: 10.1038/sj.cgt.7700632. [DOI] [PubMed] [Google Scholar]
- 19.Mizuguchi H, Kay MA. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 1998;9:2577–2583. doi: 10.1089/hum.1998.9.17-2577. [DOI] [PubMed] [Google Scholar]
- 20.Kuhnel F, Schulte B, Wirth T, Woller N, Schafers S, Zender L, Manns M, Kubicka S. Protein transduction domains fused to virus receptors improve cellular virus uptake and enhance oncolysis by tumor-specific replicating vectors. J. Virol. 2004;78:13743–13754. doi: 10.1128/JVI.78.24.13743-13754.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YA, Nemunaitis J, Samuel SK, Chen P, Shen Y, Tong AW. Antitumor activity of an oncolytic adenovirus-delivered oncogene small interfering RNA. Cancer Res. 2006;66:9736–9743. doi: 10.1158/0008-5472.CAN-06-1617. [DOI] [PubMed] [Google Scholar]
- 22.Sharbati-Tehrani S, Kutz-Lohroff B, Bergbauer R, Scholven J, Einspanier R. miR-Q: a novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol. Biol. 2008;9:34. doi: 10.1186/1471-2199-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 24.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 25.Ramachandra M, Rahman A, Zou A, Vaillancourt M, Howe JA, Antelman D, Sugarman B, Demers GW, Engler H, Johnson D, et al. Re-engineering adenovirus regulatory pathways to enhance oncolytic specificity and efficacy. Nat. Biotechnol. 2001;19:1035–1041. doi: 10.1038/nbt1101-1035. [DOI] [PubMed] [Google Scholar]
- 26.Renzing J, Lane DP. p53-dependent growth arrest following calcium phosphate-mediated transfection of murine fibroblasts. Oncogene. 1995;10:1865–1868. [PubMed] [Google Scholar]
- 27.Schaack J, Maguire HF, Siddiqui A. Hepatitis B virus X protein partially substitutes for E1A transcriptional function during adenovirus infection. Virology. 1996;216:425–430. doi: 10.1006/viro.1996.0079. [DOI] [PubMed] [Google Scholar]
- 28.Bilsland AE, Merron A, Vassaux G, Keith WN. Modulation of telomerase promoter tumor selectivity in the context of oncolytic adenoviruses. Cancer Res. 2007;67:1299–1307. doi: 10.1158/0008-5472.CAN-06-3000. [DOI] [PubMed] [Google Scholar]
- 29.Goodrum FD, Ornelles DA. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothmann T, Hengstermann A, Whitaker NJ, Scheffner M, zur HH. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J. Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, et al. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell. 2004;6:611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea CC, Soria C, Bagus B, McCormick F. Heat shock phenocopies E1B-55K late functions and selectively sensitizes refractory tumor cells to ONYX-015 oncolytic viral therapy. Cancer Cell. 2005;8:61–74. doi: 10.1016/j.ccr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X, Rao XM, Gomez-Gutierrez JG, Hao H, McMasters KM, Zhou HS. Adenovirus E1B55K region is required to enhance cyclin E expression for efficient viral DNA replication. J. Virol. 2008;82:3415–3427. doi: 10.1128/JVI.01708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch P, Gatfield J, Lober C, Hobom U, Lenz-Stoppler C, Roth J, Dobbelstein M. Efficient replication of adenovirus despite the overexpression of active and nondegradable p53. Cancer Res. 2001;61:5941–5947. [PubMed] [Google Scholar]
- 36.Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung KH, Hart CC, Al Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]