Abstract
Disease-associated SNPs detected in large-scale association studies are frequently located in non-coding genomic regions, suggesting that they may be involved in transcriptional regulation. Here we describe a new strategy for detecting regulatory SNPs (rSNPs), by combining computational and experimental approaches. Whole genome ChIP-chip data for USF1 was analyzed using a novel motif finding algorithm called BCRANK. 1754 binding sites were identified and 140 candidate rSNPs were found in the predicted sites. For validating their regulatory function, seven SNPs found to be heterozygous in at least one of four human cell samples were investigated by ChIP and sequence analysis (haploChIP). In four of five cases where the SNP was predicted to affect binding, USF1 was preferentially bound to the allele containing the consensus motif. Allelic differences in binding for other proteins and histone marks further reinforced the SNPs regulatory potential. Moreover, for one of these SNPs, H3K36me3 and POLR2A levels at neighboring heterozygous SNPs indicated effects on transcription. Our strategy, which is entirely based on in vivo data for both the prediction and validation steps, can identify individual binding sites at base pair resolution and predict rSNPs. Overall, this approach can help to pinpoint the causative SNPs in complex disorders where the associated haplotypes are located in regulatory regions. Availability: BCRANK is available from Bioconductor (http://www.bioconductor.org/).
INTRODUCTION
Human genetic variation underlies a majority of phenotypic differences between individuals, including susceptibility to disease. The most common form of genetic variation are the single nucleotide polymorphisms (SNPs), of which there are 9–10 million variants with minor allele frequency MAF ≥0.05, in the human genome (1). Large international efforts, like the HapMap project, have identified and genotyped 25–35% of these common SNPs in several populations (1). A significant fraction of human genes display differences in expression between individuals and between populations (2,3). Furthermore, gene expression patterns are heritable and their variation is genetically determined both in cis and trans (2,4–6). Some of the observed differences could arise due to environmental or physiological, rather than genetic factors, but by comparing alleles within the same cellular context, clear allele-specific gene expression differences have been observed (7,8).
Quantitative differences in gene expression are at least partially responsible for phenotypic variation between individuals, including susceptibility to disease and drug responsiveness. A common characteristic among disease-associated SNPs in complex diseases is that there are several predisposing loci, preferentially in non-coding intronic or intragenic sequences, each with a small effect (9). In some instances, differences in gene expression of a nearby gene have been observed between the predisposing and the other allele (10). This implies that quantitative rather than qualitative differences in gene expression could be important in common diseases, further suggesting that causative genetic variants may occur within the regulatory sequences.
Functional sequences not coding for proteins can be divided in various groups, e.g. non-coding RNAs, sequences regulating splicing, and with a significant fraction corresponding to those devoted to transcriptional regulation i.e. promoters, enhancers, silencers, etc. (11,12). Our understanding of the human transcriptional regulatory sequences is rather limited, but the recent large-scale technologies ChIP-chip (13) and ChIP-seq (14) have dramatically increased our knowledge of the non-coding fraction of our genome (11,15,16). However, the impact of normal genetic variation in transcriptional regulatory sequences is largely unknown and very few studies have used unbiased in vivo approaches to investigate this issue (17–19).
Large amounts of information concerning the location of common genetic variants and regulatory sequences have accumulated in recent years, but no clear strategies to combine them are currently available. Here we present novel methods both for prediction and validation of DNA-binding sites. For binding site prediction we have developed a novel algorithm called ‘predicting Binding site Consensus from RANKed sequences’ (BCRANK). BCRANK identifies short consensus sequences that occur more frequently among the top scoring regions, when compared to regions with lower enrichment. The algorithm is based on a heuristic search strategy that requires an initial guess as input, and it can either be used for scoring of known motifs or for ab initio prediction of DNA-binding sites. Here we applied the method to whole genome ChIP-chip data for USF1 (16) to predict TF-binding sites. The BCRANK predictions were compared to results of two other motif search programs, MDscan (20) and DRIM (21), which also take the rank of DNA sequences into account. Our results show that BCRANK has advantages over the other methods, the most important being that it does not require the motif length to be known a priori. A further advantage is that BCRANK is available from Bioconductor, which makes it easy to use and to include into various analysis pipelines.
Recently, generation of in vivo transcription factor binding maps has received great attention (11,22,23). In most cases, downstream analysis has focused on identifying or inferring the genes regulated by a given transcription factor. However, we believe such transcription factor binding maps are valuable resources to increase our knowledge of the effects of genetic variation at regulatory sequences and its impact on gene expression and human disease. To the best of our knowledge, the method presented here represents the first attempt to directly use in vivo transcription factor binding data as a source for predicting regulatory SNPs (rSNPs).
More specifically, our strategy to verify the predicted binding sites and to identify potential rSNPs consists of haploChIP experiments to detect allelic differences in protein–DNA interactions. We identify heterozygous SNPs in our predicted binding sites and discriminate between the alleles, using the two alleles as internal controls for each other. In this approach, binding to the distinct sequence in each allele is just affected by the sequence itself and not by trans effects or environmental differences. By performing the same experiments for various other proteins, we do not only fulfill the initial aim to validate predicted binding sites at base pair resolution, but also investigate how other proteins may be correlated with the binding event. Furthermore, we examine how USF1 binding can affect transcription.
METHODS
ChIP-chip data
USF1 ChIP-chip data was obtained from an experiment using the Affymetrix GeneChip Human Tiling 2.0R Array set (seven arrays set). The raw microarray data is available from ArrayExpress with accession number E-TABM-314. BCRANK works best when used on a relatively large number of regions where DNA-binding sites are much more frequently occurring in the top of the list when compared to the bottom. Therefore, we relaxed the cut-offs, as compared to the previous study (16), to obtain the top 5211 regions and extended each region to 1500 bp centered on a peak in the ChIP-chip data.
BCRANK
BCRANK is implemented in R and available as an open source package from Bioconductor (http://www.bioconductor.org/). BCRANK performs a breadth-first search through the space of consensus sequences to detect a solution that is optimal with respect to a scoring function. Optionally the search steps can be omitted, which is useful when the aim is to rank previously established motifs by their BCRANK scores. The behavior of the algorithm is mainly determined by the scoring function, the definition of neighborhood (sequences similar to a consensus sequence) and by the initial starting point for the search. Moreover, two optional penalties, P1 and P2, can give relatively higher scores to motifs that are believed to be more important. P1 penalizes consensus sequences with many redundant bases, i.e. other bases than A, C, G or T. P2 ensures that the resulting consensus will not be frequently occurring as a repetitive element in the enriched regions. For implementation details, see Supplementary Data.
Sequencing
From the 140 candidate SNPs, we selected those with heterozygosity above 0.1. This stringent cut-off was used due to our limited number of samples, which makes it unlikely to detect heterozygous cases for SNPs with low heterozygosity values. There were 23 SNPs above the threshold and all of them were sequenced in our four samples (HepG2, HT29, Colon1 and Colon2). Seven of these SNPs were found to be heterozygous in at least one of the samples. For each of the seven SNPs, ChIPs and genomic DNAs were obtained as previously described (24). Predicted USF1-binding sites containing SNPs were selected and regions spanning such sites were PCR amplified, using ChIP and genomic DNAs as templates. PCR products were purified by ExoSap (USB, General Electrics) and used for sequencing reactions according to ABI Prism BigDye Terminator 3.1 Kit (Applied Biosystems) instructions. Sequences were obtained on an ABI 3700 automated sequencer (Applied Biosystems). Electrophenograms were visualized using Sequence Scanner v1.0 (Applied Biosystems).
Allele quantification and statistical test
The heights of two peaks (peak signal) corresponding to a heterozygous SNP were obtained using Sequence Scanner v1.0 in several replicates for each SNP and ChIP or genomic DNA. Direct sequencing has previously been used for quantitative genotyping and quantitative measurement of allelic specific gene expression (25,26). If s1 and s2 are the peak signals for the two alleles of a SNP, then the allele signal s1/(s1 + s2) is a value measuring the relative allele abundance ranging from 0 (homozygous for allele2) to 1 (homozygous for allele1). We consistently selected s1 to be the allele containing the predicted USF1-binding sequence. For each SNP, allele signals were calculated as above for all of the replicates. Each sample was examined using at least three biological replicates. A two tailed t-test was used to determine whether the allele signals were significantly different for ChIP and genomic DNA.
RESULTS
BCRANK predicts TF-binding sites
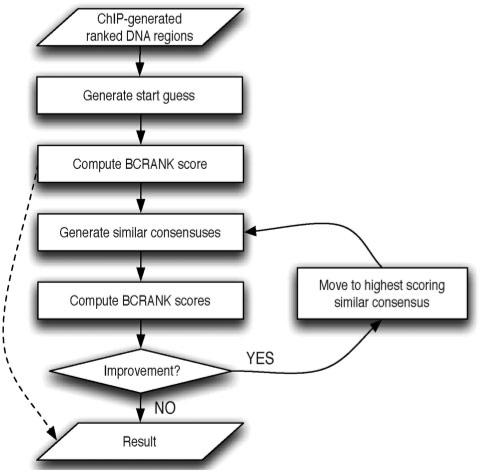
BCRANK takes a list of genomic regions ranked by ChIP-enrichment as input and, using a heuristic search strategy, outputs short DNA sequences that are overrepresented among the top scoring regions. The workflow is outlined in Figure 1. First a scoring function is applied to an initial short consensus, about 10 bases or so, typically generated by random sampling of IUPAC nucleotide symbols. Then all consensus sequences with similarity to the start guess are evaluated using the same function and the one with highest score is kept as the starting point for the next iteration. Once the algorithm can no longer find any higher scoring similar consensus, the algorithm terminates and the locally optimal consensus is reported as a result. In order to increase the chance of detecting the globally optimal solution, the algorithm may be restarted several times using different random starting points. BCRANK is described in more detail in ‘Methods’ section and Supplementary Data.
Figure 1.
Overview of the BCRANK algorithm. A file containing DNA sequences, ranked by ChIP-enrichment, is given as input. Then a consensus sequence is generated, either at random or by manual selection, and its BCRANK score is computed. Optionally, BCRANK can be used to assign scores to previously known consensus sequences, and in such case the algorithm stops here, indicated by dotted line in the figure. Otherwise the algorithm will continue to optimize the consensus by constantly moving to a similar consensus with a higher BCRANK score until no further improvement is possible and a locally optimal solution is reported. The chance of finding the globally optimal solution can be increased by re-starting BCRANK several times with different random start guesses.
We ran BCRANK with 25 random restarts on a set of 5211 regions ranked by USF1 ChIP-chip signal from a whole-genome experiment on the human liver cell HepG2 (16), and the results are presented in Figure S1a. The highest scoring consensus was CACGTGAC, which is similar to what has been previously reported as USF1-binding motif by us and others (24,27,28). Interestingly, we detected CGGAAG as the second highest scoring consensus. This is similar to the binding sequence for GABPA, a protein we previously showed by ChIP to frequently bind USF1 positive regions (16), suggesting that BCRANK is capable of detecting binding sites also for transcription factors cooperating with the investigated protein. Usually the local optimum is found after a quite small number of search steps. For the top scoring result, 12 iterations were required to move from the start guess GDYBYCTKDK and arrive at the resulting CACGTGAC (see Figure S1b), and the maximum number of iterations required throughout all 25 restarts was 14. As with any heuristic search method it is not possible to know that a global optimum has been found, but a very strong indicator is that the same top-scoring motif is found from different random restarts.
We compared the results of BCRANK to two other de novo motif search methods, MDscan (20) and DRIM (21), that also take ranked DNA sequences as input. BCRANK starts from a motif with an initial length that can then be extended or shortened throughout the search steps. MDscan and DRIM do not extend and shorten motifs in the same way as BCRANK. MDscan only searches for motifs of a specified length and DRIM performs an exhaustive search on all sequences of an initial length and then uses promising motifs as seeds that may be expanded or shortened. We ran the methods with three different length parameters, 6, 8 and 10 bases. To evaluate the predictions we considered how frequent the E-box (CACGTG), the previously established USF1-binding sequence, occurred in the predicted sites. We also investigated how many of the bindings were in the 2518 stringent and 3771 relaxed USF1 regions as defined in our previous study (16). The results are summarized in Supplementary Data and Supplementary Table S1. BCRANK predicts CACGTGAC as the USF1-binding sequence regardless of the length of the start guess, while the results from the other methods are highly dependent on length parameter used. When certain length parameters are used the other methods performed equally well as BCRANK, according to the criteria above, but never better. Our results demonstrate that BCRANK is a method that can successfully identify binding motifs from whole-genome experimental data. Furthermore, BCRANK does not require the motif length to be known a priori, which we believe is a great benefit. All three methods output a number of alternative motifs that may be relevant. For example, the second highest scoring consensus predicted BCRANK has high similarity to a GABPA-binding motif, a protein shown to frequently bind to USF1 enriched regions (23). MDscan instead reports variations of the USF1-binding sequence and DRIM detects CG rich sequences (see Supplementary Table S1). For DRIM, the run time grows rapidly with the motif length because of the exhaustive search for motif seeds.
We matched our 1757 BCRANK predicted USF1-binding sites to build 126 of dbSNP (29) and found 114 SNPs within two bases of a CACGTGAC sequence. Another 26 SNPs were detected by considering such cases where the reference genome contains the non-consensus allele of a USF1-binding site, yielding a total of 140 SNPs inside or just outside a predicted USF1-binding sequence. They are all presented in a Supplementary Data file. Of our 140 candidate regulatory SNPs, 86 (61%) and 110 (79%) were included in the USF1 stringent and relaxed regions, respectively. Supplementary Figure S2a shows that 29% of these SNPs are within 1 kb of a transcription start site (TSS) of a protein coding gene, and that more than half of the SNPs are close to the TSS of a less well defined transcript. We also observed that many of the 140 candidate SNPs are located in the CG dinucleotide of the USF1 consensus binding sequence (Supplementary Figure S2b). In most of these cases the alternative allele of C/G is a T/A, suggesting that during evolution some of the cytosines at these CG dinucleotides were methylated and therefore more prone to mutation by deamination (30).
USF1 allele-specific binding validates BCRANK predictions
Of our 140 SNPs, we identified 23 SNPs with average heterozygosity above 0.1 and all of them were sequenced in the four different cell samples (see ‘Methods’ section). Seven SNPs were heterozygous in at least one of the samples, and these were named SNP1 through SNP7. All of them were in the USF1 stringent data set except one, SNP4, which was in the relaxed set. Five of them were located inside the predicted core binding sequence CACGTGAC. The two remaining (SNP2 and SNP5) were located just outside the core sequence. They can be seen as negative controls in this experiment since we hypothesize that a base change at those positions should not affect USF1 binding. qPCR analysis of USF1 and USF2 binding for all seven SNPs was in good agreement with the ChIP-chip data. We observed more than a 2-fold USF1 and USF2 ChIP signal over background in all cases, with the exception of SNP4 (see Supplementary Figure S3). As indicated above, this SNP did not pass our stringent ChIP-chip cut off, and accordingly the qPCR enrichments were 1.3–1.5-fold.
We first sequenced ChIP material for USF1 and detected significant allelic differences in four of the five SNPs located in the core binding site. No allelic differences were detected for our negative controls, SNP2 and SNP5. To further investigate the functional effects of the SNPs, we also sequenced ChIP material for additional regulatory proteins (e.g. USF2, POLR2A, H3K4me3, H3K27me3, H3ac and H3Cter) in the seven SNPs. The overall sequencing results are summarized in Table 1, Supplementary Table S2 and Figure 2.
Table 1.
Seven heterozygous SNPs in USF1 bound regions
| Name | SNP label | Sequencea | Heterozygous in |
|---|---|---|---|
| SNP1 | rs1867760 | AA[T/C]ACGTGACCC | HepG2, HT29 |
| SNP2 | rs2754775 | A[C/A]CACGTGACCA | HepG2 |
| SNP3 | rs16875109 | CTCA[T/C]GTGACCT | Colon1 |
| SNP4 | rs1544702 | CTCAC[G/A]TGACAT | Colon2 |
| SNP5 | rs4787645 | AGCACGTGAC[G/A]T | Colon1 |
| SNP6 | rs11696955 | GAC[A/G]CGTGACTT | HepG2 |
| SNP7 | rs9920753 | TTCACGTG[A/T]CAA | Colon2 |
The underlined bases are predicted USF1-binding sites. The last column indicates the samples where the SNP was heterozygous. Additional information about these SNPs, including ChIP-analysis results, is available in Supplementary Table S2.
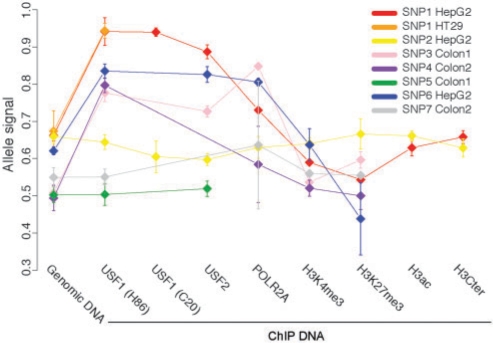
Figure 2.
Summary of the seven heterozygous SNPs. Each SNP is represented by a distinct color. SNP1 was the only one to be heterozygous in two samples (HepG2 and HT29) and it is therefore represented by two colors. Each point in the plot shows the average allele signal over all replicates in a given sample, for all proteins analyzed by ChIP. The interquartile range is displayed by error bars.
Allelic differences in USFs and POLR2A binding identifies putative regulatory SNPs
SNP1 is heterozygous both in HepG2 and HT29 and located in the first base of the core predicted USF1 site, at the third base of the sequence AA[C/T]ACGTGACCC. When sequencing the same region in USF1 ChIP material from the two cell lines we only found the C allele, which corresponds to the consensus USF1-binding sequence (see Figure 3A). This observation strongly indicates that (i) USF1 binds to the exact predicted binding site both in HepG2 and HT29 and (ii) USF1 binding occurs on the allele with the consensus and not on the other. We obtained the same results for two different USF1 antibodies, C20 and H86. We then extended the investigations by also examining USF2 and markers associated with chromatin state, H3K4me3, H3K27me3, H3ac, H3Cter and unphosphorylated POLR2A. As seen in Figure 3A, USF2 shows the same pattern as USF1, which is likely explained by the established fact that USF1 and USF2 often bind their target as heterodimers (31). For the other investigated factors, the clearest results are that POLR2A levels are augmented on the USF1 bound allele whereas H3K27me3 enrichment displays the opposite pattern with slightly increased levels for the allele not bound by USF1 (see Figure 3A). We thus show allele specific preference for POLR2A and H3K27me3. To get a statistical support for these findings, we sequenced at least three replicates of each sample and quantified the difference between the two alleles. This allowed us to compute P-values for the signals in ChIP samples compared to genomic DNA using a t-test (see ‘Methods’ section). We conclude that USF1, USF2 and POLR2A ratios were significantly increased when compared to genomic DNA whereas the H3K27me3 ratio was significantly decreased, all with P < 0.001 (see Figure 3B). The H3K4me3 signal showed a slight but significant decrease on the USF1 bound allele. At present we have no biological explanation for this observation.
Figure 3.
Sequencing results for SNP1 (rs1867760), which is in the sequence AA[T/C]ACGTGACCC. (A) Sequencing of SNP1 in various samples extracted from HepG2 cells. The SNP is heterozygous in HepG2 genomic DNA since both the C-allele (blue) and T-allele (red) show a peak at the third position (see top row). In USF1 and USF2 ChIP DNA the C-allele gives much higher signal than the T-allele. (B) Standard box-and-whisker plots showing the allele signals for SNP1 in HepG2 and HT29. Above each box are P-values from a t-test, indicating whether the allele signal in ChIP DNA is significantly different from the allele signal in genomic DNA. High allele signals are obtained for samples with higher C-allele peaks when compared to the corresponding T-allele peaks. See ‘Methods’ section for descriptions of allele signals and statistical testing. (C) Quantification of sequencing results for two SNPs at −91 (red boxes) and +1987 (blue boxes) bases from SNP1, respectively. The orange lines indicate the average allele signal for SNP1 in each sample.
The lack of significant effects for some of the examined factors could either indicate that both alleles are present in the ChIP DNA, or that none of them is present and we are just sequencing the background DNA. For SNP1 there is no enrichment of H3ac (see Supplementary Figure S4) and this explains why we get the same sequencing results for H3ac ChIP and genomic DNA. In general, we expect H3ac to be enriched at some distance from USF1 since we have previously seen that USF1 binds at promoters of genes while the H3ac peaks are located about 500 to 1 kb downstream of the TSS. H3K4me3 has been shown to display a similar signal around TSSs (11,14), which could explain why it does not either give any significant effect for most of the examined SNPs.
Accurate prediction of transcription factor binding sites facilitates distinction between potential regulatory SNPs
We hypothesize that SNPs in proximity of a functional SNP could also show allele specific binding if located close enough to the truly causative SNP. If this is the case, the binding site information could help in selecting the best candidate functional SNP. We investigated this for SNP1, where we detected yet two more SNPs, at −91 and +1987 bases, respectively, from SNP1. SNP1-91 is heterozygous in HepG2 and HT29, while SNP1 + 1987 is heterozygous only in HT29. While we did not see any allele specific binding for any of the investigated proteins for SNP1 + 1987, the SNP at -91 bp displays a similar allele specific binding pattern as SNP1 (see Table 1 and Supplementary Table S2). There are several possible explanations for the results observed at SNP1-91: either only one of the SNP1 or SNP1-91 are functional and the other looks similar just because of proximity, or both of them are functional. However, several lines of evidence suggest that SNP1 is functional while SNP1-91 is not. First, DNA fragments for ChIP are randomly generated by sonication to an average size of around 200–300 bp, it is expected that most ChIP enriched fragments will contain both SNPs although only one of them is truly affecting the USF1 binding. This also explains why we did not see similar results for SNP1 + 1987. Second, the sequence around SNP1-91, TAGAG[T/C]GTGGGT, does not even remotely resemble an E-box further strengthening the hypothesis that SNP1, rather than SNP1-91, is bound by USF1. Finally, our results show that in the allelic binding differences for USF1, USF2, POLR2A and H3K27me3 the effects are weaker for SNP1-91 than SNP1, which is precisely what we expect to see if SNP1 is indeed the causative SNP.
Allele-specific USF1 binding also occurs in normal tissue
Having obtained encouraging results in cell lines, we attempted to detect additional rSNPs by sequencing DNA from normal colon tissue from two different individuals, because of the similarities of USF1-binding profiles in liver and colon (32). We found three additional SNPs (SNP3, SNP4 and SNP5) in the predicted USF1 sites that were heterozygous in at least one of the colon samples. SNPs 3 and 4 were inside the core consensus CACGTGAC while SNP5 was just outside. The results for SNP3 and SNP4 show that USF1 is preferentially bound to the allele with a CACGTGAC sequence. However, in these cases the differences are not as large as for SNP1, which could indicate that USF1 may also bind to the other allele but with a lower frequency. For SNP3, POLR2A is also showing a significant effect (see Figure 4). For SNP4, POLR2A levels are slightly elevated on the USF1 bound allele but the results were not significant. In any case, results for SNP4 should be taken with caution, since as mentioned, this region might be weakly bound by USFs as indicated both by array and qPCR measurements. SNPs 3 and 4 were also tested for H3K4me3 and H3K27me3 but we did not see any significant effect.
Figure 4.
Sequencing results for SNP3 (rs16875109), which is in the sequence CTCA[T/C]GTGACCT. Standard box-and-whisker plots showing quantification results for SNP3 in the Colon1 sample. At the top of each box are P-values from a t-test, indicating whether the allele signal in ChIP DNA is significantly different from that in genomic DNA. High allele signals are obtained for samples with higher C-allele peaks when compared to the corresponding T-allele peaks. See ‘Methods’ section for descriptions of allele signals and statistical testing.
SNP5 was located just outside of the consensus sequence. This was the case also for SNP2, which is heterozygous in HepG2. These two SNPs show no evidence of differences in binding between the two alleles (see Supplementary Figure S5). Although this finding suggests a polymorphism may be located just outside the consensus without having any major effect, we should be careful about drawing conclusions from negative examples. Nevertheless, the results in SNP2 and SNP5 serve as a control since ChIP and genomic DNA signals were virtually identical in both cases.
Additional candidate regulatory SNPs on minor alleles
We hypothesized that more rSNPs could be identified by assuming that the reference genome might contain the allele not bound by USF1, for some SNPs. Therefore, we considered all sequences with one mismatch from the USF1 consensus and scanned for SNPs where the minor allele was CACGTGAC. This resulted in 26 additional candidates and three of these were found to be heterozygous in at least one of our four samples (i.e. HepG2, HT29, two colon samples), all of them inside the core consensus CACGTGAC. For one of the predictions, SNP6, heterozygous in HepG2, USFs and POLR2A levels were elevated on the USF1 consensus allele, while H3K27me3 levels were slightly lower than genomic DNA. SNP7 did not show any significant effect.
Some additional information for these two SNPs can be gained from qPCR ChIP results (see Supplementary Figure S3). SNP6 is heterozygous in HepG2 but homozygous for the non-consensus USF1 sequence in HT29. Interestingly, clear USF binding was observed (more than 5-fold) in HepG2 cells while lower than background levels were obtained for HT29, clearly reinforcing the conclusions derived from sequencing results. On the other hand, for SNP7, HepG2 and HT29 cells were homozygous for the USF1 consensus and non-consensus sequences respectively. In this case, qPCR signals over 2-fold could be observed in both cell lines without major differences between the cell types, in agreement with the lack of allelic differences in the heterozygous colon sample. This suggests that this region is indeed bound by USF proteins but that SNP7, which occurs outside the CACGTG core, is not affecting the interaction.
Allelic differences in transcriptional organization around SNP1
When evaluating the genomic location of the investigated SNPs displaying allele-specific protein interactions, we observed that none of them was in proximity of the TSS of a well-characterized human gene. To increase the chances of detecting such type of SNPs, we genotyped 1 million SNPs in HepG2 using an Illumina array, but we did not find any additional heterozygous SNPs in the predicted USF1-binding sites. This made it difficult to explore the effects of our SNPs on gene expression, as has been previously done, by investigating allelic differences on RNA levels typically at exonic SNPs (19). In any case RNA might be affected by post-transcriptional regulation and is perhaps not the best indicator of transcription for our purposes.
We instead used an alternative approach for investigating the allelic transcription around our candidate SNPs. By using ChIP-seq POLR2A data from HepG2 (unpublished), we found indications of transcription initiation near SNP1 and SNP6 (data not shown). For these two SNPs we then used H3K36me3 as a well-recognized indicator of transcription rates and allele specific transcription (33) that should not be affected by post-transcriptional events. qPCR analysis for H3K36me3 around SNP1 and SNP6 further suggested that transcription is only ongoing near SNP1 (data not shown), so only that SNP was further examined.
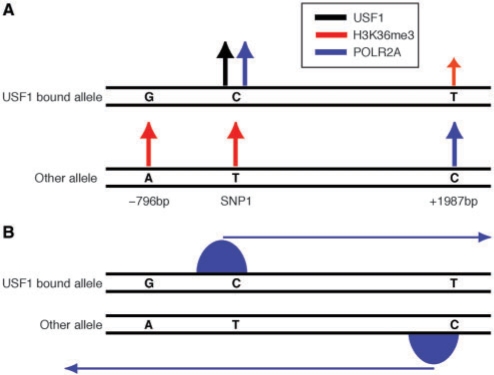
We investigated two SNPs near SNP1, at –796 and +1987 bp, respectively. Of these, only SNP1-796 was heterozygous in HepG2, so our experiments were instead performed in HT29 where both of them were heterozygous. SNP1-796, SNP1 and SNP1 + 1987 are found within a region of high linkage disequilibrium so the haplotypes can be inferred with high confidence. The two haplotypes are GCT and ATC. From before we know that USF1 is bound to the C allele of SNP1 (see Figure 3), i.e. the GCT haplotype. Next, we examined H3K36me3 at the three SNPs (see Figure S6). Interestingly, H3K36me3 showed significantly higher enrichment on the ATC allele for SNP1-796 and SNP1. For SNP1 + 1987 the opposite pattern was found with higher, although not significant, H3K36me3 enrichment on the GCT allele. The results suggest that transcripts from both strands are being generated in the region. This idea is further supported by the ChIP-seq data where distinct POLR2A peaks are found both near SNP1 and SNP1 + 1987. We therefore investigated POLR2A at those locations. Our results show that POLR2A is significantly higher on the USF1 bound allele for SNP1. For SNP1 + 1987, the opposite pattern is found with higher enrichment on the other allele (see Figure S6). The results for POLR2A and H3K36me3 are summarized in Figure 5A.
Figure 5.
Transcription organization in a region surrounding SNP1. (A) Summary of sequencing results for SNP1-796, SNP1 and SNP1 + 1987. The two alleles are shown separately. The arrows indicate positions with differential allelic enrichment of USF1, H3K36me3 and POLR2A and are placed above the allele showing higher enrichment. Differential enrichment of H3K36me3 was not significant for SNP1 + 1987 bp and is therefore indicated by smaller arrow. (B) A model explaining the transcription organization in the region. The blue peaks indicate positions with high POLR2A. The arrows show the direction of transcription.
The model in Figure 5B can explain how transcription is organized in this region. We hypothesize that for the GCT haplotype, USF1 is binding to the C allele of SNP1 and activates transcription in the downstream direction. This transcription reduces the activity of transcription in the opposite direction from a TSS located near SNP1 + 1987. The opposite patterns are expected for the ATC haplotype. However, we cannot fully exclude the possibility that the SNP at +1987 bp has some functional relevance on its own, by for instance affecting the binding of some other transcription factor. We also attempted to study RNA levels but detected no allelic differences. We have already pointed out that RNA levels are not the best measures of transcription since the levels are affected by RNA stability and other processes.
DISCUSSION
Recent developments in high throughput technologies open new possibilities for combining and analyzing data from different sources. By ChIP-chip and ChIP-seq we can now query the whole human genome for a given protein. Methods such as BCRANK can predict thousands of binding sites from these genome-wide experiments, and many of these are likely to be functionally important. Using the presented strategy, we identified seven heterozygous SNPs in the immediate neighborhood of predicted USF1-binding sites in the studied cell types. In four of the five cases where the SNP was inside the core sequence, we observed clear effects of USF-binding, and in 2/2 SNPs just outside, there was no effect. Thus, we were successful in predicting and validating TF-binding sites at base pair resolution. Furthermore, for one of these SNPs, we could observe allelic effects on transcription at heterozygous neighboring SNPs. Our results therefore suggest that SNPs affecting USF1 binding can result in allelic differences in gene expression. However, since none of our SNPs was in a gene promoter we could not investigate the downstream effects on well-characterized transcripts for protein coding genes. The distal localization of the SNPs does not exclude that they are functional. Recent large-scale transcription factor binding maps have uncovered that most TFs, especially those with cell-type specific functions, bind mainly at sites far from genes (11,23). Moreover, these distal binding sites very often display several features characteristic of enhancers, and are more cell-type specific and functionally relevant during development and at establishing cell-type specific transcription programs (22,34,35). Promoters on the other hand often show the same epigenetic features in different cell types. However, with current technologies it is difficult to infer the gene/s regulated by a given enhancer, and therefore, in the case of our study, it would be challenging to link our rSNPs to the genes they might affect. Another possibility is that some of the SNPs, e.g. SNP1 identified in this study, might occur close to transcription start sites of uncharacterized coding or non-coding transcripts, which seem to be more frequent than previously anticipated (36).
In the literature there are two major types of strategies to identify regulatory genetic variation. We believe our method adds novelty and that it has several advantages compared to the previous ones. One approach uses large-scale technologies in order to detect allele-specific transcription (37–39), histone modifications (17) or POLR2A binding (19). Although the throughput in these studies is higher than ours, they essentially do not detect rSNPs but rather identify genes with allelic differences in their expression patterns. In those cases where haploChIP experiments were part of the experimental setup (17,19), the SNPs showing allelic-differences in protein binding can only be seen as reporters, since the investigated proteins lack any sequence-specificity in their binding. Importantly, the true rSNPs in those studies can simply be in linkage disequilibrium with the detected SNPs, as exemplified by −91 SNP1 in our study, or even at much larger distance e.g. within an enhancer. Our strategy identifies SNPs located in sites bound by sequence specific TFs, so the chances are much higher that our predicted SNPs are truly regulatory. The hypothesis that a SNP within a TF-binding site will affect the protein–DNA interaction is very intuitive, but has rarely been shown using in vivo experiments. One limitation with our approach is the relatively low number of validated rSNPs, but as discussed below, there are several ways to increase our throughput. A second approach of rSNP detection largely relies on bioinformatics predictions (40–44). A number of interesting genes are selected, based on various criteria, e.g. allelic-differences in gene expression, association to human disease, or evolutionary conservation. Then the promoter regions of those genes are scanned for putative TF-binding sites. Subsequently, SNPs found within the predicted binding sites are expected to affect the binding and are considered as potential rSNPs. However, the initial TF-binding sites are typically predicted based on in silico methods, which are known to result in many false positives (45). A further disadvantage is that these methods do not give any information as to which cell type or tissue is relevant for a given rSNP. Finally, if validation of the detected rSNPs is performed, which is not always the case in these methods, this is often done using in vitro methods (i.e. EMSA), which might not reflect the in vivo situation. Our method only takes into account regions bound in vivo by a TF. In this way, we dramatically reduce the number of false positives and also gain information about which cell type/s should be investigated for a particular SNP. In addition, since we validate our candidate SNPs by an in vivo method (i.e. haploChIP), the regulatory potential of the reported SNPs should be largely increased.
Prediction of rSNPs using ChIP data requires thorough analysis strategies since there is a risk of false prediction in cases when there are several nearby SNPs. Without binding site information it would be much harder for us to predict which of the SNPs plays a regulatory role. This point is important and it indicates that a SNP can be seen as having functional consequences like changes in POLR2A, H3K27me3, USFs, etc. that could explain the potential changes in expression, but without being the most likely rSNP. This is exemplified by the SNP located −91 bp from SNP1 (Figure 3C). Therefore, our strategy may help identify rSNPs in cases where multiple SNPs on a haplotype with high linkage disequilibrium are all associated to a disease, expression of a gene or some other phenotype.
Here we have demonstrated a proof of principle that our strategy can be used for detection of rSNPs. The number of experimentally tested SNPs is relatively low mainly due to the low number of genotyped samples. However, the throughput of our strategy can be increased at least in two ways: first, by genotyping a larger panel of cell lines or individuals the chances of detecting heterozygous SNPs inside predicted binding sites will largely increase, and so will also the candidate functional SNPs to be tested. Second, our approach can in principle be applied to any available transcription factor binding data generated by ChIP-chip or ChIP-seq, taking in consideration the cell type where the data was generated so it matches the samples used to screen for heterozygous SNPs. The current amount of TF-binding data available from public repositories, such as GEO (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), is already very large and expected to grow at an increasing rate due to recent next-generation sequencing approaches. Therefore, our strategy can provide an inexpensive and easily adaptable way for many laboratories to effectively screen their own or public data for detection of regulatory genetic variation. Finally, using appropriate cohorts of patient and control samples for a certain disease, we hypothesize that our strategy could facilitate the detection of disease causing regulatory SNPs.
In summary, we have presented a strategy that successfully can be used for detection of regulatory SNPs throughout the whole human genome. We believe that this approach could prove to be an important complement to the ever-increasing amount of data generated by large-scale association studies, and that it could help pinpoint the causative SNPs when they are located in regulatory regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Councils for natural science and for medicine, The Cancer Foundation, Markus Borgström Foundation, Swedish Diabetes association and The Family Ernfors Fund (to C.W.); Swedish Foundation for Strategic Research, The Knut and Alice Wallenberg Foundation, Uppsala University and the Swedish University of Agricultural Sciences (to J.K.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 5.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 6.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckland PR. Allele-specific gene expression differences in humans. Hum. Mol. Genet. 2004;13(Spec No 2):R255–R260. doi: 10.1093/hmg/ddh227. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proc. Natl Acad. Sci. USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 16.Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadota M, Yang HH, Hu N, Wang C, Hu Y, Taylor PR, Buetow KH, Lee MP. Allele-specific chromatin immunoprecipitation studies show genetic influence on chromatin state in human genome. PLoS Genet. 2007;3:e81. doi: 10.1371/journal.pgen.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 19.Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat. Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 20.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 21.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 23.Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD, et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum. Mol. Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 25.Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–1591. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu P, Soder GJ, Sanfiorenzo VJ, Wang L, Greene JR, Fritz MA, Cai XY. Quantification of single nucleotide polymorphisms by automated DNA sequencing. Biochem. Biophys. Res. Commun. 2003;309:331–338. doi: 10.1016/j.bbrc.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells—a model, its experimental support and implications. Mutat. Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 31.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 32.Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF, et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 38.Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000006. e1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KC, Koka V, Dias J, Gurd S, Martin NW, Mallmin H, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–127. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mottagui-Tabar S, Faghihi MA, Mizuno Y, Engstrom PG, Lenhard B, Wasserman WW, Wahlestedt C. Identification of functional SNPs in the 5-prime flanking sequences of human genes. BMC Genomics. 2005;6:18. doi: 10.1186/1471-2164-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GuhaThakurta D, Xie T, Anand M, Edwards SW, Li G, Wang SS, Schadt EE. Cis-regulatory variations: a study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics. 2006;7:235. doi: 10.1186/1471-2164-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milani L, Gupta M, Andersen M, Dhar S, Fryknas M, Isaksson A, Larsson R, Syvanen AC. Allelic imbalance in gene expression as a guide to cis-acting regulatory single nucleotide polymorphisms in cancer cells. Nucleic Acids Res. 2007;35:e34. doi: 10.1093/nar/gkl1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P, Lenhard B, Wasserman WW, Odeberg J. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput. Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim BC, Kim WY, Park D, Chung WH, Shin KS, Bhak J. SNP@Promoter: a database of human SNPs (single nucleotide polymorphisms) within the putative promoter regions. BMC Bioinformatics. 2008;9(Suppl. 1):S2. doi: 10.1186/1471-2105-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.