Abstract
DNA methylation is a key regulator of gene transcription. Alterations in DNA methylation patterns are common in most cancers, occur early in carcinogenesis and can be detected in body fluids. Reliable and sensitive quantitative assays are required to improve the diagnostic role of methylation in the management of cancer patients. Here we present an optimized procedure, based on differential-high resolution melting analysis (D-HRMA), for the rapid and accurate quantification of methylated DNA. Two sets of primers are used in a single tube for the simultaneous amplification of the methylated (M) and unmethylated (Um) DNA sequences in D-HRMA. After HRM, differential fluorescence was calculated at the specific melting temperature after automatic subtraction of UM-DNA fluorescence. Quantification was calculated by interpolation on an external standard curve generated by serial dilutions of M-DNA. To optimize the protocol, nine primer sets were accurately selected on the basis of the number of CpG on promoters of hTERT and Bcl2 genes. The use of optimized D-HRMA allowed us to detect up to 0.025% M-DNA. D-HRMA results of DNA from 85 bladder cancers were comparable to those obtained with real time quantitative methylation specific PCR. In addition, D-HRMA appears suitable for rapid and efficient measurements in ‘in vitro’ experiments on methylation patterns after treatment with demethylating drugs.
INTRODUCTION
Epigenetic modifications are defined as heritable information other than nucleotide sequence, including DNA methylation and histone deacetylation. These mechanisms can regulate a wide range of physiological and pathological processes (1). DNA methylation occurs by the covalent addition of a methyl group at the 5′ of the cytosine ring, resulting in 5-methylcytosine (2). In mammalian DNA, 5-methylcytosine is found in approximately 4% of genomic DNA, primarily at cytosine–guanosine dinucleotides (CpGs). Such CpG sites are found more frequently in small stretches of DNA, called CpG islands. These islands are typically found in or near promoter regions of genes (3). In contrast to the bulk of genomic DNA, in which most CpG sites are heavily methylated, CpG islands of normal somatic cells remain unmethylated, allowing gene expression to occur (2). Hypermethylation of CpG islands results in either downregulation or complete abrogation of gene-associated expression, playing a role comparable to inactivating mutations or deletions (1). Tumor suppressor genes, genes encoding cell adhesion molecules and growth-regulatory proteins, are often silenced in malignancies by DNA hypermethylation (4). Promoter hypermethylation can therefore constitute the initial hit in many cancers (5). For these reasons the study of DNA methylation shows great promise in the future management of cancer patients. Potentially, DNA methylation should provide relevant information in the classification of tumors, depending on their methylation status. Such classification might be of use in determining patient prognosis or potential response to therapy. However the most attractive application of DNA methylation is probably as a specific marker for the early detection of cancer. It is now clear that aberrant DNA methylation is an early event in tumor development and that hypermethylated sequences can be detected in apparently normal epithelia largely before the appearance of cancer (6). The development of PCR techniques for the detection of methylated DNA, in particular methylation-specific PCR (MSP) (7), has allowed methylated sequences to be detected in human biofluids containing small amounts of tumor-derived DNA, as recently reviewed (8). However, to be used as a cancer marker, DNA methylation requires fine mapping and quantitative determinations.
Among the several approaches proposed for quantitative assessment of DNA methylation [for details see (9)], real time qPCR (quantitative methylation specific PCR: qMSP) appears particularly suitable. As in traditional qualitative MSP, the first step of qMSP includes DNA treatment with sodium bisulfite that generates differences in methylation-dependent sequences at CpG dinucleotides, by converting unmethylated cytosine residues to uracil, while methylated cytosine is unmodified. The methylation profile of target sequences can then be detected with primer sets and fluorescent probes, specific for methylated and unmethylated CpGs. The term MethyLight (10) indicates a flexible platform of possible fluorescence-based qMSP assays, determined by the alternative combinations of primer-probe sets specific for methylated or unmethylated sequences.
High resolution melting (HRMA), an extension of previous DNA dissociation or melting analysis, was recently introduced as a technique for genotyping single-nucleotide polymorphisms (11). This closed-tube method is made possible by heteroduplex-detecting DNA dyes that can be used at saturating concentrations without inhibiting PCR (12). These third generation fluorescent dsDNA dyes have low toxicity in an amplification reaction and can therefore be used at higher concentrations for greater saturation of the dsDNA sample that collects fluorescent signals with much greater optical and thermal precision (11,12). There are already for HRMA several applications in the diagnosis of human diseases (13). Very recently HRMA was also proposed as a rapid and sensitive technique for the assessment of DNA methylation (14,15) for the diagnosis of genetic imprinting disorders (16,17), BRCA1 inactivation in breast cancer (18) and MGMT and APC methylation in colorectal cancer (19).
Here we describe the optimization of a protocol for the quantitative evaluation of DNA methylation based on the differential analysis of fluorescence during HRMA. Our study focused on a preliminary test to determine the best conditions for the assay on two different genes: hTERT that codifies for the telomerase catalytic subunit and the anti-apoptotic gene Bcl2. Both genes have been found to be frequently methylated in bladder cancer (20). The optimized protocols were then applied to the measurement in biological samples and the results were compared with those obtained with a MethyLight assay.
MATERIALS AND METHODS
Bladder cancer samples
A sample of fresh tumor tissue was obtained with a cold cup biopsy forceps from 85 patients undergoing transurethral resection of bladder cancer. Tissue samples were immediately steeped in RNAlater (Qiagen, Germany) and stored according to the manufacturer's instructions. DNA was extracted by QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions.
Cell line
The adrenal cancer cell line H295R was cultured in Ham's-F12:DMEM (1:1) medium with ITS (insulin, transferring and selenium), 2 mM glutamine, antibiotics and 10% fetal bovine serum in 5% CO2. Cells were treated with the demethylating agent 5-Aza-2′-deoxycytidine (5-Aza) 24 h after the seeding with a concentration of 5 μM for 1, 3 and 6 days. Cells were collected and centrifuged at 10 000 × g at room temperature for 2 min and stored at –80°C, until the DNA extraction. DNA was extracted by QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions and stored at –80°C. DNA concentration was estimated with NanoDrop 1000 (NanoDrop Technologies).
Bisulfite treatment
DNA (500 ng) obtained from cell lines or tissue samples was submitted to bisulfite modification using the EpiTect Bisulfite Kit (Qiagen) following the manufacturer's protocol. Bisulfite-treated DNA was resuspended in 40 μl elution buffer and 1 μl was used for D-HRMA and MethyLight, respectively. For each experiment, CpG Genome Universal Methylated and Unmethylated DNA (Chemicon International Inc.) were used as positive (100% methylated) and negative (0% methylated) controls. After bisulfite treatment, DNA was immediately submitted to D-HRMA and MethyLight analyses. Since accurate quantification of DNA after bisulfite treatment was not possible due to its high degradation, the presence of amplifiable DNA was tested by real time PCR using a primer pair and a TaqMan™ probe for the bisulfite converted sequence of a non-CpG-containing region of β-actin gene (see ‘MethyLight’ section for details), as previously described (20). All samples provided a correct amplification plot with a relatively constant Ct value of 26.0 ± 3.1 (mean ± SD) and therefore were considered suitable for D-HRMA and MethyLight assays. For β-actin, the sequence of primers was (Forw) 5′-TGGTGATGGAGGAGGTTTAGTAAGT and (Rev) 5′-AACCAATAAAACCTACTCCTCCCTTAA, while TaqMan probe was Fam-5′-ACCACCACCCAACACACAATAACAAACACA.
hTERT and Bcl2 primers for HRMA
Analysis of the hTERT gene (Genebank: AF325900) with MethylPrimer Express V1.0 software (Applied Biosystems) revealed two CpG islands: island #1 from −4771 to −4334 and island #2 from −2016 to −1151. Island #2 was schematically divided into two sequences (A from −2016 to −1532 and B from −1415 to 1151). Three couples of primers were designed on sequence A and three sets on sequence B, to generate amplicons with a variable number of CpG dinucleotides. Accordingly, primer pairs were named hTERT-3A, hTERT-11A, hTERT-13A in sequence A and hTERT-7B, hTERT-15B and hTERT-21B in sequence B, on the basis of target sequence, CpG numbers and localization. For each sequence we designed separated couples of primers for the methylated and unmethylated form, with comparable annealing temperature. Primers sequences and amplicon lengths are reported in Table 1.
Table 1.
Primer sets used for the amplification of methylated and unmethylated genes
| Gene name and sequence area | Primer sets and annealing T | CpGs in amplified sequence | CpGs in forward primer | CpGs in reverse primer | Total CpGs in amplicons | Amplicon size (bp) | CpG density (bp/CpG) | Forward primers | Reverse primers |
|---|---|---|---|---|---|---|---|---|---|
| hTERT (Sequence A) −2016 to −1532 | hTERT-3 60°C | 3 | 2 | 2 | 7 | 110 | 15.7 | M 5′AGGTTAGCGGTTAAAGGGTC U 5′ATTAGGTTAGTGGTTAAAGGGTT | M 5′CCAACGAAAAAAAATCGAAT U 5′ACCCCAACAAAAAAAAATCAAAT |
| hTERT-11 60°C | 11 | 2 | 2 | 15 | 116 | 7.7 | M 5′GAGGTATTTCGGGAGGTTTC U 5′GGGAGGTATTTTGGGAGGTTTT | M 5′GAACACCACGAATACCGAA U 5′AAACACCACAAATACCAAACA | |
| hTERT-13 60°C | 13 | 4 | 3 | 20 | 142 | 7.1 | M 5′TCGTTTTTAGTCGCGTTTAC U 5′GGTTTGTTTTTAGTTGTGTTTAT | M 5′CGTACGACGACCCTTTAAC U 5′TACATACAACAACCCTTTAACC | |
| hTERT (Sequence B) −1415 to −1151 | hTERT-7B 60°C | 7 | 3 | 4 | 14 | 116 | 8.3 | M 5′GGATTCGCGGGTATAGACGT U 5′GGATTTGTGGGTATAGATGTTTAG | M 5′CGAAATCCGCGCGAAA U 5′CCAAATCCACACAAAAAAAAC |
| hTERT-15B 58°C | 15 | 2 | 3 | 20 | 177 | 8.8 | M 5′GGGATTCGGGTATTCGTTTTGT U 5′GGGATTTGGGTATTTGTTTTG | M 5′AACGCTACCTAAAACTCGCGC U 5′AACACTACCTAAAACTCACAC | |
| hTERT-21B 58°C | 21 | 3 | 3 | 27 | 232 | 8.6 | M 5′TGGATTCGCGGGTATAGACGT U 5′TGGATTTGTGGGTATAGATGT | M 5′AACGCTACCTAAAACTCGCGC U 5′AACACTACCTAAAACTCACAC | |
| Bcl2 −1 to +263 | Bcl2-7 57°C | 7 | 4 | 2 | 13 | 89 | 6.8 | M 5′TCGTATTTCGGGATTCGGTC U 5′TTGTATTTTGGGATTTGGTT | M 5′ACCTAAACGCAAACCCCGC U 5′AACTAAACACAAACCCCAC |
| Bcl2-12 58°C | 12 | 2 | 3 | 17 | 175 | 10.3 | M 5′GGGTACGATAATCGGGAGATAGTGA U 5′GGGTATGATAATTGGGAGATAGTGA | M 5′CCGAATCCCGAAATACGACTAAA U 5′CCAAATCCCAAAATACAACTA AAA | |
| Bcl2-17 58°C | 17 | 2 | 3 | 22 | 231 | 10.5 | M 5′GATGGCGTACGTTGGGAGAA U 5′GATGGTGTATGTTGGGAGAA | M 5′ACAACCGAAATCTACAACGACGAA U 5′ACAACCAAAATCTACAACAACAAA |
For Bcl2 gene (Genebank: NM_000633) we identified a single CpG island localized between the 5′UTR and the first exon (from –1 to +263). In this sequence three couples of primers were designed to generate amplicons containing 7, 12 and 17 CpGs and indicated as Bcl2-7, Bcl2-12 and Bcl2-17, respectively (Table 1).
All sets of primers for the methylated and unmethylated forms were tested in separated MSP to verify amplification performances and to check their capacity to amplify selectively the unmethylated and methylated sequences, respectively (data not shown).
D-HRMA
HRMA was carried out on a Rotor-Gene™ 6000 (Corbett Research). PCR was performed in 10 μl volume containing 1× buffer, 1.5 mM MgCl2, 1 mM each dNTPs, 300 nM of each primer, 5 μM of SYTO 9 (Invitrogen), 0.04 U TaqGold (Applied Biosystems) and 1 μl of bisulfite modified DNA template. The amplification protocol was 15 min at 95°C, then 50 cycles of 30 s at 95°C, 30 s at annealing temperature, 30 s at 72°C and a final step of 30 min at 72°C. HRMA was performed at 95°C for 5 min, 40°C for 1 min and with a ramping from 65°C to 95°C rising by 0.1°C/s. Melting curves were normalized using the HRMA software before and after the major fluorescence decrease. A differential profile was then evaluated for each sample by comparing fluorescence at the melting point against the value of fluorescence of the negative control (unmethylated DNA). All experiments were performed in duplicate.
Since reproducibility is an important issue in any quantitative determination, we tested the intra- and inter-assay variability of D-HRMA using three primers sets (hTERT-3A, hTERT-7B and Bcl2-7) in three bladder cancer samples carrying variable methylation levels. To evaluate intra-assay reproducibility, 10 replicates of the same bisulfite-treated DNAs were measured in a single D-HRMA experiment. In addition, using the same samples and primers, we repeated the measurement in seven consecutive experiments to determine inter-assay variability.
hTERT and Bcl2 MethylLight assays
MethyLight reaction was performed in 12.5 μl final volume consisting in 1× quantiTect Probe PCR Master Mix (Qiagen), 200 mM of each probe, 600 nM of forward and reverse primers, 1 μl of bisulfite-treated DNA. The primers and probe sets were the same as previously described (20). Amplification was carried out at 95°C for 10 min, followed by 50 cycles at 95°C for 15 s, 60°C for 1 min in a Rotor-Gene™ 3000 (Corbett Research). To evaluate the percentage of methylation status, each sample was normalized referring to a non-CpG containing region of β-actin gene. The relative level of methylation was determined by the 2–ΔΔCt method (21), according to the following formula in which ΔΔCt = unknown sample (Cttarget gene – Ctβ-actin) – 100% methylated DNA (Cttarget gene – Ctβ-actin).
RESULTS
Linearity of D-HRMA
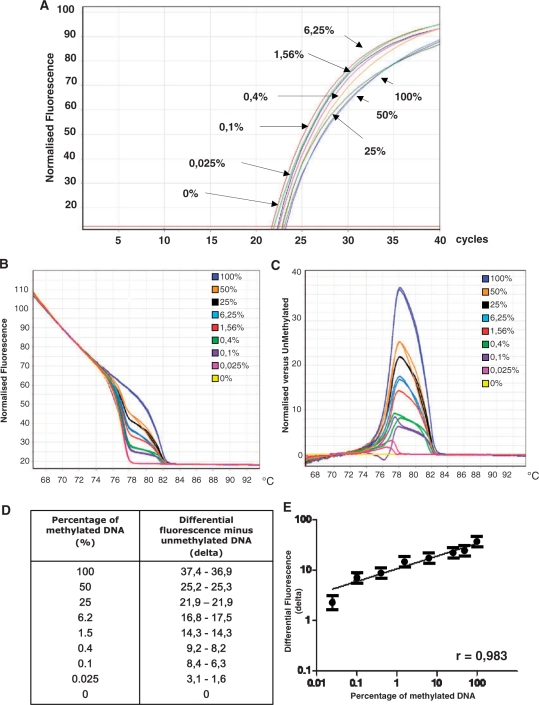
Linearity tests were performed by D-HRMA using the nine primer sets. We prepared serial dilutions of methylated and unmethylated DNA standards to create reconstituted samples with a constant DNA quantity (12.5 ng) but containing 100%, 50%, 25%, 6.25%, 1.56%, 0.4%, 0.1%, 0.025% and 0% methylated DNA. Each dilution was then amplified with primer sets for hTERT and Bcl2 and submitted to D-HRMA, as previously described. The obtained amplification plots indicated that all dilutions were amplified with comparable Ct values (Figure 1A). Through the graph of normalized fluorescence we verified the HRMA profile of the different amplicons (Figure 1B). Then a differential graph was generated by normalizing each HRMA profile against unmethylated DNA (Figure 1C). The maximal height of the peak was detected at the melting temperature of each amplification product. Differential analysis generated peaks with variable height, due to the difference in fluorescence between unmethylated DNA and other dilutions. The highest peak corresponded to the fully methylated DNA (100%). The height of the other peaks decreased proportionally in samples containing progressively lower methylation percentages. Through the specific software it was possible to extrapolate the value of differential fluorescence peak for each dilution (Figure 1D). These values were then plotted against the dilution factor to generate a linear calibration curve (Figure 1E).
Figure 1.
Example of the flow chart for D-HRMA. (A) Amplification plots of serial dilutions of methylated DNA (from 100% to 0%) were amplified with primers for methylated and unmethylated forms (primer set hTERT-7B) in the same tubes. All the dilutions were amplified with comparable amplification plots (B) HRMA profile of the same samples. (C) Fluorescence of each sample was normalized as differential signal against unmethylated control. (D) Values of differential fluorescence obtained at the melting temperature for each dilution. (E) Values of differential fluorescence were plotted against the percentage of methylation of each dilution to generate a typical standard curve. All the experiments were performed in duplicate.
Performances of D-HRMA with different primer sets
HRMA was repeated with all primer sets for hTERT and Bcl2 genes to compare the different performances of the assays in relation to the specific features of the amplicons (i.e. length in bp and number of CpGs). To evidence differences among primer sets, standard curves were generated plotting methylated DNA concentrations versus the maximal height of differential peaks, at the respective melting temperature. We found that all primer sets were able to generate a linear dose–response even if with a differential capacity to discriminate among percentages of DNA methylation and with a different operating range. All the curves were compared for their slope, correlation coefficient and number of correctly fitted points.
The slope of standard curves is the expression of the proportionality between dose (percentage of methylated DNA) and signal (differential of fluorescence). In this sense hTERT-3A primer set showed the best slope that was nearly parallel to the ideal 45° angle and therefore particularly suitable for accurate measurements. Unfortunately, the relatively low number of CpGs amplified with this primer set (seven CpGs: three in the internal amplified sequence and four covered by the two primers) appeared less sensitive in the detection of low levels of DNA methylation, since 0.1% and 0.025% were not detectable. This result seems to indicate that D-HRMA needs a major number of modified CpGs to reach good levels of sensitivity. Accordingly, when the hTERT-11A primer set (11 + 4 = 15 total CpG) was used, we obtained a slight reduction in the slope but an improvement in sensitivity, since 0.025% methylated DNA was clearly distinguishable from unmethylated DNA. When the number of amplifiable CpGs was further increased with primer set hTERT-13A (13 + 7 = 20 total CpGs), the slope tended to be dramatically reduced with a consistent loss of discriminating capacity. In addition, the 0.025% concentration was not detectable (Figure 2A–C).
Figure 2.
Diagrams of normalized fluorescence of methylated DNA dilutions normalized against unmethylated control fluorescence (left panels) and corresponding plotted standard curve (right panels) for hTERT primer sets in sequence A of gene promoter (top: hTERT-3A; medium: hTERT-11A; bottom: hTERT-13A). Slope and coefficient of correlation were reported for each standard curve.
To confirm these findings and to exclude possible bias due to DNA sequence, the experiment was repeated on the other sequence (Sequence B) of the same gene. Primer set hTERT-7B (7 + 7 = 14 total CpGs) had the best slope and the maximal sensitivity in comparison to hTERT-15B (15 + 5 = 10 total CpGs) and hTERT-21B (21 + 6 = 27 total CpGs) primer sets (data not shown). Finally, to exclude the possible gene-to-gene bias, we repeated the same test using a sequence on the Bcl2 CpG island. The results indicated once again that the best assay performances were obtained with primer sets containing an intermediate number of CpG repeats. The Bcl2-7 primer set (7 + 6 = 13 total CpGs) had cumulatively the best performance in comparison to Bcl2-12 (12 + 5 = 17 total CpGs) and Bcl2-17 (17 + 5 = 22 total CpGs) (data not shown). All parameters of fitting curves obtained after D-HRMA with the different primer sets were reported in Table 2.
Table 2.
Parameters of fitting curves obtained after D-HRMA with the different primer sets
| Gene name and sequence | Primer sets | Linear equation | Coefficient of correlation | Minimum detectable percentage of methylated DNA (%) |
|---|---|---|---|---|
| hTERT Sequence A | hTERT-3A | Y = 0.4740x − 0.8460 | 0.9939 | 0.4 |
| hTERT-11A | Y = 0.3029x + 0.0114 | 0.9853 | 0.025 | |
| hTERT-13A | Y = 0.0852x + 1.1250 | 0.9602 | 0.1 | |
| hTERT Sequence B | hTERT-7B | Y = 0.2637x + 0.2265 | 0.9828 | 0.025 |
| hTERT-15B | Y = 0.1803x + 0.7131 | 0.9289 | 0.1 | |
| hTERT-21B | Y = 0.1042x + 1.0659 | 0.9889 | 0.4 | |
| Bcl2 | Bcl2-7 | Y = 0.3727x − 0.1966 | 0.9808 | 0.025 |
| Bcl2-12 | Y = 0.2266x − 0.0903 | 0.9126 | 0.025 | |
| Bcl2-17 | Y = 0.0281x + 1.432 | 0.7899 | 0.1 |
The number of CpGs, independently of the DNA sequence, seems to influence the discriminative capacity of each assay, represented by the slope of the respective standard curves. As reported in Figure 3, the slope of the standard curves generated by the nine primer sets is inversely related to the number of total CpGs included in the respective amplicon (Figure 3).
Figure 3.
Inverse linear relationship between the number of CpGs in sequences (for primers and internal sequences see Table 1) amplified by the nine primer sets (x-axis) and the slope of standard curves generated by serial dilutions of methylated DNA (y-axis). The number of CpGs, independent of the DNA sequence, seems to influence the discriminative capacity of each assay, represented by the slope of respective standard curves (see Table 2 for values).
D-HRMA and bladder cancer methylation
After these preliminary assessments and to verify the accuracy of D-HRMA in quantifying the amount of methylated DNA in unknown samples, we used all sets of primers to determine the concentration of methylated alleles in DNA extracted from 85 human bladder cancers. Methylation was also measured in the same samples with a conventional MethyLight method, based on real time PCR and TaqMan probes. We evaluated the methylation status with D-HRMA as percentage referring to a standard curve whereas MethyLight results were calculated as relative quantification with the 2–ΔΔCt method (see ‘Material and Methods’ section for details).
Our data indicated that D-HRMA and MethyLight are highly concordant when the best performing primer sets (hTERT-3A, hTERT-7B and Bcl2-7) were used: taking into account positive or negative samples with both methods, the agreement was 91% for hTERT-3A, 96% for hTER-7B and 99% for Bcl2-7. The agreement between D-HRMA and MethyLight was reinforced by the comparability of numerical results obtained by the two methods (Figure 4A–C). The use of other primer sets (hTERT-11, hTERT-13, hTERT-15, hTERT-21) reduced dramatically the correlation with MethyLight results (r = 0.114, r = 0.358, r = 0.122, r = 0.379) (data not shown).
Figure 4.
Relationships between D-HRMA and MethyLight results in DNA from 85 bladder cancers, using three primer-probe sets (from the top, hTERT-3A, hTERT-7B and Bcl2-7). The levels of DNA methylation obtained by D-HRMA (y-axis) are reported as percentage of methylated DNA whereas MethyLight results (x-axis) are expressed as relative concentrations calculated with the 2–ΔΔCt method (see ‘Materials and Methods’ section for details). Each sample was measured in duplicate. Note the logarithmic scale.
Reproducibility of D-HRMA
We calculated intra-assay variability as coefficient of variation (%) at three different levels of methylation and using three primer sets (hTERT-3A, hTERT-7B and Bcl2-7). In Table 3 we reported the mean methylation percentage, its standard deviation and the coefficient of variation. According to methylation levels we found that intra-assay variability ranged from 4.4% to 34%. Inter-assay variability ranged from 7.4% to 48%.
Table 3.
Intra- and inter-assay variability of D-HMRA in three bladder cancer samples
| Samples | hTERT-3A |
hTERT-7B |
Bcl2-7 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intra-assaya |
Inter-assayb |
Intra-assay |
Inter-assay |
Intra-assay |
Inter-assay |
|||||||||||||
| Mean | SD | CV | Mean | SD | CV | Mean | SD | CV | Mean | SD | CV | Mean | SD | CV | Mean | SD | CV | |
| #1 | 57.1 | 16.4 | 28 | 60.1 | 19.9 | 33 | 88.2 | 14.3 | 16 | 86.7 | 18.7 | 21 | 0 | – | – | 0 | – | – |
| #2 | 13.8 | 4.7 | 33 | 19.4 | 9.3 | 48 | 71.5 | 19.6 | 27 | 73.9 | 17.6 | 23 | 90.9 | 4.0 | 4.4 | 88.4 | 6.6 | 7.4 |
| #3 | 2.0 | 0.7 | 35 | 3.3 | 1.3 | 41 | 64.5 | 11.8 | 18 | 62.4 | 16.6 | 26 | 93.9 | 4.0 | 4.3 | 93.0 | 11.2 | 12 |
Mean indicates mean percentage of DNA methylation measured by D-HRMA (%); SD: standard deviation of the mean; CV: coefficient of variation (%).
aIntra-assay variability was calculated in 10 replicates of three samples measured in the same experiment.
bInter-assay variability was calculated in three samples measured in seven different assays.
D-HRMA and 5-Aza treatment
The accurate measurement of methylated DNA concentration may also be useful when the effect of a demethylating agent is tested in ‘in vitro’ experiments. To test the capacity of D-HRMA to measure slight variations in DNA methylation in these experimental conditions, we used the hTERT-3, hTERT-7 and Bcl2-7 primer sets to verify the effects of 5-Aza-2-deoxycytine treatment on the human cell-line H295R in which hTERT and Bcl2 genes were methylated. As reported in Figure 5, D-HRMA was able to demonstrate the progressive demethylating effects by progressive reduction of Bcl2 methylated DNA in cellular extracts. Similar results were obtained for hTERT (data not shown).
Figure 5.
Measurement of methylation status of Bcl2 gene in the adrenal cancer cell line H295R exposed to the demethylating agent 5-Aza-2-deoxycitidine for 1, 3 and 6 days, using D-HRMA. (A) HRMA profile of DNA extracted from H295R cells on different days of treatment and control unmethylated (0%) DNA. (B) Fluorescence of each sample was normalized as differential signal against unmethylated control. (C) Percentage of DNA methylation in cells on different days of treatment. In this example, Bcl2-7 primer set was used. All the experiments were performed in duplicate.
DISCUSSION
DNA methylation appears to be a relevant biomarker in the diagnosis of cancer. This epigenetic modification has potential advantages over genetic tests, since methylation occurs widely throughout cancer cells and, in most cases, always affects the same promoter regions (22).
The introduction of highly sensitive techniques, such as the qMSP has enhanced the clinical relevance of methylation studies. Quantitative techniques for the detection of abnormal methylation patterns have potential advantages for the correct use of methylated DNA as a cancer biomarker. First, aberrant DNA methylation of some genes also occurs in non-malignant epithelia in physiological and pathological conditions and tends to progressively increase with age (23). Secondly, levels of DNA methylation may be related to tumor stage, grade and progression.
After the first studies on the use of melting analysis to detect methylated DNA (24,25), more recent papers have demonstrated that HRMA can be a sensitive, easy and inexpensive method to analyze DNA methylation (14,15,19). However, in the set-up of the assay, strategies for primer design can deeply influence the reliability of HRMA for methylation studies. Primers can be designed to overlap CpG dinucleotides or not. If primers anneal to a sequence lacking in CpG, methylated and unmethylated DNA is simultaneously amplified during PCR cycling and HRMA can distinguish between them on the basis of melting profile. Probably this would be the best approach, but in most cases this primer design is not allowed by gene sequence. In addition, preferential amplification of one allele cannot be excluded.
Wojdacz and Dobrovic (14) demonstrated that HRM can provide a sensitive and linear assay for a high-throughput assessment of methylation using a single primer set, specific for methylated DNA, when annealing temperature was chosen to amplify both methylated and unmethylated DNA simultaneously. The main limitation of this method is the bias deriving from the use a single primer pair, which is expected to preferentially amplify one of the two sequences (unmethylated/methylated). This is a critical aspect, since even limited sample-to-sample variations can deeply influence the reliability of the assay. The problem appears even more important when results from different assay sessions must be compared.
We tried to introduce a novel approach based on the use of two sets of primers in a single tube, for the simultaneous amplification of the methylated and unmethylated forms of the same DNA sequence. Even if the homologous primer pairs were chosen to have comparable annealing temperatures, methylated and unmethylated DNA cannot be proved to be amplified with similar efficiency. However, at least in the best-performing primer sets, we can postulate that the single tube approach introduces a sort of competitive amplification between the two amplifiable targets that should maintain their reciprocal concentrations constant across the amplification. This improvement seems to reduce the bias deriving from preferential amplification of unmethylated or unmethylated alleles, as demonstrated by the consistency of our results.
After HRMA the two molecular forms of methylated and unmethylated DNA can be distinguished through the different melting profile. Quantification was calculated by interpolation on a standard curve generated with serial dilution of methylated and unmethylated DNA. For each concentration we plotted the height of differential peaks obtained after automatic subtraction of fluorescence corresponding to unmethylated DNA. A similar approach was used by Lorente et al. (26) who applied a conventional melting analysis with SybrGreen, enabling them to detect 5% methylated DNA. With the use of an optimized protocol of HRMA we were able to detect 0.025% methylated DNA in reconstituted samples. This is a theoretical sensitivity, valuable for clinical studies. The high levels of sensitivity are particularly important when HRMA is used to detect traces of cancer DNA in complex human matrices (blood, urine, stool, etc.). In addition, since the amount of methylated DNA is not predictable in an unknown sample, linearity should be guaranteed in a wide range of possible concentrations. For these reasons, we tried to optimize our test to satisfy both these requirements.
Using different genes and different methylated sequences, we tested the influence of CpG number as amplicon length on HRMA performances. Our data seem to indicate that the best proportionality between methylated DNA and fluorescence signal, as well as the maximal sensitivity, are obtained when the number of CpGs in amplicons is low. In the three sets of experiments, our cumulative results indicated that a total of 13–15 CpGs in the amplified products guaranteed good linearity and sensitivity. When the number of CpGs is lower, the detection limit seems to get worse. Conversely, a higher number of CpGs tends to reduce the slope and sensitivity of the standard curve.
We cannot exclude that the length of amplicons may influence D-HRMA performance, since a major number of amplified CpGs are obviously connected to an increase in the extent of amplicons. However, the big differences in D-HRMA performances between amplicons of comparable length (hTERT-3A versus hTERT-11A or hTERT-11A versus hTERT-7B) seem to indicate that the dimensions of PCR products are less relevant. Similarly, the density of CpGs expressed as mean base pair per CpG in amplified sequence, did not appear to influence D-HRMA. A clear example emerged from the evaluation of hTERT primer sets in sequence B of the promoter: the three sets had the same CpG density (8.3, 8.8 and 8.6, respectively) but showed very different behavior.
In conclusion, our results seem to demonstrate that HRM can directly provide a quantitative assay for estimating the concentration of methylated DNA in any biological sample. We demonstrated that in standardized conditions numerical data deriving from D-HRMA are linear in a wide range of DNA methylation percentage. Reproducibility and sensitivity of the assay are absolutely compatible with a diagnostic use of the method. In addition, the use of best-performing sets of primers allowed the accurate measurement of methylated DNA in bladder cancer biopsies, as shown by the high levels of correlation with data obtained with a quantitative reference method like real time MethyLight. Finally, the use of an external reference curve, generated by serial dilutions of methylated DNA, is particularly relevant to keep under control the experimental performances over the time. The evaluation of reference parameters, like curve slope, its intercept and coefficient of correlation are parameters that can monitor experimental variations between experiments. In addition, D-HRMA appears suitable for rapid and efficient measurement in ‘in vitro’ experiments of modification of methylation patterns after treatments with demethylating drugs. However, the use of D-HRMA for quantitative purposes requires an accurate optimization of the choice of the best panel of primers to be used.
FUNDING
Grant from the Italian Ministry of University and Research (COFIN 2006).
Conflict of interest statement. None declared.
REFERENCES
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Strathdee G, Brown R. Aberrant DNA methylation in cancer: potential clinical interventions. Expert Rev. Mol. Med. 2002;4:1–17. doi: 10.1017/S1462399402004222. [DOI] [PubMed] [Google Scholar]
- 5.Baylin SB. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2:S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 6.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat. Rev. Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 7.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shames DS, Minna JD, Gazdar AF. Methods for detecting DNA methylation in tumors: from bench to bedside. Cancer Lett. 2007;251:187–198. doi: 10.1016/j.canlet.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Brena RM, Huang TH, Plass C. Quantitative assessment of DNA methylation: potential applications for disease diagnosis, classification, and prognosis in clinical settings. J. Mol. Med. 2006;84:365–377. doi: 10.1007/s00109-005-0034-0. [DOI] [PubMed] [Google Scholar]
- 10.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:e32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 12.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 13.Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp. Mol. Pathol. 2008;85:50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive melting analysis after real time- methylation specific pcr (smart-msp: high-throughput and probe-free quantitative dna methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White HE, Hall VJ, Cross NC. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin. Chem. 2007;53:1960–1962. doi: 10.1373/clinchem.2007.093351. [DOI] [PubMed] [Google Scholar]
- 17.Wojdacz TK, Dobrovic A, Algar EM. Rapid detection of methylation change at H19 in human imprinting disorders using methylation-sensitive high-resolution melting. Hum. Mutat. 2008;29:1255–1260. doi: 10.1002/humu.20779. [DOI] [PubMed] [Google Scholar]
- 18.Snell C, Krypuy M, Wong EM, kConFab investigators. Loughrey MB, Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balic M, Pichler M, Strutz J, Heitzer E, Ausch C, Samonigg H, Cote RJ, Dandachi N. High quality assessment of DNA methylation in archival tissues from colorectal cancer patients using quantitative high-resolution melting analysis. J. Mol. Diagn. 2009;11:102–108. doi: 10.2353/jmoldx.2009.080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich MG, Weisenberger DJ, Cheng JC, Chandrasoma S, Siegmund KD, Gonzalgo ML, Toma MI, Huland H, Yoo C, Tsai YC, et al. Detection of methylated apoptosis associated genes in urine sediments of bladder cancer patients. Clinical Cancer Res. 2004;10:7457–7465. doi: 10.1158/1078-0432.CCR-04-0930. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 23.Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin. Cancer Biol. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 24.Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin. Chem. 2001;47:1183–1189. [PubMed] [Google Scholar]
- 25.Guldberg P, Worm J, Gronbaek K. Profiling DNA methylation by melting analysis. Methods. 2002;27:121–127. doi: 10.1016/s1046-2023(02)00063-4. [DOI] [PubMed] [Google Scholar]
- 26.Lorente A, Mueller W, Urdangarín E, Lázcoz P, von Deimling A, Castresana JS. Detection of methylation in promoter sequences by melting curve analysis-based semiquantitative real time PCR. BMC Cancer. 2008;8:61. doi: 10.1186/1471-2407-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]