Abstract
Methylation of CpG islands associated with genes can affect the expression of the proximal gene, and methylation of non-associated CpG islands correlates to genomic instability. This epigenetic modification has been shown to be important in many pathologies, from development and disease to cancer. We report the development of a novel high-resolution microarray that detects the methylation status of over 25 000 CpG islands in the human genome. Experiments were performed to demonstrate low system noise in the methodology and that the array probes have a high signal to noise ratio. Methylation measurements between different cell lines were validated demonstrating the accuracy of measurement. We then identified alterations in CpG islands, both those associated with gene promoters, as well as non-promoter-associated islands in a set of breast and ovarian tumors. We demonstrate that this methodology accurately identifies methylation profiles in cancer and in principle it can differentiate any CpG methylation alterations and can be adapted to analyze other species.
INTRODUCTION
It has become increasingly clear how epigenetic modification can affect the structure and the expression of genes encoded in the DNA. One such modification is the methylation of cytosines that are 5′ to guanines, so-called CpG dinucleotides. Found scattered across the genome, although at a lower than expected frequency, CpG dinucleotides also cluster into what have been termed CpG islands. The definition of a CpG islands differs somewhat based on the algorithm used for identification, two commonly used algorithms being Gardiner-Garden and Frommer (1) and Takai-Jones (2). The islands identified can be classified as falling into two distinct classes, those that are overlapping or proximal (within 2000 bp) to the transcription start site (TSS) of genes and those that are not associated with any transcription start site (non-TSS) for an obvious gene. Most CpG islands proximal to the TSS of genes (TSS–CGIs) are largely unmethylated normally, and methylation of these islands, as can occur during tumorigenesis, has been shown to correlate highly to the suppression of transcription (3). Of the non-TSS CpG islands (non-TSS–CGIs) in the genome, many of these are proximal or inclusive to repetitive sequences, and are generally heavily methylated in normal tissue (4,5). However, during tumorigenesis hypomethylation occurs at these islands (4,5), which can result in the expression of certain repeats (6,7). Interestingly, this hypomethylation correlates to the severity of some cancers (8,9) and DNA breakage and genome instability (10).
Under certain circumstances, which can occur in pathologies such as cancer, imprinting, development, tissue specificity and X-chromosome inactivation, TSS–CGIs can be heavily methylated (11). Specifically, in cancer, methylation of islands proximal to tumor suppressor genes such as p16, RASSF1A, BRCA1, is a frequent event (12–14). Since the analysis of such genes was previously done one at a time using bisulfite sequencing, the value of accurate high-throughput methods is obvious.
Several higher throughput methods, many array based, have been developed to identify CpG methylation in the genome for several different species including human and plants. An indirect approach compares expression analysis of 5-aza-cytidine treated to untreated cells (15). Early methods utilized fragments cloned from CpG island libraries (16). Illumina Inc. has developed an array-like procedure based on their bead platform (17). Several other approaches have adopted more standard array platforms utilizing DNA precipitations for human and arabidopsis with either methyl-binding proteins or antibodies that recognize methyl cytosine (ChIP-chip) (18–21). Other methods utilize methylation sensitive restriction endonucleases with or without fractionation of the genome (22–25). More recently, sequencing methods have been developed using the newest generation sequencers such as the Roche Genome Sequencer FLX (454 Lifesciences technology) (26). Although these methods are likely to replace array-based method they are presently prohibitively expensive to analyze the entire genome of large sets of samples. It is more likely that for the time being, many islands will be analyzed for few samples or few islands will be analyzed for many samples.
We have developed a method to profile genome-wide methylation that is similar to the HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) assay (27) and MASS, which utilizes the enzyme McrBC (28), but we have made modifications to increase the methylation detection limits as well as the classes of islands analyzed. We have analyzed cell lines and validated measurements of methylation with bisulfite sequencing. We then went on to develop methods, which allow us to analyze tumors with unmatched normals, thereby accessing any samples in our tumor collection. This methodology will be useful to identify methylation events in the field of cancer as well as other fields such as development, aging, imprinting, etc.
METHODS
Materials
Enzymes, MspI, McrBC, T4 DNA ligase, were supplied by new England Biolabs. Primers were supplied by Sigma Genosys. Cot1 DNA and tRNA were supplied by invitrogen. The Megaprime−labeling kit, Cy3-conjugated dCTP, and Cy5-conjugated dCTP were supplied by Amersham-Bioscience. Taq polymerase [Eppendorf mastermix (2.5X)] was supplied by Eppendorf. Centricon YM-30 filters were supplied by Amicon and formamide was supplied by Amresco. Phenol:chloroform was supplied by Sigma. NimbleGen photoprint arrays were synthesized by NimbleGen Systems Inc. Design of the array was described previously (29).
Samples
Cell lines, SKBR3, Huh7, PANC1, were acquired through ATCC and grown according to specified conditions. The cell line chp-skn-1 is a primary fibroblast cell line cultured from a skin sample provided by an anonymous donor, cultured under the following conditions: DMEM + 20% FBS, Penn/Strep, and non-essential amino acids. Tumor DNA from 11 patients with advanced ovarian carcinomas who were treated at the Department of Gynecological Oncology at The Norwegian Radium Hospital (NRH) during the period May 1992 to February 2003 were included in this study. The collection is approved by the Regional ethical review board (Reference No: S-01127). Tumor DNA from 28 patients treated for localized breast cancer at the Norwegian Radium Hospital (NRH) from 1995 to 1998 was included in this project. The samples were collected under an informed consent and the project approved by the local REK/IRB (30). A small number of samples 12 breast tumors and 12 normals and 7 ovarian normals were obtained from The Cooperative Human Tissue Network, a repository of tumor material run by the National Institutes of Health.
Methylation array and detection
Coverage
Our approach to map genome-wide methylation involves tiling all the predicted CpG islands. All annotated CpG islands were obtained from the UCSC genome browser. These islands were predicted using the published Gardiner-Garden and Frommer (1) definition and involves the following criteria: length ≥200 bp, %GC ≥ 50%, observed/expected CpG ≥ 0.6. There are ∼26 219 CpG islands in the range of 200–2000 bp in the genome. These islands are well covered by Msp I restriction fragmentation. Arrays were manufactured by Nimblegen Systems Inc. using the 390K format to the following specifications. The CpG island annotation from human genome build 33 (hg17) was used to design a 50-mer tiling array. The 50 mers were shifted on either side of the island sequence coordinates to evenly distribute the island. The 390 K format has 3 67 658 available features which would not fit all islands with a 50-mer tiling. Therefore, we made a cutoff on the islands to be represented based on size, with only CpG islands of size 200–2000 bp being assayed. The array represents classical CpG islands and does not include imprint control regions or other non-island promoters known to be methylated. Background hybridization signal could be high with probes of high GC content since by definition these probes will be. Therefore, control probes, which are not in an MspI representation, were designed to represent background signal, and these probes were used to calculate signal to noise (see Supplementary Figure 2). Array design, probe sequences and further annotation are available on line http://www.ncbi.nlm.nih.gov/geo/, dataset number GSE15801.
Sample preparation and hybridization
Representations have been described previously (29), with the following changes. The primary restriction endonuclease used is MspI. After the digestion the following linkers were ligated (MspI24mer CAGCATCGAGACTGAACGCAGCAG, and MspI12mer CGCTGCTGCGTT. The 12 mer is not phosphorylated and does not ligate. After ligation the material is cleaned by phenol chloroform, precipitated, centrifuged and resuspended. The material is divided in two, half being digested by the endonuclease McrBC and the other half being mock digested according to specification by New England Biolabs. The digestion time is 3 h. As few as four 250 μl tubes were used for each sample pair for amplification of the representation each with a 100 μl volume reaction. The cycle conditions were 95°C for 1 min, 72°C for 3 min, for 15 cycles, followed by a 10-min extension at 72°C. The contents of the tubes for each pair were pooled when completed. Representations were cleaned by phenol:chloroform extraction, precipitated, resuspended and the concentration determined. Representations were run on a gel to check for content, the McrBC digested representation being ∼100–150 bp shorter on average than the Mock. DNA was labeled as described with minor changes (29). Briefly, 2 μg of DNA template was placed (dissolved in TE at pH 8) in a 0.2 ml PCR tube. Five microliters of random nonomers (Sigma Genosys) were added brought up to 25 μl with dH2O, and mixed. The tubes were placed in Tetrad at 100°C for 5 min, then on ice for 5 min. To this 5 μl of NEB Buffer 2, 5 μl of dNTPs (0.6 nm dCTP, 1.2-nm dATP, dTTP, dGTP), 5 μl of label (Cy3-dCTP or Cy5-dCTP) from GE Healthcare, 2 μl of NEB Klenow fragment and 2 μl dH2O were added. Procedures for hybridization and washing were followed as reported previously (29) with the exception that oven temperature for hybridization was increased to 50°C. In general two hybridizations were performed for all samples (carried out as dye swaps to decrease variation of labeled nucleotide incorporation) with the exception for samples analyzed in Figure 2 where four hybridizations were performed.
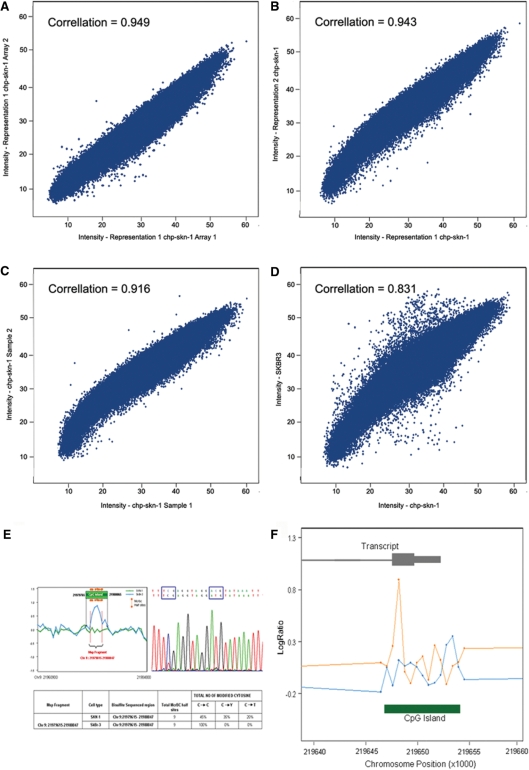
Figure 2.
Comparison of methylation analysis of cell lines. (A–C) shows the comparison of a normal fibroblast used to produce representations from two separate aliquots of DNA and the representations were analyzed and the intensity compared to each other by scatter plot; the intensities of each being on one and the other axis, and the correlation calculated: (A) being replicate hybs, (B) being technical replicates and (C) being biological replicates. (D) The comparison of the breast cancer cell line SKBR3 and the normal female fibroblast cell line as a scatter plot of the intensities from the SKBR3 experiment as the y-axis and the intensities of chp-skn-1 on the x-axis, and the correlation shown. (E) An example of bisulfite validation for a fragment found methylated in the tumor and not in the normal at a CpG island at chr 9:2197965–21980065, which lies upstream of the p14 gene. The original data showing the ratio differences is shown with the position of the fragment. To the right is the electropherogram identifying a region of cytosines that do not convert and at the bottom is a short table identifying the genomic position and number of McrBC site. (F) shows data for the cell line Huh7 compared to the normal cell line chp-skn-1 for the p16 gene CpG island graphed by genomic position. The x-axis is the genomic position of the probes and the y-axis is the ratio of two experiments, blue being chp-skn-1 and orange being Huh7. Inset above is shown is the relation of the transcript and inset below is the relation of the CpG island to the probes.
Data analysis and statistics
Microarray images were scanned on GenePix 4000B scanner and data extracted using Nimblescan software (Nimblegen Systems Inc.). For each probe, the geometric mean of the ratios (GeoMeanRatio) of McrBc and control treated samples were then calculated for each experiment and its associated dye swap. The GeoMeanRatios of all the samples in a dataset were then normalized using quantile normalization method (31). The normalized ratios for each experiment were then collapsed to get one value for all probes in every MspI fragment using the median polish model. The collapsed data were then used for further analysis (32). Collapsed fragment data were then subjected to Welch's two-sample t-test for identifying significant differences between tumors and normals. P-values were corrected for multiple testing by controlling false discovery rate (FDR) (Benjamini-Hochberg algorithm) (33) implemented in R [multitest package (34)]. Significance threshold was set at 0.001 for breast and 0.05 for the smaller ovarian set.
General computations and statistics were performed in Python (35), S-Plus and R (32).
Bisulfite sequencing
Probes were identified with differing methylation between the two samples, SKBR3 and chp-skn-1. DNAs from the two samples were treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA). PCR primers were designed to flank the corresponding MspI fragments using MethPrimer (36). For two fragments, the PCR-amplified fragments were ligated and transformed using the TOPO-TA Cloning kit (Invitrogen, CA, USA); the transformant clones were picked for plasmid extraction, which were then sequenced. To increase throughput, the remaining 32 were sequenced as PCR products. Results for subcloning and direct sequencing were compared to determine the peak heights required to call heterozygotes.
McrBC PCR
The sequence of the MTSS1 CpG island was obtained, including sequence beyond the ends of the island to aid in the design of PCR primers. Genomic DNA was digested with McrBC or mock digested followed by heat killing. Ten nanograms of DNA was used for PCR with Qiagen Taq polymerase for 30 cycles. Ten microliters of this product was then used as template for an additional 15 cycles of PCR. Products were run on 2% agarose gel, and pictures were taken to illustrate the fragments that were methylated.
RESULTS
In order to further investigate the role that CpG island methylation plays in cancer, we have designed a new comprehensive CpG island microarray and have developed robust methods for its use. While sharing some similarities to previously developed methylation arrays (22,27,37), our method has several features that allow for increased CpG-island coverage and sensitivity. First, we utilized high-density oligonucleotide arrays with close to 400K features that allowed us to maximize tiling coverage of 26 219 out of 27 801 (HG17) annotated CpG islands (which includes non-promoter islands), while other CpG island arrays only contain selected promoter sequences (22,37). Second, our hybridization target, because it is made from MspI representations, enriches for CpG island sequences by 10-fold relative to total genomic DNA (based on the size or the MspI amplifiable fragment) and thus provides superior hybridization specificity. In addition since this method is representational based, very little DNA is required (as little as 50 ng) which makes this method well suited to the analysis of primary tumors. Third, each island generally corresponds to multiple MspI fragments, yielding positional information of which portion of an island is methylated. Finally, our method is based on enzymatic depletion of methylated sequences with fewer steps than other methods (37,38); having fewer steps may be less prone to variability. The enzyme chosen for depletion, McrBC, has the unusual recognition site A/GCm(N40–3000)A/GCm and has been used by others to analyze methylation including its application to arrays (39,40). This enzyme recognizes two methyl groups and because of the varied distance and that the methyl groups can be on the same or both strands (41), a type of combinatorial recognition and cleavage occurs, which greatly increases the number of potentially methylated CpG dinucleotides that can be queried. Using in-silico analysis we calculate using McrBC (specifically the preferred distance for recognition between methyl cytosines of 40–150 bp) our methodology queries over 1 million of the 1.7 million CpG dinucleotides occurring in CpG islands, much more than can be queried by other techniques that utilize different enzymatic depletions such as HpaII and MspI (∼225 000). This gives our methodology a 4-fold increase in potential coverage over other methods. According to these calculations combined with the increased array coverage (where we include non-promoter sequences and do not discriminate against any other promoter sequences except for larger islands, see ‘Methods for array design’ section) we will have increased the level of DNA methylation that can be measured in the genome over other methods which either use restriction enzymes or limited array coverage (data not shown).
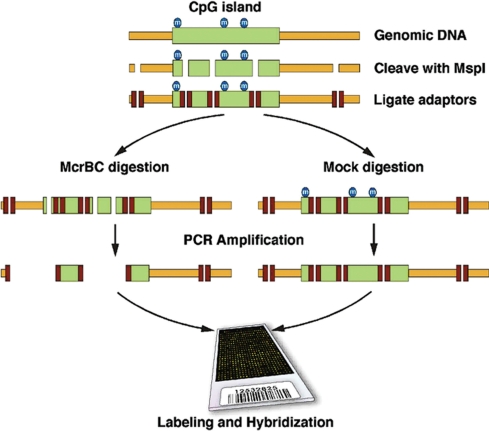
The procedure as schematized in Figure 1 involves digesting the genome with a restriction endonuclease with a CG-rich recognition sequence (MspI), and ligation of adaptors for use in a subsequent step of reducing genomic complexity. We next divide the ligation in half and deplete one-half of its methylated sequences by digestion with the methylation-specific endonuclease, McrBC (37), and mock treat the other half. In both cases, we use carefully balanced PCR conditions to size select MspI fragments and reduce the overall genome complexity as previously described (29). The McrBC-treated representation is compared to the mock-treated sample that serves as the reference for comparative hybridization to the designed oligonucleotide array.
Figure 1.
Schematic of the procedure. Shown at the top is genomic DNA with a CpG island that is methylated. The DNA is cleaved with the restriction endonuclease MspI and adaptors ligated. The ligated material is divided evenly, one half being digested with McrBC and the other half being mock digested. This material is used as template for PCR amplification and the resulting product is used for microarray comparison.
To determine if our method accurately identifies the methylation state of CpG islands, we first performed initial experiments with cell lines. To determine the level of noise in the system we performed analysis on a normal fibroblast cell line (chp-skn-1). Two cultures of chp-skn-1 were grown and DNA prepared. Representations were prepared and were used to compare hybridization results from the same representations, representation from the same sample prepared separately, as well as the chp-skn-1 grown separately. Generally two hybridizations are performed one being a dye swap to decrease variation but in this case four hybridizations were performed for all conditions; however to demonstrate how little noise there is in the system, results and correlation from a single hybridization were compared graphically shown in Figure 2A–C. The close correlation demonstrates the small variation in the representational process, the labeling and the hybridization. We also found little variation among different lots of McrBC (data not shown). Included on the array are probes that are not in a representation but have moderate CpG density. Using the average intensity for these probes in the mock channel as the background component we measured signal to noise for the probes on the array. The average signal to noise for probes on the array was 10.39.
We then performed a comparison of the breast cancer cell line SKBR3 with chp-skn-1, shown in Figure 2D (with a correlation of 0.831, which demonstrates that there are differences detected between two different samples as compared to the control experiments shown in Figure 2A–C. In the scatter plot (Figure 2D), points off the central diagonal represent fragments detecting differential methylation between the two samples. To evaluate the accuracy of measurements 25 fragments identified with methylation differences between chp-skn-1 and SKBR3, and nine fragments with no detection of methylation were selected for validation by sodium bisulfite sequencing (42). Analysis of one representative fragment is shown in Figure 2E. In each of the 28 fragments sequenced CpG dinucleotides inclusive ofo McrBC sites were mapped and their methylation state identified in the two samples (Supplementary Figure 1 for five non-gene islands, five non-CpG island regions and 10 gene-associated islands; Supplementary Table 1 which has data for all 28 fragments). Fragments were selected based on varying the difference in the array ratios measured for both samples. By doing so we were able to determine at what level accurate measurements of methylation can be determined. At a difference around 0.3 the measurements may be incorrect as can be seen by two fragments, which had a difference in ratio of 0.317. Any ratio difference above this level was found to be methylated in one of the samples. To determine if our method could accurately identify known tumor suppressor CpG island methylation, we then analyzed the hepatocellular cancer cell line HuH7 with known methylation of the p16 promoter (43). In Figure 2F we show the detection of methylation of the region of the p16 gene CpG island commonly methylated and correlated with decreased gene transcription.
Finally, we compared the results of our methodology on the two cell lines HPDE and PANC1 with results from the same two cell lines reported by Sato and colleagues using a different method and different array design (44). Although there were some differences in detection between the two methods they were in good agreement (68%) for islands methylated in the samples. Of the 34 interesting islands listed and validated, 26 were represented on our array (Supplementary Table 2, panel A). Of the 26, 17 regions were properly identified with methylation in PANC1 or both. Four regions identified as methylated by Sato and colleagues were not detected in our assay. There was disagreement in four regions. One gene RELN, which was reported as not methylated in either sample by Sato and colleagues, was found methylated in PANC1 cells. Interestingly, this gene is frequently methylated and silenced in pancreatic tumors (45), suggesting that our cell line may have genetically drifted over time apart from that used by Sato and colleagues. With our method we can obtain positional information for the region of the CpG island methylated and for all methylation events the region is very close to the TSS, the region critical for suppression of transcription. To determine the accuracy of methylation detection as well as the positional prediction we performed sodium bisulfite sequencing on the fragment, which was detected as methylated in the RELN CpG island in the PANC1 cell line. Interestingly, we found that the CpG island fragment at position chr7:103223731–103223823 is methylated (Supplementary Table 2, panel B) demonstrating that the method can accurately identify methylation and that positional information is also available. In addition we have determined that our line of PANC1 cells has deviated from the line used by SATO and colleagues.
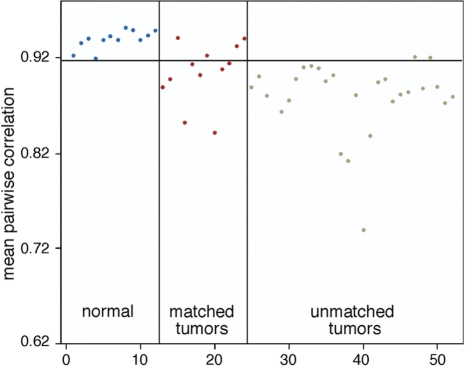
The standard method to analyze tumor-methylation profiling is to utilize matched normal-tumor samples, and we have done this for 12 breast tumor and normal pairs. As with many tumor banks, ours contain many unmatched tumor samples. To determine if unmatched tumor-normal pairs could be analyzed first we compared our results obtained with matched samples to those obtained with unmatched samples as a set and measured the degree of common alterations. We developed statistical criteria for identifying CpG-islands most significantly altered between tumor and normal (see ‘Methods for details’ section). Our analysis identified considerable overlap (30%) of CpG-islands significantly altered (Supplementary Table 3) between a matched tumor-normal set (12 normals, 12 tumors) compared to an unmatched set (12 normals, 28 tumors). Since all tumor samples are different, similarity among a set could be a measure of tumor selection. We determined how different all 40 tumors were from the normal samples. We computed pair-wise correlations between each normal sample and the other normals and each tumor sample to each normal. From this data we then computed the mean correlation between each sample and the normal samples. The mean correlation from normals to normals is 0.93, from tumors to normals is .88 (P < 10E–017). These data (plotted in Figure 3) show that tumors are different from matched or unmatched normals. However, while some tumors are vastly different from normal several are more normal like in their methylation profile. In the future it will be interesting to have more detailed clinical information to determine if methylation profile can determine specific clinical parameters. Thus, we moved on to the analysis of a larger set of unmatched tumors and normals.
Figure 3.
The mean correlation for each sample with the normal samples (mean of 12 correlations for each tumor and 11 correlations for each normal) is shown. The 12 samples to the left of the vertical line are the normals. The 40 to the right are tumors. The horizontal line is drawn at the minimum mean correlation for the normals with normals. It is apparent that the tumors are on the whole distinct from the normals in comparison with each other (P < 10E–017).
We then examined the set of 40 breast tumors, compared to 12 normal breast samples and 11 ovarian tumors compared to 7 normal ovarian samples (33). Using the statistical criteria developed (detailed in the ‘Methods’ section) we obtained a list of 916 significant alterations in breast cancer and 151 in ovarian cancer and have listed these by their genomic location (see Supplementary Table 4 for entire list and Table 2 for a summary of whether the alterations were found with more methylation or less methylation than normal, based on specific classes, non-promoter associated or promoter associated as well as non-island regions). In Table 1 we highlight genes selected based on functional information, many of which have previously been documented to alter methylation in cancer again demonstrating the ability of this methodology to detect CpG methylation. Genes identified hypermethylated in both ovarian and breast tumors include several HOX genes and protocadherins, known to be methylated in many tumor types (46–49). In addition to genes known to undergo methylation we have found new targets of methylation. For example, we detected for promoter methylation of a micro-RNA gene has-mir-9-3, occurring in more than half the breast tumors. This miRNA has been shown to be downregulated transcriptionally in thyroid cancers (50).
Table 2.
Summary of significant methylation changes listed in Supplementary Table 4 for breast cancera
| TSS associated (%) |
Non-TSS associated (%) |
|||||
|---|---|---|---|---|---|---|
| Methylated | Demethylated | Total | Methylated | Demethylated | Total | |
| CGI (801) | 166 (77.93) | 47 (22.07) | 213 | 517 (87.92) | 71 (12.07) | 588 |
| Non CGI (115) | 3 (33.33) | 6 (66.64) | 9 | 23 (21.70) | 83 (78.30) | 106 |
aAll methylation and demethylation events from Supplemental Table 4 were broken down into categories of associated to a gene (TSS associated) or not (Non-TSS associated), and methylated or demethylated within each of these groups. Finally the probes are divided based on inclusiveness to a CpG island or the probes that are non-CGI probes. The total number is given for breast cancer comparison as well as the conversion to percentages in parentheses.
Table 1.
Short list of genes selected from Supplementary Table 4 with altered CpG islandsa
| GeneName | Name | Location | Importance |
|---|---|---|---|
| Methylated | |||
| EPHA2 | Ephrin receptor A2 | chr1_16196139_16227870 | With E cadherin, mult roles in cancer |
| HOXD11 | Homeodomain protein | chr2_176795292_176795763 | Development and differentiation |
| HOXD9 | Homeodomain protein | chr2_176807548_176807855 | Development and differentiation |
| PPM1G | Protein phos'tase 1G | chr2_27544151_27545066 | Cell cycle progression |
| RAB6B | Ras oncogene family | chr3_135097550_135098100 | G protein regulation of proliferation |
| MFI2 | Melanoma-associated Ag 2 | chr3_198245561_198245644 | Melanoma tumor progression |
| IL17RB | Interleukin 17 receptor B | chr3_53855151_53855185 | With HOXB13 progression in breast cancer |
| HHIP | Hedgehog interacting protein | chr4_145923993_145924160 | Metastasis in panc cancer |
| SFRP2 | Secreted frizzled related protein 2 | chr4_155067811_155068195 | Methylated is marker for colon cancer |
| CRMP1 | Collapsin response mediator prot 1 | chr4_6010436_6010699 | Wnt pathway, lung met suppression |
| PKD2 | Polycystic kidney disease 2 | chr4_89285337_89285745 | Cell cycle |
| PCDHGA10 | Protocadherin gamma, subfam A | chr5_140768066_140768556 | Cell adhesion |
| HOXA2 | Homeodomain protein | chr7_26919212_26919376 | Development and differentiation |
| HOXA3 | Homeodomain protein | chr7_26942376_26942852 | Development and differentiation |
| EPDR1 | Ependymin related protein 1 | chr7_37728939_37729281 | Ca++ dependent cell adhesion |
| TAC1 | Tachykinin, precursor 1 | chr7_97005815_97006071 | Inflam prohormones; low act'y in brst cncr |
| MTSS1 | Metastasis suppressor 1 | chr8_125810238_125810819 | Metastasis suppressor |
| TNFRSF10D | Tumor necrosis factor superfamily | chr8_23077244_23077520 | Progresssion neuroblastoma/breast cancer |
| CUGBP1 | CUG triplet rpt, RNA bind prot | chr11_47531208_47531279 | Reg p21, cell cycle control |
| NFYB | Nuc. transcr. fctr Y beta | chr12_103034912_103035336 | p53 cell cycle |
| *PCDH8 | Protocadherin 8 | chr13_52320446_52320832 | Cell adhesion |
| ONECUT1 | Onecut homeobox domain 1 | chr15_50874380_50874668 | Reg.expression of FOXA2 |
| CDH8 | Cadherin 8, type 2 | chr16_60627191_60627579 | Cell adhesion; freq deleted in brst cncr |
| PPP1R14A | Protein phos'tase 1 reg subunit 14A | chr19_43439132_43439444 | Reg act'y RAS and ERK in tumor lines |
| IL28A/B | Interleukin 23 interferon like | chr19_44447259_44447674 | Inhibit tumor proliferation |
| FOXA2 | Forkhead box A2 | chr20_22514937_22515431 | Lung/prostate cancer |
| PLCG1 | Phospholipase C gamma 1 | chr20_39198472_39198934 | Implicated in cancer and metastatsis |
| RTEL1 | Regulator telemere elong.helicase | chr20_61758790_61759467 | Req for telomer elong.; chr breaks/loss |
| PRR5 | Proline rich 5 (renal) | chr22_43445548_43445907 | Downreg in breast cancer |
| SHOX | Short stature homeobox | chrX_551021_551222 | Reg of proliferation and viability |
| Demethylated | |||
| BRDT | Bromo domain prot testis spec. | chr1_92126308_92126790 | Spermatogenesis |
| RRM2 | Ribonucleotide reductase M2 | chr2_10212057_10212966 | Chemotherapy target |
| BCL11A | B cell lymphoma protein 11A | chr2_60694090_60694350 | Cofactor w/ SIRT1 in transcrip'n regulation |
| DUB3 | Deubiquinating enzyme 3 | chr4_9040795_9041453 | Proliferation |
| RHOG | Ras homolog gene family G | chr11_3819507_3820119 | Apoptosis, migration |
| SULT1A1 | Sulfotransferase family | chr16_28542367_28542407 | Response to therapy and prog. brst cancer |
| BECN1 | BCL2 interacting protein | chr17_38229845_38229898 | Multiple roles in cancer |
| STRA13 | Stimulated by retinoic acid 13 | chr17_77574908_77575250 | Role in cell cycle and carcinogenesis |
| MAPK1 | Map kinase 1/ERK | chr22_20546877_20547317 | Proliferation |
aGenes of possible functional interest were tabulated from the two categories based on TSS association. The gene abbreviation, full gene name, fragment location, and description of the gene are given.
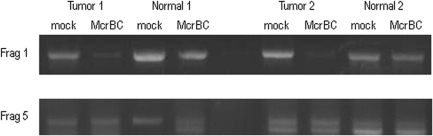
Another gene identified as methylated in the breast tumors, MTSS1 (metastasis suppressor 1) (Table 1) is known to be preferentially methylated in several cancers including breast cancer (51–53). To determine if our analytical methods identified this gene accurately, two primary tumors were identified with methylation from the dataset for further validation. Being that they were not cell lines we only had 25 ng of material that we wanted to check multiple times. Although bisulfite sequencing is generally used for validation we wished to use an alternative due to the small amount of genomic DNA remaining. Others have utilized McrBC combine with PCR to study methylation (39,54). Due to the constraints on us we chose to perform McrBC PCR (40) to validate the region of the island detected as methylated (Figure 4). The island was broken down into five fragments where fragment one overlaps the TSS up to fragment five that is the farthest away from the TSS. For tumors that were suspect to have methylation, fragment 1, which overlaps the gene TSS could not be amplified in the tumor samples (due to digestion by McrBC demonstrating methylation) as compared to the normal, which was not digested by McrBC (no methylation). Fragment 5 that is far from the TSS is amplified in all samples including McrBC digested due to a lack of methylation and no cleavage by McrBC, demonstrating that the MTSS1 CpG island is methylated in primary tumors.
Figure 4.
McrBC PCR of two different fragments of the MTSS1 CpG island for tumors identified of having methylation of this island compared to matched normal samples. Fragment 1 encompasses the MspI fragment we have identified as being methylated and overlaps the gene TSS. Fragment 5, we do not detect methylation. Both Normal and Tumor were digested with McrBC or mock digested for both matched pairs.
DISCUSSION
We have designed a methylation array and developed methods to detect CpG methylation. This methodology was performed on cell lines and measurements validated with more standard methods. Comparing the analysis of two cell lines by our methodology allowed us to test the accuracy by bisulfite sequencing fragments with differential methylation between the two cell lines. We found no errors except for one fragment that was methylated but was not detected. This could have been a failure of the McrBC or the array detection. We suspect the oligonucleotide probe since as in all hybridization-based method, there are bound to be probes that do not report well. In comparison of our method with another array-based methods for the analysis of the same samples, we found good correspondence between the two methods. One gene we found particularly interesting, RELN, is frequently silenced in pancreatic cancer. Sato and colleagues demonstrate that it is not methylated in this particular pancreatic cancer cell line but our measurements suggest that it is methylated. Judging from its frequency of silencing and our success in validation of our measurements it is likely that our version of the cell line has genetically, or in this case, epigenetically drifted from that used by Sato and colleagues. To determine if this was the case we validated our findings by bisulfite sequencing and determined that our version of the cell line had deviated and the array was correct in its measurement.
In our analysis of tumors versus normal, as expected, the tumors had more variation than the normals. However, some tumors were remarkably similar while others very different from their matched normal. More importantly, the unmatched tumors were for the most part similar in variation to normal as the matched tumors. Three breast tumors out of 40 and 2 ovarian tumors out of 11 were extremely different from the normals, which could be interesting in reference to clinical parameters or genome structure and we are investigating this further with sample sets of a larger size. We then developed statistical methods that could be used to identify those CpG islands that differ in their methylation status in the tumors as compared to the normals.
We have identified a number of CpG islands, both associated with gene TSSs or islands far from the TSS for any genes, which have altered methylation from both breast and ovarian tumors as compared to normal. However, we did not identify well-known tumor suppressors in the primary tumor dataset such as p16. The lack of known tumor suppressors could be a fault in our analysis or our methods. However, the frequency of methylation for many classically known tumor suppressors varies widely, some being as low as 15% (55–58). Using p16 as an example, analysis of the tumor samples with matched normals did not uncover tumor suppresors such as p16 and since we demonstrate the ability to detect p16 methylation in the cell line HuH7 (43) with known methylation it is likely that none of the tumors had methylation of p16. Therefore, although improvements are always possible, we feel that our technical and analytical methods are sound. Unfortunately, the tumor suppressor Rassf1a that is a common target of methylation in cancer is not measurable with the current array. The next generation array will include better design of CpG islands such as those proximal to the Rassf1A.
One disadvantage of our method of analysis is that those genes whose CGIs are found with altered methylation in few samples may not be identified. The MiR-196 loci on chromosome 17 is methylated in several of the breast cancers in our study as compared to matched normals but overall is not significant in the larger set. It has been previously shown that the miR-196 directs the cleavage of HoxB8 mRNA (59), which appears to function in myeloid differentiation (60,61), however it is possible that it plays a role in the deregulation of breast cells in becoming cancer. Presently we are further developing our statistical methods to improve our level of detection.
Of the islands associated with gene TSSs identified as methylated there are a number that have been found altered and/or have been shown by other to decrease in expression in cancer, such as MTSS1 (51–53). We found this island methylated in a number of the primary breast tumors and verified by McrBC PCR (54) that it was methylated in two of these (one such sample shown in Figure 4). Tumor suppressors that are methylated are often found in regions of loss of heterozygosity (LOH). It will be interesting to determine which of the genes found methylated are in regions of LOH. It is interesting that MTSS1 is found on the q arm of chromosome 8, which is amplified very frequently in breast cancer and ovarian tumors. It is possible that MTSS1 loss is important to the tumor growth as suggested by its role in cytoskeletal rearrangement (62) and its correlation with the disease state (51), so that in order to ensure lack of transcription within a region of genomic amplification the gene is silenced by methylation. It will be interesting to associate CGH data with methylation data and incorporate expression analysis to identify genes that are methylated and suppressed and determine if they are amplified or deleted genomically.
Of the genes identified, those that are functionally interesting should have their methylation validated by other means such as bisulfite sequencing or McrBC PCR. Those that pass can be functionally validated as candidates with possible tumor-suppressive activity. In conclusion, we have developed a powerful method to profile genome-wide DNA methylation. We have demonstrated that there is very low system noise from either the representational process or the labeling and hybridization. We then legitimatized the method's ability to detect methylation by bisulfite sequence validating over 15 fragments. We went on to analyze a number of samples that have been analyzed by others either by bisulfite sequencing or by other genome-wide approaches for methylation detection (43,45). In the case of Maeta et al., we have validated specific methylation events and in the case of Sato et al. we have reproduced the majority of their findings using our methodology. We then used this method to develop methods for the analysis of tumors with unmatched normals, which will be of interest to the greater community since many tumor banks do not have matched normals. We plan on using this methodology to identify methylation events that correlate to clinical parameters to determine if tumor sub-classification can be achieved, markers from this type of analysis being very valuable. In addition, this methodology will have utility in the study of other pathologies, such as imprinting, development, or tissue specificity, which all are affected by epigenetic modifications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health and National Cancer Institute [K01CA93634-01 to R.L.]; Department of Defense [W81XWH-05-1-0068 to R.L.] as well as by grants awarded to Michael Wigler from the Simons Foundation and the Breast Cancer Research Foundation. Funding for open access charge: awarded to to RL and Michael Wigler from Philips Research North America.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Scott Powers, Scott Lowe, Michael Wigler and Michael Zhang for critical comments on the manuscript. We also thank Christopher Johns for assistance in design of the arrays. Some tumor samples were supplied by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Other investigators may have received samples from these same tissues.
REFERENCES
- 1.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 2.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 5.Bird AP. The relationship of DNA methylation to cancer. Cancer Surv. 1996;28:87–101. [PubMed] [Google Scholar]
- 6.Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2) J. Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 8.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J. Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 9.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int. J. Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002;132:2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 11.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 12.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 13.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 14.Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 15.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr. Mol. Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 16.Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibikova M, Lin Z, Zhou L, Chudinz E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 19.Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH, Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 22.Adrien LR, Schlecht NF, Kawachi N, Smith RV, Brandwein-Gensler M, Massimi A, Chen S, Prystowsky MB, Childs G, Belbin TJ. Classification of DNA methylation patterns in tumor cell genomes using a CpG island microarray. Cytogenet. Genome Res. 2006;114:16–23. doi: 10.1159/000091923. [DOI] [PubMed] [Google Scholar]
- 23.Balog RP, de Souza YE, Tang HM, DeMasellis GM, Gao B, Avila A, Gaban DJ, Mittelman D, Minna JD, Luebke KJ, et al. Parallel assessment of CpG methylation by two-color hybridization with oligonucleotide arrays. Anal. Biochem. 2002;309:301–310. doi: 10.1016/s0003-2697(02)00294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sato M, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- 25.Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: an application toward deciphering epigenetic signatures of breast cancer. Clin. Cancer Res. 2000;6:1432–1438. [PubMed] [Google Scholar]
- 26.Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 27.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim AE, Thorne NP, Baird K, Barbosa-Morais NL, Tavare S, Collins VP, Wyllie AH, Arends MJ, Brenton JD. MMASS: an optimized array-based method for assessing CpG island methylation. Nucleic Acids Res. 2006;34:e136. doi: 10.1093/nar/gkl551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes HG, Lingjærde OC, Strømberg M, Wiedswang G, Kvalheim G, Kåresen R, et al. Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol. Oncology. 2007;1:160–171. doi: 10.1016/j.molonc.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 32.R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2007. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 34.Pollard K, Dudoit S, van der Laan MJUC. Multiple testing procedures: r multtest package and applications to genomics. Berkeley Div Biostat Working Paper Ser. 2004:164. [Google Scholar]
- 35.Van Rossum G, Drake FL., Jr . The Python Language Reference Manual. UK: Network Theory Ltd; 2003. p. 144. [Google Scholar]
- 36.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 37.Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 38.Costello JF, Smiraglia DJ, Plass C. Restriction landmark genome scanning. Methods. 2002;27:144–149. doi: 10.1016/s1046-2023(02)00067-1. [DOI] [PubMed] [Google Scholar]
- 39.Chotai KA, Payne SJ. A rapid, PCR based test for differential molecular diagnosis of Prader-Willi and Angelman syndromes. J. Med. Genet. 1998;35:472–475. doi: 10.1136/jmg.35.6.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada Y, Watanabe H, Miura F, Soejima H, Uchiyama M, Iwasaka T, Mukai T, Sakaki Y, Ito T. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–266. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 42.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci.USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeta Y, Shiota G, Okano J, Murawaki Y. Effect of promoter methylation of the p16 gene on phosphorylation of retinoblastoma gene product and growth of hepatocellular carcinoma cells. Tumour Biol. 2005;26:300–305. doi: 10.1159/000089288. [DOI] [PubMed] [Google Scholar]
- 44.Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, Hidalgo M, Goggins M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 47.Strathdee G, Holyoake TL, Sim A, Parker A, Oscier DG, Melo JV, Meyer S, Eden T, Dickinson AM, Mountford JC, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin. Cancer Res. 2007;13:5048–5055. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 48.Waha A, Guntner S, Huang TH, Yan PS, Arslan B, Pietsch T, Wiestler OD, Waha A. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying J, Gao Z, Li H, Srivastava G, Murray PG, Goh HK, Lim CY, Wang Y, Marafioti T, Mason DY, et al. Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br. J. Haematol. 2007;136:829–832. doi: 10.1111/j.1365-2141.2007.06512.x. [DOI] [PubMed] [Google Scholar]
- 50.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, et al. Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin. Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utikal J, Gratchev A, Muller-Molinet I, Oerther S, Kzhyshkowska J, Arens N, Grobholz R, Kannookadan S, Goerdt S. The expression of metastasis suppressor MIM/MTSS1 is regulated by DNA methylation. Int. J. Cancer. 2006;119:2287–2293. doi: 10.1002/ijc.22106. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Liu J, Smith E, Zhou K, Liao J, Yang GY, Tan M, Zhan X. Downregulation of missing in metastasis gene (MIM) is associated with the progression of bladder transitional carcinomas. Cancer Invest. 2007;25:79–86. doi: 10.1080/07357900701205457. [DOI] [PubMed] [Google Scholar]
- 54.Tryndyak V, Kovalchuk O, Pogribny IP. Identification of differentially methylated sites within unmethylated DNA domains in normal and cancer cells. Anal. Biochem. 2006;356:202–207. doi: 10.1016/j.ab.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 55.Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 56.Brenner AJ, Paladugu A, Wang H, Olopade OI, Dreyling MH, Aldaz CM. Preferential loss of expression of p16(INK4a) rather than p19(ARF) in breast cancer. Clin. Cancer Res. 1996;2:1993–1998. [PubMed] [Google Scholar]
- 57.Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001;61:3105–3109. [PubMed] [Google Scholar]
- 58.Silva J, Silva JM, Dominguez G, Garcia JM, Cantos B, Rodriguez R, Larrondo FJ, Provencio M, Espana P, Bonilla F. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J. Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 59.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 60.Fujino T, Yamazaki Y, Largaespada DA, Jenkins NA, Copeland NG, Hirokawa K, Nakamura T. Inhibition of myeloid differentiation by Hoxa9, Hoxb8, and Meis homeobox genes. Exp. Hematol. 2001;29:856–863. doi: 10.1016/s0301-472x(01)00655-5. [DOI] [PubMed] [Google Scholar]
- 61.Knoepfler PS, Sykes DB, Pasillas M, Kamps MP. HoxB8 requires its Pbx-interaction motif to block differentiation of primary myeloid progenitors and of most cell line models of myeloid differentiation. Oncogene. 2001;20:5440–5448. doi: 10.1038/sj.onc.1204710. [DOI] [PubMed] [Google Scholar]
- 62.Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.