Abstract
An LC-MS-NMR platform is demonstrated, which combines two innovations in microscale analysis, nanoSplitter LC–MS and microdroplet NMR, for the identification of unknown compounds found at low concentrations in complex sample matrixes as frequently encountered in metabolomics or natural products discovery. The nano-Splitter provides the high sensitivity of nanoelectrospray MS while allowing 98% of the HPLC effluent from a large-bore LC column to be collected and concentrated for NMR. Microdroplet NMR is a droplet microfluidic NMR loading method providing severalfold higher sample efficiency than conventional flow injection methods. Performing NMR offline from LC-UV-MS accommodates the disparity between MS and NMR in their sample mass and time requirements, as well as allowing NMR spectra to be requested retrospectively, after review of the LC–MS data. Interpretable 1D NMR spectra were obtained from analytes at the 200-ng level, in 1 h/well automated NMR data acquisitions. The system also showed excellent intra- and interdetector reproducibility with retention time RSD values less than 2% and sample recovery on the order of 93%. When applied to a cyanobacterial extract showing antibacterial activity, the platform recognized several previously known metabolites, down to the 1% level, in a single 30-μg injection, and prioritized one unknown for further study.
Over the past two decades, considerable efforts have been dedicated to the hyphenation of high-performance liquid chromatography (HPLC), mass spectrometry (MS), and nuclear magnetic resonance spectroscopy (NMR).1–4 This combination of technologies has emerged as an extremely powerful tool for the detection, identification, and quantitation of known, and more importantly unknown, compounds in complex clinical and pharmaceutical matrixes as well as in natural product extracts.5 Definitive identification of such unknowns is essential in the discovery of new biomarkers or drug candidates, and in the characterization of drug metabolites. However, compounds in complex matrixes generally require extensive separation and consequently often are only available in small quantities, from the microgram to nanogram level. Such low analyte amounts are problematic in even recognizing known compounds, let alone de novo structure determination. In order to effectively couple both NMR and MS to HPLC, a number of challenges need to be overcome.
The primary challenge is the intrinsically low sensitivity of NMR relative to MS and HPLC-UV. Where MS and MS/MS analyses are completed in less than 1 s with nanograms of analyte, NMR analysis at the microgram level requires acquisition times of minutes for simple 1D spectra to days for many 2D spectra. Many methods have been developed to enhance the sensitivity of LC-NMR analysis. Stopping the LC flow is routinely used to increase the residence time of a peak of interest. The loading capacity, however, of LC columns is a major limitation; larger columns elute peaks in a larger volume, making it necessary to use large probes having much lower sensitivity. Trapping LC peaks on solid-phase extraction (SPE) cartridges or guard columns has been successful in concentrating all of the available analyte into smaller, more sensitive NMR probes.6–8
Marked improvements in NMR mass sensitivity have been achieved in recent years with the development of microcoil probes, which use very small, highly sensitive radio frequency (rf) detectors. A reduction in the rf coil diameter proportionally decreases noise in the NMR probe, improving signal-to-noise ratio (S/N) if the same amount of analyte is soluble in the smaller volume, which is typically the case with LC-purified analytes.9,10 Additionally, using a solenoidal coil provides 3-fold higher signal than a comparably sized saddle coil.11–13 Microcoil probes have shown better mass sensitivity than a 5-mm cryoprobe at a similar field (a 1-mm superconducting probe currently holds the mass sensitivity record14). Microcoil probes have major advantages in being affordable, priced similar to conventional tube probes, and they can be quickly exchanged with other probes on shared NMR instruments, and thus are more readily available.
Although SPE-NMR streamlines the traditional approach15 of purifying samples for NMR, its need to preselect LC peaks for NMR leaves strong arguments for using LC–MS with fraction collection, followed by NMR acquisition of select fractions or regions.16 Offline LC-NMR retains the potential to detect “LC–MS-silent” analytes–which lack UV chromophores or have poor MS ionization, such as many glycans and lipids. Simple 1D NMR data available from high-throughput methods are frequently needed to complement MS to definitively recognize previously known compounds or irrelevant metabolites. The interactive analysis time and expense required for de novo structure identification of unknown compounds is a limited resource in most projects; only the most promising compounds can be prioritized for this investment if high-throughput MS and NMR data can be conveniently acquired. Fraction collection is a common accessory in LC and LC–MS facilities, it can be performed routinely at the very modest cost of plates and it enables NMR data to be acquired retrospectivelyswithout repeating an LC–MS analysis–after LC–MS data are reviewed together with any subsequent analysis (e.g., bioassay). Although any automated NMR sample-loading method could in principle be used to analyze the collected fractions, flow-based automation from well plates avoids the need for sophisticated robotics and high ongoing material costs of tube-based automation or SPE collection systems.
Achieving high sensitivity in flow NMR requires attention to sample preparation and loading. The commercially available microcoil probe used in this platform, for example, requires filling a 1.5-μL NMR observed volume through a significant dead volume–6 μL from the probe inlet and 25 μL or more from a sample handler. The resulting sample efficiency (the percentage of the injected sample in the observed volume) can be low. Addressing issues of sample efficiency and dilution, several flow NMR methods have been developed17,18 including direct injection NMR,19 flow injection analysis NMR (FIA-NMR),13,18 and recently segmented flow analysis NMR (SFA-NMR)20,21 including the present microdroplet NMR.
Segmented flow analysis (SFA) has been demonstrated to be a particularly mass-sensitive, sample-efficient approach for high-throughput microcoil NMR.20 In SFA, samples are moved as a “plug” in an immiscible carrier fluid; “droplet microfluidics” is a rapidly emerging field.22 Because segmented sample plugs do not disperse into the immiscible carrier, as sample zones do in FIA, smaller sample volumes can be loaded without dilution or dispersion, providing an exponential reduction in NMR acquisition times as well as reducing consumption of both analyte and deuterated solvent. Successful implementation of SFA-NMR requires that sample plugs be moved through several meters of transfer capillary between the sample loader and the NMR probe without the sample plugs becoming fragmented or the sample adsorbing onto capillary surfaces. The microdroplet system utilizes “zero dispersion” segmented flow, based on the principle that if the carrier fluid has a favorable contact energy with the tubing wall, relative to the sample, a layer of carrier is maintained between the wall and the sample as the plugs are transported.23–25 Perfluorocarbons, which have a Teflon-like immiscibility with all common NMR solvents, may be used as carrier fluids in Teflon tubing to achieve zero-dispersion sample transfer.24,25 Based on this principle, an automated system for loading samples into a microcoil NMR probe from 96-well plates was developed and has been applied to high-throughput NMR analysis of compound libraries.20 Although the published system was optimized for rapid analysis of compound libraries in a custom-built microcoil probe, its sample loading efficiency was quite high relative to other flow-NMR methods and even NMR tubes. It has been further developed here for trace analysis applications and adapted to commercially available microcoil probes.
A second challenge in the development of an LC-MS-NMR system, after addressing the sensitivity of NMR, is the establishment of an LC–MS interface that provides fast, sensitive, and routine MS analysis, while collecting as much material as possible for NMR. When LC is coupled to MS for the analysis of biological samples, nanoelectrospray ionization is overwhelmingly preferred due to its ability to achieve optimum MS sensitivity by lowering ion suppression, increasing ionization efficiency, and minimizing sample consumption.26 Nanoelectrospray is generally limited to low-capacity narrow-bore columns that feed the entire eluent flow to the MS at a flow rate of <300 nL/min. To provide as much material as possible for NMR analysis, a normal bore as opposed to a narrow-bore column is required. Traditional packed columns have loading capacities of as much as 200 μg/injection, or even as much as 1 mg of material for new 4-mm monolithic columns. Moreover, normal-bore columns are justifiably considered easier to use, have more reproducible retention times, and can tolerate injection of larger volumes of relatively less clean samples, such as reaction mixtures or biological fluids.
An LC-MS-NMR platform would thus ideally couple normal-bore HPLC (2−4-mm columns) with nanoelectrospray ionization, in order to provide enough material for the NMR while maximizing the sensitivity of MS. With these considerations in mind, we have recently developed an interface, termed the nanoSplitter, that accomplishes this goal by delivering a small fraction of the HPLC effluent (<0.1%) to the MS through a novel concentric split design while maintaining the chromatographic integrity of the LC–MS system.27,28 When compared to conventional LC electrospray ionization MS, the nanoSplitter interface showed an average improvement of 10-fold in concentration sensitivity and 1000-fold in mass sensitivity.29
In combining the optimal technologies of the automated microdroplet NMR loading system for microcoil NMR analysis and the nanoSplitter interface for nanoelectrospray LC–MS analysis, a highly sensitive, offline LC-UV-MS-microcoil NMR platform was developed for the trace analysis and structural characterization of compounds in complex matrixes. This apparently simple approach chooses and adapts two LC–MS and NMR methodologies providing the highest sensitivity when used together, and which fit naturally into the workflow of (traditionally separate) LC–MS and NMR facilities.
In the approach described here, 98% of the large-bore column HPLC effluent is directed to a fraction collector for subsequent NMR and bioassay studies while the remaining 2% is directed to the nanoSplitter for nanospray LC–MS analysis. To evaluate this system, a series of experiments testing separation, fraction collection, preconcentration, and microcoil NMR acquisition were performed on a mixture of four commercial drugs (cycloheximide, indapamide, digitoxin, taxol). The system offers impressive LODs, at the 50-ng level for NMR, excellent reproducibility (RSD = 1.17%), and sample recovery on the order of 93%. Finally, a bioactive cyanobacterial extract was analyzed to demonstrate the system's applicability in natural product discovery. The LC-MS-NMR platform recognized four known natural products, ambiguine A, I, E, and hapalindole H, from a single 30-μg LC injection of cyanobacterial crude extract LC and, most impressively, identified one LC–MS peak as a novel bioactive compound. This illustrates the system's significant potential in natural product discovery as well as its potential in metabolomics and other fields requiring trace analysis of components of complex mixtures.
EXPERIMENTAL METHODS
Chemicals
Deuterated solvents were obtained from Cambridge Isotope Laboratories (Andover, MA). HPLC-grade acetonitrile (99.9%) and methanol were from Fisher Scientific (Pittsburgh, PA). Water was purified by using a Milli-Q Plus system (Millipore, MA). Fluorocarbon FC-43 was from 3 M Corp. (St. Paul, MN). Cycloheximide, indapamide, digitoxin, paclitaxel (taxol), and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Materials
Teflon capillaries and tubing were obtained from Cole-Parmer (Vernon Hills, IL). PEEK capillaries, unions, in-line filters, and adapters were from Upchurch (Oak Harbor, WA). The 96-well low retention PCR plates were obtained from Nunc (part 240600, Rochester, NY).
HPLC
Chromatographic separations and fraction collection were performed on an HPLC system consisting of a binary pump, an autosampler, a UV–vis diode-array detector (Agilent 1100 series), and a fraction collector (Agilent 1200 series) controlled by Agilent ChemStation (version B.02.01) software. The HPLC column used for standards was a 4.6 × 150 mm HPLC column (Agilent Zorbax C-18SB 3.5 μm), and for cyanobacterial extracts, a 4.6 × 250 mm HPLC column (Waters Atlantis C18 5 μm) was used. A restriction valve was used to split the flow from the LC column with ∼98% of the flow to the UV-vis diode-array detector and eventually to the fraction collector, and the other ∼2% of the flow to the nanoSplitter. The delay volume between the UV detector and the fraction collector was determined to be 71 μL. The chromatographic methods are described below.
NanoSplitter Interface and Mass Spectrometer
The nanoSplitter consists of a splitter (FSMUAS1.5, Valco Instruments Co. Inc., Houston, TX), a micro flow-through connector (Valco Instruments Co. Inc.), a needle valve (86041, Alltech, Deerfield, IL), and an XYZ positioner (FP-2 Newport, Irvine, CA). These components are fastened to a rail-and-mount system (9742 (M), New Focus, Inc., Sunnyvale, CA). The fused-silica emitters, obtained from New Objective (Woburn, MA), had an inner diameter of 20 μm with a tip (distal coated) of 10-μm inner diameter. The high-voltage connection was made by attaching a clip to the emitter. The split ratio was adjusted using the needle valve to obtain optimal electrospray. The flow into the MS was ∼200 nL/min and was measured by collecting flow with electro-spray voltage off. More details regarding the design and construction of the nanoSplitter can be found in the previous publications.27,29,30
Both MS and MS/MS spectra were acquired on a Finnigan LCQ classic quadrupole ion trap (San Jose, CA) controlled by Xcalibur software (version 1.3).
Microdroplet NMR (Zero Dispersion Segmented Flow Analysis NMR)
The automated system for loading samples from 96-well plates into the microcoil NMR probe consists of a Gilson (Middleton, WI) model 215 sample handler and a sample loader, model HTSL-1100, from Protasis Corp. (Marlborough, MA). The Gilson sample handler drew sample plugs from the 96-well plate into the HTSL sample loop. The sample plugs were formed by alternately drawing the immiscible fluorocarbon FC43, the sample, and then more FC. Additionally, wash plugs of clean solvent were drawn between samples. The HTSL sample loader consists of an LC injection valve with sample loop, a high-pressure pump, and a microprocessor controller. It was used to deliver sample plugs from the sample loop to the microcoil probe via a 3-m-long transfer line of 150-μm-i.d. Teflon tubing. Gilson and HTSL automation was controlled using Varian VAST automation programming on the spectrometer host computer (Sparc Ultra 5, Solaris 8, VNMR 6.1 C NMR software). NMR acquisition setup macros were written to automatically detect and position an arriving sample and to set up standard spectra of samples. In addition, four sample handler programs (Tcl scripts) were written to (1) form a train of three samples and hold it in the needle line, (2) draw a train from the needle line into the sample loop, and (3) change samples by triggering the sample loader to run until stopped by the autodetection macro. The fourth script (4) was run once on the first train to initialize the sample queue by moving this sample train half of the distance from the sample loop to the NMR probe. Additional details on the design, construction, and characterization of the automated segmented flow analysis NMR system can be found in our previous publication.20
The microcoil probe used in this study was an ICG capLC microflow probe manufactured by Magnetic Resonance Microsensors (MRM, Savoy, IL) and distributed by its parent company, Protasis Corp. This probe has an observed volume (Vobs) of 1.5 μL as determined by SFA of small plugs, in a fill volume of 6 μL, through 75-μm inlet and outlet capillaries. The probe was internally coated with fluorooctylsilane for use with microdroplet NMR.
NMR spectra were acquired on a Varian (Palo Alto, CA) Inova spectrometer with an 11.7-T (500 MHz) actively shielded magnet; the data were processed and analyzed with VNMR version 6.1C software.
System Reproducibility and Sample Recovery
A test mixture comprising equimolar quantities (0.65 mM) of cycloheximide, indapamide, digitoxin, and taxol dissolved in 30% acetonitrile/70% water (v/v) was used for testing system reproducibility and compound recovery. A 100-μL volume of this mixture (containing 24.1 μg indapamide) was injected onto a 150 × 4.6 mm i.d., Agilent Zorbax SB-C18 column (3.5 μm) (Wilmington, DE). The HPLC was operated at a flow rate of 1 mL/min and in gradient elution mode. Mobile phase A was water with 0.1% (v/v) formic acid, and mobile phase B was acetonitrile with 0.085% (v/v) formic acid. Mobile phase B was held at 30% for the first minute and then increased linearly to 95% over 15 min. Subsequently, mobile phase B was held at 95% for 4 min, giving a total run time of 20 min. The eluents were monitored by the UV-DAD at 210 nm with DAD spectra acquired every 0.5 s. The LCQ-classic ion trap was operated in positive ion mode. The fraction collector was operated in either a time-based or the peak-based mode as indicated, and the fractions were collected into a 96-well plate with a maximum collection volume of 250 μL/well. After evaporating the solvents in the wells, all the wells containing indapamide were washed with acetonitrile (with 5% DMSO) and the solutions were then pooled and transferred into a well in another plate. The solvent in the well was evaporated again and the material in the well was dissolved in 5 μL of deuterated DMSO containing 15.5 mM caffeine, used as internal standard for quantitation. The solution of the mixture in the well (3.5 μL out of 5 μL) was then loaded by the automated microdroplet NMR system into the microcoil NMR probe for NMR analysis. The quantitative NMR spectra were acquired at ambient temperature (22.5 °C) with 500 transients, 8000-Hz spectrum width, auto gain, water presaturation and, for this quantitative analysis, a 90° pulse and an additional 30-s relaxation beyond the 2-s acquisition time. The above process was repeated six times to test the reproducibility of the LC-MS-NMR system.
Additionally, to determine indapamide recovered from the LC, 24.1 μg of indapamide was dissolved in 5 μL of deuterated DMSO containing 15.5 mM caffeine and the resulting mixture was added into a well of a 96-well plate. The solution in the well (3.5 μL out of 5 μL) was also loaded by the automated sample loading system into the microcoil NMR probe for NMR analysis. The NMR spectrum was then acquired under the same conditions as above. The entire process was again repeated six times.
Detection Limit and Linearity
A solution of 250 ng of indapamide in 50 μL of 30% acetonitrile/70% water (v/v) was injected onto the Zorbax column. The LC, UV, MS, and fraction collection methods were the same as above. The indapamide fraction was collected, dried, and resuspended in 5 μL of deuterated DMSO. A 3.5-μL aliquot of the solution was loaded by the automated sample loading system into the microcoil probe for NMR analysis. The NMR spectra were acquired at ambient temperature with 1200 transients (1 h), 8000-Hz width, 30° tip angle, fixed gain (max 60), and 1-s water presaturation after the 2-s acquisition time. The indapamide resonance was integrated as in the recovery determination above. The procedure was applied for the recovery of 250 ng, 500 ng, 750 ng, 1 μg, 1.5 μg, and 15 μg indapamide in 50 μL of 30% acetonitrile/70% water (v/v) to test the linearity of the system's performance.
Natural Products Characterization
A crude extract of cyanobacteria, strain Fischerella ambigua, was prefractionated by silica gel chromatography into 6 fractions (eluted with a step gradient of CH2Cl2 and MeOH solvent mixtures). Fraction 6 (eluted from the silica gel with 100% CH2Cl2) showed activity in a proteasome inhibition assay and was used to demonstrate the LC-MS-NMR system's applicability to drug discovery from natural products.
A solution of 30 μg of the bioactive fraction in 30 μL of methanol, spiked with 300 ng of taxol, was loaded onto a 250 × 4.6 mm i.d., Waters Atlantis C18 column (5 μm) (Milford, MA). The HPLC was operated in gradient elution mode at a flow rate of 1 mL/min. Mobile phase A was water, and mobile phase B was methanol. Mobile phase B was held at 80% for the first minute and then was increased linearly to 90% in 50 min. After that, mobile phase B was increased linearly to 100% in 4 min and then decreased to 80% in 1 min and held at 80% for another 4 min, which gave a total run time of 60 min. The effluents were monitored by the UV-DAD at 254 nm with DAD spectra acquired every 0.5 s and by the LCQ-classic ion trap operating in positive ion mode. The MS/MS acquisition was executed in data-dependent mode. In addition, the fraction collector was operated in peak-based mode and the fractions were collected into a 96-well plate with a maximum collection volume of 250 μL/well. After evaporating the solvents in the wells, all the wells containing the peaks of interest were resuspended, washed with methanol (with 5% DMSO as keeper), and transferred into another plate, pooling fractions of the same peak. The solvent in each well was evaporated again and the dried material was dissolved in 5 μL of deuterated DMSO. Subsequently, the solution in each well (3.5 μL out of 5 μL) was transferred by the automated sample loading system into the microcoil NMR probe for NMR analysis. The NMR spectra were acquired at ambient temperature with 2000 transients (2 h), 8000-Hz width, auto gain, and 1-s water presaturation after a 2-s acquisition time.
In addition, two more LC runs (same LC, fraction collection and MS methods as the above run) were performed with a total loading of 80 μg of the bioactive fraction onto the Atlantis column. Fractions containing the same chromatographic peaks from separate runs were pooled together for microcoil NMR analysis as described above.
RESULTS AND DISCUSSION
The goal of this work was to implement a high-throughput LC-MS-NMR platform, which would provide the highest possible sensitivity for the structural identification of unknowns in complex sample matrixes. A schematic of this platform is shown in Figure 1. As discussed previously, two innovative established techniques, the nanoSplitter for LC–MS and the microplug automated sample loading method for offline microcoil LC-NMR, were combined offline in tandem to best complement each detector's optimal working conditions. Successful integration of these techniques requires the preservation of the optimal performance of each individual system's components and can be shown by overall system reproducibility, quantitative transfer of collected fractions for NMR analysis, and satisfactory limits of detection and dynamic range. Subsequently, the system will be demonstrated in the recognition of components in a natural product extract and prioritizing the unknowns identified for subsequent structural determination.
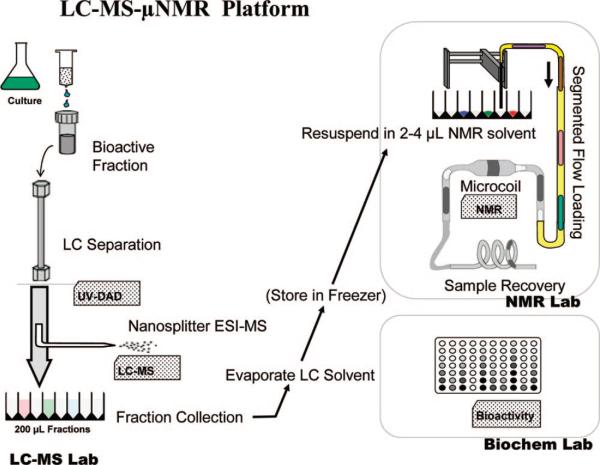
Figure 1.
Schematic diagram of the LC-MS-NMR platform, as applied to natural product discovery. The complex sample (bioactive fraction) is separated using high-resolution LC with UV and MS data acquired online. 98% of the eluent is directed to a UV-guided fraction collection. Fractions are concentrated by drying and may be stored. For NMR, fractions are resuspended in a small volume (2−5 μL) of deuterated solvent and loaded into a microcoil NMR probe, with an observed volume of 1−2 μL, using microplate automation. Samples are recovered after NMR analysis for additional analyses, archival or bioassay.
Correlation of UV, MS and NMR Data
An implicit requirement of any LC-MS-NMR system is that the NMR spectra can be correlated with features in the UV and MS chromatograms. This is very important in analysis of uncharacterized complex samples, so the system can reliably correlate the NMR spectra to specific time points in the chromatographic separation with enough confidence to confirm when signals are not seen on the other detectors, for example, if sample components lack UV chromophores or have poor MS ionization, such as glycans and lipids. Because the UV detector guides the fraction collection in this current implementation, in order to correlate the NMR data acquired for each fraction with its MS data, any variation in retention time between UV and MS chromatographic peaks must be negligible, relative to peak width.
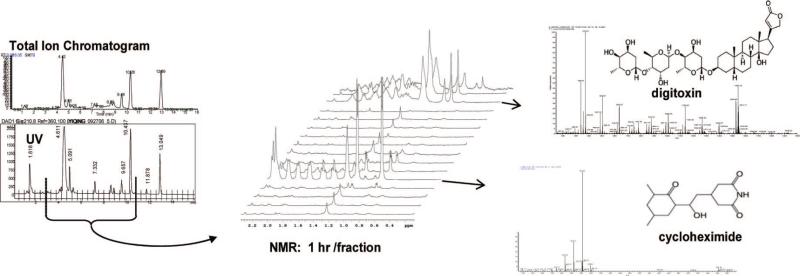
The system's performance was tested with the analysis of a mixture of the commercially available drugs cycloheximide, digitoxin, indapamide, and taxol. Figure 2 shows a comparison of the MS total ion chromatogram (TIC) and the HPLC UV chromatogram monitored at 210 nm. Minor peaks seen are either degradants from the analytes being dissolved in methanol or impurities. The tic marks on the UV chromatogram indicate the time-based fractions collected. The NMR section of Figure 2 shows a stacked plot of a representative region of the NMR spectra (0.2−2.2 ppm) for the indicated fractions. The point where each compound is eluted is indicated, as is its corresponding MS. Time-based collection, as shown, provides data similar to online LC-NMR. In LC-MS-NMR, the NMR acquisition time is optimized by targeting specific features of interest, and peak-based fraction collection is generally preferred.
Figure 2.
LC-MS-NMR data shown for a standard mixture of cycloheximide, digitoxin, and taxol. At left an MS TIC is compared with a UV chromatogram to show preservation of chromatographic integrity and peak retention times. The red tic marks on the UV trace indicate the time-based fractions collected, with the indicated fractions shown as stacked NMR spectra in the center. At right are MS spectra corresponding to the UV and NMR data shown.
Retention times between the UV and MS chromatograms were compared and, in six repetitions of the analysis, MS and UV peaks aligned to within less than 0.1 min. Systematic differences were minimized by timing the MS acquisition start with that of the LC injection. Variations in retention times were minimal over the entire chromatogram and were significantly less than the widths of the LC and MS peaks being compared. Additionally, each NMR spectrum corresponded accurately to both the UV and MS fraction with which it was correlated, demonstrating the ability of all three detectors in the system to simultaneously and reproducibly detect all compounds in the entire chromatogram.
Sample Recovery
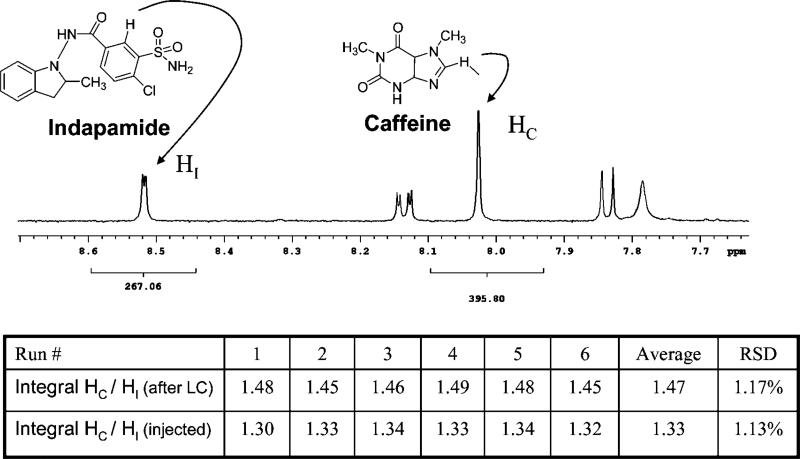
An additional requirement for integration of LC–MS and NMR is near-quantitative recovery of compounds from LC–MS and transfer to NMR. Because NMR sensitivity and data quality depend primarily on the amount of compound, the steps of fraction collection, concentration, transfer, and reconstitution are critical, especially when low-level compounds are of interest. Experiments were thus performed to compare the amount of a standard injected onto the column with the amount collected and recovered for NMR. The comparison was made using quantitative NMR,31 which is a precise method for comparing the concentrations of two analytes based on the property that the area of each NMR peak, under appropriate experimental conditions, is directly proportional (within 2%) to the number of the corresponding nuclei. Indapamide, added to the standard mixture above, was used as the standard for injection and recovery; the amounts injected and recovered were each compared to a quantitative addition of caffeine used as the reference standard for quantitative NMR.
To determine recovery, an aliquot of 24.1 μg of indapamide was dissolved and injected into the HPLC. Its peak fraction was collected and resuspended in 5 μL of DMSO-d6 with quantitative addition of 15.5 mM caffeine as an internal standard. This mixture was then transferred by the automated segmented flow sample loading system into the microcoil probe, and quantitative spectra were acquired. In the 1H NMR spectra, a 1-proton peak from caffeine (8.02 ppm) and a 1-proton peak from indapamide (8.52 ppm) were integrated and compared. The longitudinal relaxation times, T1, of these caffeine and indapamide resonances were 6 and 1.5 s, respectively, and the NMR relaxation delay was 30 s. The ratio of the indapamide and caffeine integrals was 1.47 with an RSD of 1.2% over six repetitions. This low RSD shows consistency and reproducibility in the HPLC recovery and NMR sample handling process. To compare this with the amount of indapamide loaded onto the LC column, an identical 24.1-μg aliquot of indapamide was dissolved in the same caffeine-spiked DMSO-d6 standard as the dried fraction-collected sample and loaded to NMR using the same automated protocol, to normalize any NMR system losses. In those measurements, the ratio between the integrals of the caffeine and indapamide resonances was 1.33, with an RSD of 1.1% over six repetitions. Therefore, as shown in Figure 3 based on the two integral ratios obtained (1.33 and 1.47) and the split ratio of LC flow (97.5% of the LC flow goes to fraction collector), the recovery from LC loading, separation, fraction collection, drying, and resuspension was 92.8%.
Figure 3.
Quantitative NMR spectrum of indapamide and caffeine indicating the NMR peaks integrated to determine recovery and reproducibility of the LC-MS-NMR platform. The values of integrals from six repetitions are tabulated.
This experiment clearly demonstrates the recovery and reproducibility of the nanoSplitter MS/microdroplet NMR approach. Results with other LC methods may of course vary with the method used for the analyte of interest.
Limit of Detection (LOD) and Dynamic Range
For hyphenated methods, the limit of detection is generally defined by the performance of the less-sensitive detector. Various interpretations of the “limit of detection” in NMR span several orders of magnitude, depending on the sample (natural line width and multiplicity), the instrument (magnetic field strength and probe type), and the information sought from NMR. There can be as much as a 1000-fold difference in the amount of sample required for a simple confirmation of a proposed structure (1D NMR) or a challenging de novo structure determination (heteronuclear 2D NMR).
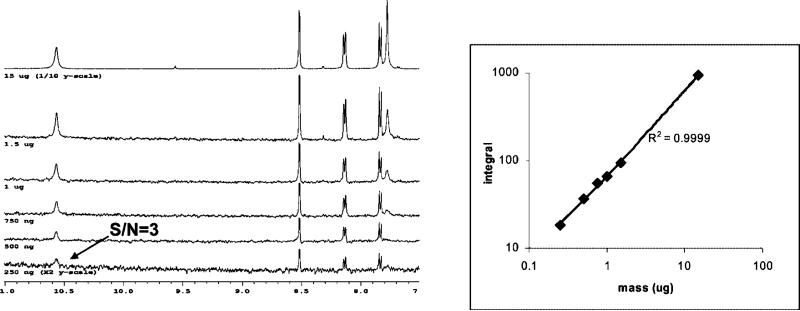
For the LC-MS-NMR system in natural product applications, the LOD would appropriately be the amount of a minor component necessary to obtain an interpretable NMR spectrum, suitable for dereplication against a library of NMR spectra, under typical acquisition conditions. We thus performed experiments to determine the minimum amount of indapamide that, when spiked into the standard mixture above, would generate a 1H NMR spectrum with a S/N of 3 for the smallest peak (10.6 ppm) in a 1-h NMR acquisition. This definition would apply to a high-throughput LC-NMR analysis of 12−48 fractions in an overnight or weekend, on our 500-MHz NMR spectrometer. LC separations were performed by loading 250 ng, 500 ng, 750 ng, 1 μg, 1.5 μg, or 15 μg of indapamide onto the LC column. After fraction collection, LC solvent evaporation, deuterated solvent resuspension, and NMR acquisition, the 250-ng (0.68 nmol) sample provided an S/N of 3 for the 10.6 ppm peak in 1 h. Therefore, 0.68 nmol is a reasonable expectation of the platform's limit of detection for the high-throughput characterization of unknowns. If a specific LC peak were of interest, a similar S/N could be expected from a 16-h overnight acquisition from 1/4 the amount of material, 0.17 nmol. In targeted overnight analyses, NMR acquisition of a single well obtained interpretable 1D spectra from 50 ng of taxol (58 pmol) and HMBC spectra from 35 μg. These data will be published elsewhere (manuscript in preparation).
A second critical property of an analytical system is dynamic range. The complexity of a natural product extract requires analysis of many unknown compounds present in concentrations ranging over many orders of magnitude, and NMR provides an estimate of concentration to evaluate potency. To assess linear dynamic range, a representative resonance (8.5 ppm) was integrated for each of the above six loadings and is plotted in Figure 4. The R2 value of the series is 0.9999, which primarily indicates that sample recovery is constant over this loading mass range, given the established quantitative linearity of NMR.
Figure 4.
LOD following the linearity of 8.5 ppm indapamide peak over six concentrations and resulting concentration curve. (Left) Regions of the NMR spectra of indapamide indicating the smallest peak, used to determine LOD. (Right) The plot of NMR integrals of indapamide (as in Figure 3) versus amount loaded onto the LC column, showing linear dynamic range over the six concentrations analyzed.
The above results show that a routine 1-h NMR acquisition with the automated microdroplet NMR system can detect and quantitate analytes from over 10 μg down to less than 300 ng with confidence. With the demonstration of reproducibility, recovery, sensitivity, and dynamic range above, the applicability of the system to the characterization of natural products and identification of components will be described next.
Identification of Metabolites in Extracts of Cyanobacteria
Following confirmation of the reproducibility, recovery, and dynamic range of the LC-MS-NMR system, its practical utility toward the identification and characterization of natural product unknowns in cyanobacteria was examined. A particularly compelling need for trace-level chemical analysis is seen in the field of drug discovery from natural products. Natural products and their derivatives have long played an important role in drug discovery; 61% of the 877 small-molecule drug candidates developed during the period of 1981−2002 can be traced to or were inspired by natural products.32,33 These compounds are traditionally discovered by “activity-guided fractionation”5 where, when an active extract is found, it is separated chromatographically and fractions are tested again for activity in the bioassay. The active fraction is then separated again using an orthogonal separation method, and fractions are reassayed, until a pure compound is obtained. This series of separations is then scaled up and repeated to purify enough of the active component, typically several milligrams, for its structural identification and potency determination in a quantitative activity assay. However, because many active components involve the rediscovery of known compounds, this approach to the discovery of lead candidates from natural products can be time-consuming and costly. The combination of LC–MS and NMR data has been shown to be valuable in “dereplication”5,34–elimination of known compounds from further investigation and prioritization of likely unknowns for the expensive steps of scale-up and structure determination.
The ability to obtain LC-MS-NMR data of submicrogram-level compounds in a complex sample can streamline the traditional bioactivity-guided fractionation approach to natural products discovery, obviating the need to perform scale-up purification of milligrams of the active component after it is isolated if it is already known.5 A convenient LC-MS-NMR system could thus reduce this high overhead of purifying large amounts of redundant compounds and thereby accelerate drug discovery from promising natural sources.
Cyanobacteria are unique phyla that grow in competitive niches and, as a result, are promising sources of bioactive compounds.35 However, their slow growth rate in culture and low biomass yield have made them prohibitively expensive and time-consuming to search for natural products by traditional methods, which require milligram amounts of material for identification. Successful characterization of active metabolites from cyanobacteria would thus establish significant practical advantages of the microgram-sensitivity LC-MS-NMR platform described herein.
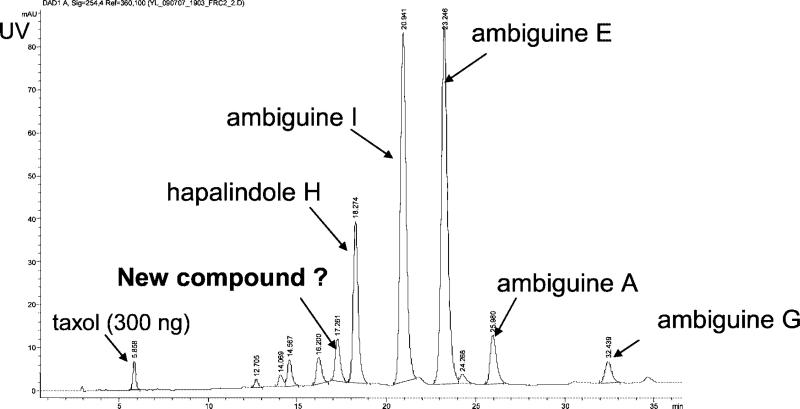
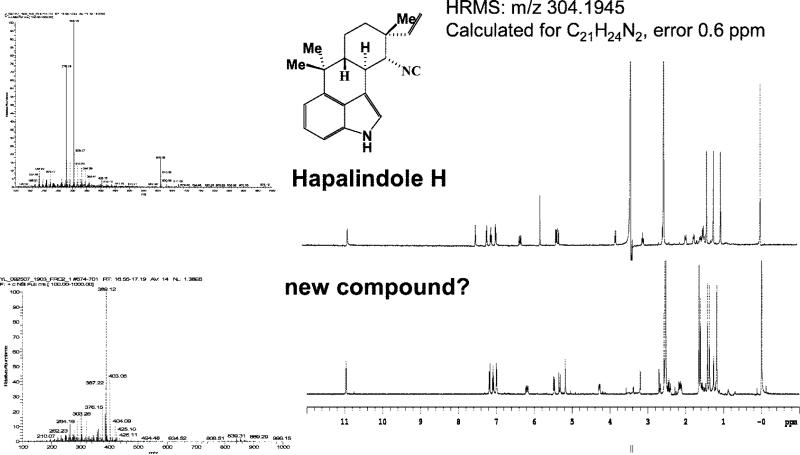
An extract of the cyanobacterium F. ambigua (Utex 1903) showed antibacterial activity against Myobacterium tuberculosis. The most-active fraction of an initial silica-gel solid-phase extraction, eluted with 100% dichloromethane (“fraction 6”), was collected, and 30 μg of this bioactive extract was subjected to LC-MS-NMR analysis. In the spectra shown in Figures 5, 6, and 7, the fraction was spiked with 300 ng of taxol as an internal standard. Based on the UV chromatogram of the separation shown in Figure 5, 12 peak fractions were collected, in addition to the taxol standard, and prepared for μNMR analysis. Each fraction then underwent a 2-h 1H NMR acquisition in an overnight autosampler run. The four largest LC peaks were readily recognized as four known isonitrile-containing indole alkaloids (isonitriles of ambiguines A, E, and I, and hapalindole H) by comparing the experimental MS and 1H NMR spectra with published data.36–41
Figure 5.
UV chromatogram of the separation of a bioactive cyanobacteria extract analyzed with the LC-MS-NMR system, indicating known and unknown compounds found.
Figure 6.
(Top) MS and NMR spectra of the 18.3-min LC peak of Figure 5, identified from the literature as hapalindole H. (Bottom) MS and NMR spectra of the 17.3-min LC peak of Figure 5 not found in the literature or natural product databases. It was therefore prioritized for detailed structure studies by scale-up purification.
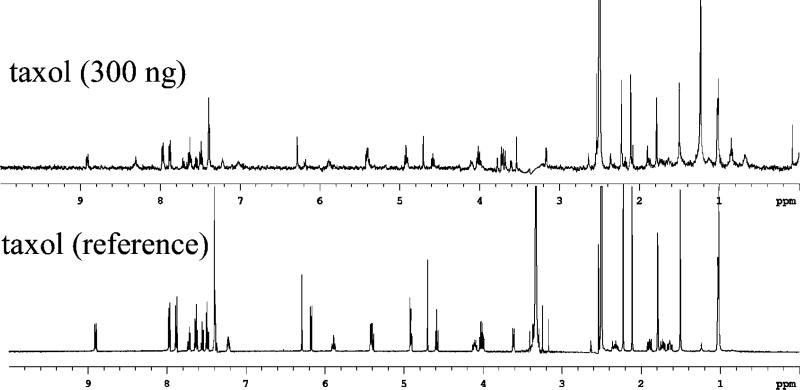
Figure 7.
(Top) The 1-h NMR spectrum of LC fraction recovering 300 ng of taxol spiked into cyanobacterial extract shown in Figure 5 (5.9-min peak) (water, 3.3 ppm, suppressed by presaturation and solvent subtraction). (Bottom) The reference 1H NMR spectrum, of 700 μL of 1 mg/mL taxol in DMSO-d6 acquired using an inverse probe, 16 transients.
From the LC–MS and 1H NMR data shown above, the value of having sufficient data to recognize and eliminate known compounds from further consideration can be illustrated. In addition to the four known compounds identified, the MS and NMR data for one peak, indicated as an unknown compound in Figure 5, had some similarities to the known ambiguines but was not found in the literature or natural product databases. It was therefore prioritized for further study. A scaled-up growth (3 L, 32 days) yielded 0.85 mg of this product for rigorous de novo structure determination, including X-ray crystallography and conventional NMR on a 900-MHz cryoprobe, to establish the novel ambiguine K isonitrile (manuscript in preparation). A related compound, ambiguine L isonitrile, was also found in the scaled-up growth. This successful example illustrates how the microanalytical capabilities of the LC-MS-NMR system can prioritize samples for scale-up, avoiding the four known compounds and streamlining the natural products drug discovery effort.
The internal standard spiked into the extract confirms the limit of detection in complex matrixes. The 2-h 1H NMR acquisition of the collected taxol (0.35 nmol) shows a clear 1-to-1 correspondence of peaks with a reference spectrum, as shown in Figure 7. Some minor peaks attributed to its known degradation in methanol can be seen. From this result, it can be reasonably assumed that any fraction that cannot generate an interpretable 1H NMR spectrum in 2 h using this system contains significantly less than 0.35 nmol of a pure compound. These data suggest that, by loading only 30 μg onto the column, the system can report components down to 1% of the total column loading. Alternatively, for 4-mm packed columns loaded at their 200-μg capacity, this number goes down to 0.2%, to below 0.01% for 4-mm monolithic columns with capacities of up to 3 mg or more.
Significantly lower limits of detection can be obtained by pooling LC runs. Noting that the LC separation time of ∼1 h is considerably less than the NMR analysis time (e.g., 1 h/fraction for multiple fractions), it is practical and time-efficient to perform multiple LC separations, pooling the fraction of interest. Off-line LC-NMR lends itself to this approach, which has been described in LC-SPE-NMR.42,43 Tripling the amount of material subsequently triples the S/N or reduces the time required to obtain comparable S/N values by nearly 10-fold. If a specific peak is of interest, column loadings may be frequently increased significantly without broadening or contaminating the peaks of interest. From just two injections of the active cyanobacterial extract (total 80 μg), the S/N of similar NMR spectra for all fractions previously collected were doubled (data not shown).
As demonstrated above, compared to traditional approaches for natural product analysis, the greatly reduced amounts of material and instrument time required by this LC-MS-NMR system can gather sufficient data for dereplication during activity-guided fractionation. This system therefore has the potential to eliminate much of the time and expense of large-scale purification of previously known leads, allowing resources and effort to be focused on the characterization of novel active compounds discovered during screening and thereby substantially shortening and streamlining the natural product discovery process.
CONCLUSIONS
An LC-MS-NMR platform has been demonstrated, using an approach that accommodates the large disparities in the sample mass and time requirements of MS and NMR. The nanoSplitter LC–MS method can collect an analyte for NMR, while improving MS sensitivity and maintaining chromatographic resolution. An offline approach to NMR permits all of the analyte available in each LC peak to be concentrated into the most sensitive NMR probe readily available and to allocate NMR analysis time intelligently among the most relevant LC peaks. The collection of LC fractions into 96-well plates is readily available in many laboratories, inexpensive enough to use routinely, and enables LC-NMR to be obtained retrospectively. Microdroplet NMR samples can be recovered for reanalysis, archival, or bioassay.
The combined MS and NMR system performed well in routine performance tests of recovery and reproducibility. Any validated LC method used with the microplate automation can be expected to perform equally as well. The dynamic range was from the NMR mass limit of detection of 250 ng up to the linear capacity of the LC separation. Higher capacity columns would also lower the concentration limit of detection, from compounds at the 1% level in the example above (30 μg loaded), to 0.2% for 4-mm packed columns loaded at their 200-μg capacity, to below 0.01% for 4-mm monolithic columns with capacities of 3 mg or more. Severalfold lower limits of detection can readily be obtained by pooling LC collections, which is a much less time-consuming process relative to NMR data acquisition of many samples.
Applied to natural products, this new microanalytical platform could record LC–MS and NMR data during the discovery phase of bioactivity-guided fractionation. The recorded data were sufficient for dereplication where four LC peaks were recognized as known compounds, focusing time and effort on a druglike compound not found in databases or literature. This capability streamlines the process of natural product discovery and has the potential to reinvigorate the field by making feasible sources that were too limited or slow-growing for traditional discovery methods. Beyond natural products, this LC-MS-NMR platform promises to be similarly applicable in a variety of fields that rely on identification of trace components of complex mixtures, ranging from environmental remediation to metabolite identification in metabolomics as well as pharmaceutical DMPK, toxicology, and ADME studies.
ACKNOWLEDGMENT
We gratefully acknowledge the support of NIH RO1 GM075856 for the development of the microscale analytical platform for natural product characterization (Y.L., S.S., R.K., J.O., P.V.) and to R01 CA102536 and R01 CA69390 (S.S., P.V.). We are also indebted to Protasis and MRM for loan of the microcoil probe used in this work, and to Varian for assistance in automation programming. This is publication no. 926 from the Barnett Institute.
References
- 1.Silva Elipe MV. Anal. Chim. Acta. 2003;497:1–25. [Google Scholar]
- 2.Murakami T, Fukutsu N, Kondo J, Kawasaki T, Kusu F. J. Chromatogr., A. 2008;1181:67–76. doi: 10.1016/j.chroma.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 3.Norwood DL, Mullis JO, Feinberg TN. Sep. Sci. Technol. . 2007;8:189–235. [Google Scholar]
- 4.Weber B, Hartmann B, Stoeckigt D, Schreiber K, Roloff M, Bertram H-J, Schmidt CO. J. Agric. Food Chem. 2006;54:274–278. doi: 10.1021/jf051606f. [DOI] [PubMed] [Google Scholar]
- 5.Bobzin SC, Yang S, Kasten TP. J. Chromatogr., B: Biomed. Sci. Appl. 2000;748:259–267. doi: 10.1016/s0378-4347(00)00289-9. [DOI] [PubMed] [Google Scholar]
- 6.Wilson SR, Malerod H, Petersen D, Rise F, Lundanes E, Greibrokk T. J. Sep. Sci. 2007;30:322–328. doi: 10.1002/jssc.200600238. [DOI] [PubMed] [Google Scholar]
- 7.Djukovic D, Liu S, Henry I, Tobias B, Raftery D. Anal. Chem. 2006;78:7154–7160. doi: 10.1021/ac0605748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert M, Wolfender JL, Staerk D, Christensen SB, Hostettmann K, Jaroszewski JW. Anal. Chem. 2007;79:727–735. doi: 10.1021/ac0616963. [DOI] [PubMed] [Google Scholar]
- 9.Olson DL, Norcross JA, O'Neil-Johnson M, Molitor PF, Detlefsen DJ, Wilson AG, Peck TL. Anal. Chem. 2004;76:2966–2974. doi: 10.1021/ac035426l. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder FC, Gronquist M. Angew. Chem., Int. Ed. 2006;45:7122–7131. doi: 10.1002/anie.200601789. [DOI] [PubMed] [Google Scholar]
- 11.Hoult DI, Richards RE. J. Magn. Reson. 1976;24:71–85. doi: 10.1016/j.jmr.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Jayawickrama Dimuthu A, Sweedler Jonathan V. J. Chromatogr., A. 2003;1000:819–840. doi: 10.1016/s0021-9673(03)00447-3. [DOI] [PubMed] [Google Scholar]
- 13.Jansma A, Chuan T, Albrecht RW, Olson DL, Peck TL, Geierstanger BH. Anal. Chem. 2005;77:6509–6515. doi: 10.1021/ac050936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brey WW, Edison AS, Nast RE, Rocca JR, Saha S, Withers RS. J. Magn. Reson. 2006;179:290–293. doi: 10.1016/j.jmr.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Glauser G, Guillarme D, Grata E, Boccard J, Thiocone A, Carrupt PA, Veuthey JL, Rudaz S, Wolfender JL. J. Chromatogr., A. 2008;1180:90–98. doi: 10.1016/j.chroma.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Plumb RS, Ayrton J, Dear GJ, Sweatman BC, Ismail IM. Rapid Commun. Mass Spectrom. 1999;13:845–854. doi: 10.1002/(SICI)1097-0231(19990530)13:10<845::AID-RCM556>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Keifer PA. Annu. Rep. NMR Spectrosc. 2007;62:1–47. [Google Scholar]
- 18.Keifer PA. Curr. Opin. Chem. Biol. 2003;7:388–394. doi: 10.1016/s1367-5931(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 19.Keifer PA. Prog. Drug Res. 2000;55:137–211. doi: 10.1007/978-3-0348-8385-6_5. [DOI] [PubMed] [Google Scholar]
- 20.Kautz RA, Goetzinger WK, Karger BL. J. Comb. Chem. 2005;7:14–20. doi: 10.1021/cc0498940. [DOI] [PubMed] [Google Scholar]
- 21.Lacey ME, Sweedler JV, Larive CK, Pipe AJ, Farrant RD. J. Magn. Reson. 2001;153:215–222. doi: 10.1006/jmre.2001.2443. [DOI] [PubMed] [Google Scholar]
- 22.Teh S-Y, Lin R, Hung L-H, Lee AP. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 23.Nord LK. B Anal. Chim. Acta. 1984:233–249. [Google Scholar]
- 24.Patton CJ. Anal. Instrum. Handb. 1997;15:3–155. [Google Scholar]
- 25.Curcio M, Roeraade J. Anal. Chem. 2003;75:1–7. doi: 10.1021/ac0204146. [DOI] [PubMed] [Google Scholar]
- 26.Wilm M, Mann M. Anal. Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 27.Gangl ET, Annan MM, Spooner N, Vouros P. Anal. Chem. 2001;73:5635–5644. doi: 10.1021/ac010501i. [DOI] [PubMed] [Google Scholar]
- 28.Schiavo S, Ebbel E, Sharma S, Matson W, Kristal BS, Hersch S, Vouros P. Anal. Chem. 2008;80:5912–5923. doi: 10.1021/ac800507y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews CL, Yu CP, Yang E, Vouros P. J. Chromatogr., A. 2004;1053:151–159. [PubMed] [Google Scholar]
- 30.Schiavo S, Ebbel E, Sharma S, Matson W, Kristal BS, Hersch S, Vouros P. Anal. Chem. doi: 10.1021/ac800507y. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao G, Kaut R, Peng S, Cui G, Giese RW. J. Chromatogr., A. 2007;1138:305–308. doi: 10.1016/j.chroma.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Newman D, Cragg G, Snader K. J. Nat. Prod. 2003;7:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 33.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 34.Cordell GA, Shin YG. Pure Appl. Chem. 1999;71:1089–1094. [Google Scholar]
- 35.Clardy J, Walsh C. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 36.Raveh A, Carmeli S. J. Nat. Prod. 2007;70:196–201. doi: 10.1021/np060495r. [DOI] [PubMed] [Google Scholar]
- 37.Smitka TA, B R, Doolin L, Jones ND, Deeter JB, Yoshida WY, Prinsep MR, Moore RE, Patterson GML. J. Org. Chem. 1992;57:857–861. [Google Scholar]
- 38.Klein DD,D, Braekman JC, Hoffmann L, Demoulin V. J. Nat. Prod. 1995;58:1781–1785. doi: 10.1021/np9900324. [DOI] [PubMed] [Google Scholar]
- 39.Moore REC,C, Patterson GML. J. Am. Chem. Soc. 1984;106:6456–6457. [Google Scholar]
- 40.Park AMRE, Patterson GML. Tetrahedron Lett. 1992;33:3257–3260. [Google Scholar]
- 41.Stratmann KM,RE, Bonjouklian R, Deeter JB, Patterson GML, Shaffer S, Smith CD, Smitka TA. J. Am. Chem. Soc. 1994;116:9935–9942. [Google Scholar]
- 42.Xu F, Alexander A. J. Magn. Reson. Chem. 2005;43:776–782. doi: 10.1002/mrc.1617. [DOI] [PubMed] [Google Scholar]
- 43.Exarchou V, Krucker M, van Beek TA, Vervoort J, Gerothanassis IP, Albert K. Magn. Reson. Chem. 2005;43:681–687. doi: 10.1002/mrc.1632. [DOI] [PubMed] [Google Scholar]