Abstract
Background
To facilitate in the identification of gene products important in regulating renal glomerular structure and function, we have produced an annotated transcriptome database for normal human glomeruli using the SAGE approach.
Description
The database contains 22,907 unique SAGE tag sequences, with a total tag count of 48,905. For each SAGE tag, the ratio of its frequency in glomeruli relative to that in 115 non-glomerular tissues or cells, a measure of transcript enrichment in glomeruli, was calculated. A total of 133 SAGE tags representing well-characterized transcripts were enriched 10-fold or more in glomeruli compared to other tissues. Comparison of data from this study with a previous human glomerular Sau3A-anchored SAGE library reveals that 47 of the highly enriched transcripts are common to both libraries. Among these are the SAGE tags representing many podocyte-predominant transcripts like WT-1, podocin and synaptopodin. Enrichment of podocyte transcript tags SAGE library indicates that other SAGE tags observed at much higher frequencies in this glomerular compared to non-glomerular SAGE libraries are likely to be glomerulus-predominant. A higher level of mRNA expression for 19 transcripts represented by glomerulus-enriched SAGE tags was verified by RT-PCR comparing glomeruli to lung, liver and spleen.
Conclusion
The database can be retrieved from, or interrogated online at http://cgap.nci.nih.gov/SAGE. The annotated database is also provided as an additional file with gene identification for 9,022, and matches to the human genome or transcript homologs in other species for 1,433 tags. It should be a useful tool for in silico mining of glomerular gene expression.
Background
Renal glomeruli are highly specialized capillary tufts that produce a nearly protein-free ultrafiltrate of plasma at a rate of about 30 plasma volumes daily. Several hereditary, immune-mediated and metabolic disorders cause glomerular injury, proteinuria, and can lead to renal failure. The three intrinsic glomerular cell types, podocytes, mesangial cells, and glomerular endothelial cells (EC) are highly specialized. Podocytes extend an elaborate array of actin-rich foot processes around the exterior of the glomerular capillary loops, forming a scaffold with nephrin-based filtration slit diaphragms spanning the space between adjacent foot processes [1]. Mesangial cells are pericyte-like cells that, unlike most other pericytes, form an interstitium within the intracapillary space [2]. Glomerular EC are packed with transcellular fenestrae ringed by actin [3,4]. The fenestrae serve the high glomerular capillary wall hydraulic conductivity [5], while a glycocalyx covering the glomerular EC and podocytes together with the podocyte filtration slit diaphragm impede the movement of plasma proteins [6-8] across the glomerular capillary wall.
Transcriptome and proteomic approaches are helping to define genes highly expressed and/or enriched in glomeruli [9-12]. For instance, Sau3A-anchored SAGE databases have been built with RNA extracted from microdissected nephron segments, and enrichment of several glomerular transcripts relative to other nephron segments has been reported [9]. Furthermore, many proteins uniquely expressed by, or enriched in podocytes have been identified over the past decade and their specific functions are increasingly well defined [13]. Finally, by analysis of ESTs enriched in glomeruli, ehd3 was shown to be the first transcript expressed exclusively by glomerular EC [12].
The current study sought to extend previous transcriptome-based work by building a human glomerular LongSAGE database that can be interrogated directly online. SAGE is based on the principal that a small (here 17 bp) tag sequence immediately 3' of an "anchoring" restriction site is a unique identifier of each transcript [14]. The frequency of specific SAGE tags relative to the total pool of tags reflects their abundance in the source mRNA. In silico comparison of SAGE libraries from diverse tissues can then be used to discover differential expression of transcripts [15]. The level of expression of any specific transcript can also be probed in silico by interrogating SAGE libraries with the transcript's unique SAGE tag sequence.
We report here the gene expression profile of transcripts for human glomeruli and compare them to pooled SAGE libraries for non-glomerular tissues and cells. Many of the most highly enriched glomerular transcripts reported here were previously found in a Sau3A-anchored glomerular library [9]. Nonetheless, the current SAGE database contains additional glomerulus-enriched transcript tags, and since it is NlaIII-anchored it now allows direct comparison with many non-renal SAGE libraries. The data should serve as a useful resource for investigators studying glomerular gene expression.
Construction and content
Cells and Tissues
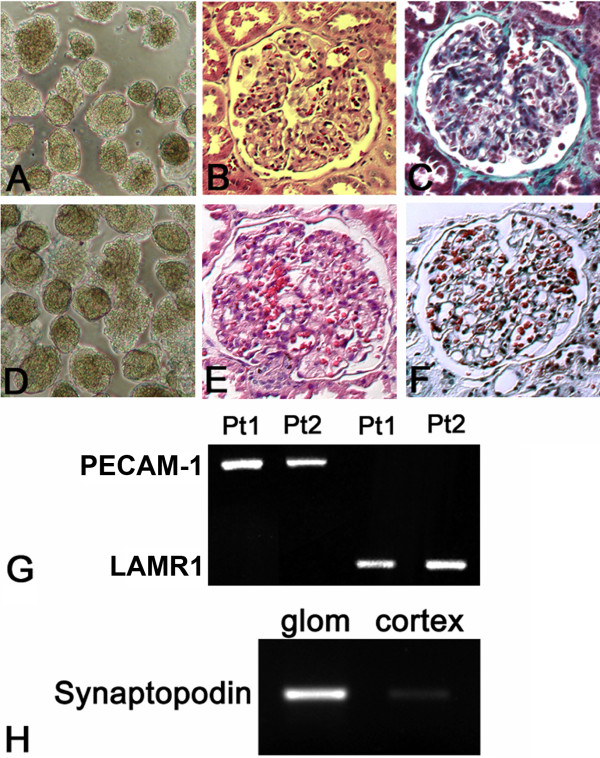
Human kidney tissue was obtained from the uninvolved portion of tumor nephrectomy specimens (Human Subjects Protocols: #6196 University of Alberta and #155/97 University of Frankfurt). Patient 1 was a 45 year old Caucasian female, patient 2 a 72 yo Caucasian male. Renal tissue was collected only from patients in whom the serum creatinine was within normal limits, and in whom diabetes mellitus, hypertension, and proteinuria were absent. Specific parameters were not collected for individual patients. The relative purity of isolated glomeruli and the normal histological appearance of kidney cortex used in this study are shown in Figure 1. That the cDNA template used for SAGE contained mRNA representing glomerular capillary endothelium is shown by RT-PCR amplification of PECAM-1 (836 bp) and the non-integrin laminin receptor LAMR1 (256 bp). Greater synaptopodin transcript abundance in glomerular, compared to whole kidney cortex mRNA from the same specimen also shows appropriate enrichment of the source mRNA in glomerular podocyte transcripts (Figure 1). Amplification of the long PECAM-1 sequence furthermore shows that the source mRNA was intact. Sufficient mRNA for construction of the SAGE library was only obtained from patient 2. The integrity of this source material was also verified by Agilent 2100 microfluidics analysis (data not shown).
Figure 1.
Human Kidney Source Material. Isolated glomeruli (A, D), Hematoxylin & Eosin Stain (B, E) and Masson Trichrome Stain (C, F) for patient 1 (A, B, C) and patient 2 (D, E, F). RT-PCR for PECAM-1 and the 67 kDa non-integrin laminin receptor LAMR1 for patient 1 (pt 1) and patient 2 (pt2) (G). Enrichment of the synaptopodin mRNA abundance, determined by RT-PCR, in glomeruli relative to whole kidney cortex from patient 2 (H).
Human Glomerular SAGE Library Construction
For SAGE, human glomeruli were isolated by sieving in ice-cold phosphate buffered saline (PBS) from the kidney of a 72-year old male using minor modifications of the protocol for rat glomeruli [16,17]. Glomeruli were immediately placed into RNA-Protect (Qiagen, Valencia, CA) followed by isolation of 4.7 μg total RNA with the RNeasy kit (Qiagen). A SAGE library was then custom-constructed by Genzyme Corporation (Framingham, MA) using the "long" SAGE protocol, producing 17 bp SAGE tags with the CATG (NlaIII) anchoring restriction site [18]. A total of 2304 clones containing concatenated ditags were sequenced, resulting in 48,926 tags. Of these, 1,361 were derived from duplicate ditags. Tags from duplicate ditags were not removed [19]. Twenty-one tags were removed as they contained ambiguous (N) nucleotides leaving 48,905 tags and 22,907 unique long SAGE tags for analysis.
Tag sequences and their absolute counts in 115 distinct "long" SAGE libraries were retrieved from cgap.nci.nih.gov/SAGE (98,944,923 tags). All "long" SAGE Libraries available before July 1, 2008 were included without selection. Tissues and cells represented include normal brain, breast, skin, pancreas, bladder, gallbladder, uterus, vein, testis, white blood cells, lung macrophages, embryonic stem cells as well as malignant tumors including colon and lung adenocarcinoma, melanoma, among others. The frequency of each tag (count/total tag number) was calculated for each of the 115 libraries and expressed as tags per million (TPM). The mean TPM for the non-glomerular libraries (Pool TPM) is reported. The frequency ratio of the glomerular: Pool TPM was then calculated to establish degree of enrichment of specific tags in glomeruli (Ratio G: P). Statistical comparison of the pooled libraries with the current glomerular SAGE library was based on Chi-square analysis using absolute tag counts [20]. Comparison to a human kidney SAGE library (SAGE_Kidney_normal_B_1, from cgap.nci.nih.gov/SAGE) was based on the short (10 bp) tag sequences.
For each SAGE tag, identification was based on the "Hs_long.best_gene.gz" database found at ftp://ftp1.nci.nih.gov/pub/SAGE/HUMAN/. The SAGE Genie algorithm for identifying the best gene match for SAGE tags was reported by Boon et al. [21]. For some tags, the Blast n algorithm at http://blast.ncbi.nlm.nih.gov/Blast.cgi, was used to match tag sequences that could not be assigned by the "Hs_long.best_gene.gz" database. For these, the SAGE tag had to be in the +/+ orientation with the corresponding mRNA or EST, and fully match the 17 bp sequence immediately 3' of the NlaIII site nearest the Poly(A)+ tail or a stretch of > 8 A's as previously reported [22]. Positive identification based on this latter search strategy is indicated in additional file 1 by asterisks.
RT-PCR Analysis
For quantitative RT-PCR, glomeruli were microdissected from distinct pre-transplant kidney biopsy specimens obtained from three separate donors aged 57, 59 and 63 at the University of Göteborg (Human Subjects Protocol #653-05). Immediately after biopsy, one half of one biopsy core was placed into 0.5 ml of ice-cold PBS containing 100 U RNAse inhibitor (RNAsin) (Applied Biosystems, Foster City, CA, USA). Four to fifteen glomeruli were isolated using a stereomicroscope (Zeiss, Jena, Germany) followed by extraction of total RNA. cDNA was generated from glomerular RNA with SuperScript™ III RT (Invitrogen, Carlsbad, CA, USA). Human kidney, spleen, lung and liver mRNA was purchased from Invitrogen/Ambion (Carlsbad, CA). Reactions without RT for each primer set served as controls. PCR cycling was performed with 100 ng template (94°C – 3 min; 35 cycles: 94°C – 30 sec; 55°C – 30 sec; 68°C – 30 sec plus 1 min for each kilobase pair (kbp) of PCR product to be amplified; 72°C – 7 min). Quantification of gene expression was performed according to the delta Ct method (DeltaCt2/DeltaCt1), as described by others [23], and by this laboratory [22].
Human Glomerular SAGE Database Content
The complete human glomerular SAGE library was deposited in the Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ repository (record GSE8114, Accession # GSM199994) and in the SAGE Genie collection http://cgap.nci.nih.gov/SAGE as "LSAGE_Kidney_Glomeruli_Normal_B_bjballer1". It consists of 22,907, unique 17 bp tag sequences and the absolute tag count for each sequence. The total tag count in the library is 48,905. The library is also appended in spreadsheet format with tag identification (additional file 1).
Utility
Retrieval of Highly Enriched Glomerular Transcripts
The transcripts most highly enriched in human glomeruli identified by this study are shown in Tables 1 and 2 and Additional files 3 and 4. Of the 22,907 tags, 291 were observed with an absolute count of 4 (81 TPM) or greater and enriched more than 10-fold relative to pooled non-kidney SAGE libraries. For 84 of these no reliable match to a known cDNA sequence was found, and a match to incompletely defined ESTs was observed for 8 others. The tags representing Aldolase B, uromodulin, glutamyl aminopeptidase, glutathione peroxidase, and SLC25A45 were excluded from this set because they were not enriched relative to whole kidney. They likely represent transcripts expressed at very high levels in contaminating tubules. Several highly expressed transcripts produced more than one unique tag, which is common and usually reflects priming from internal poly A(+) runs or alternatively spliced transcripts. After removal of such redundant tags, 133 well-characterized tags highly enriched in glomeruli were established (Tables 1 and 2 and Additional files 3 &4).
Table 1.
Transcripts represented by SAGE tags enriched 30 fold or more in glomeruli
| Symbol | Gene Name | NlaIII long Tag | Sau3A Tag | G:TPM Sau3A |
G:TPM NlaIII |
P:TPM NlaIII |
K:TPM NlaIII |
Ratio G: P |
Ratio G: K |
Link SAGE Genie |
Link Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NPHS2 | Nephrosis 2, idiopathic, steroid-resistant (podocin) | TTTCCGTGACTCATCTA | CCTCACTGAA | 1,534 | 2,287 | 0 | 48 | Infinity | 48 | NPHS2 | Ref:NPHS2 |
| SOST | Sclerosteosis | ACATATGAAAGCCTGCA | TCGAGGAGAC | 158 | 82 | 0 | 0 | Infinity | SOST | ||
| MME | Membrane metallo-endopeptidase | CTGCAGTGTTCGAGTGG | TATAAAGCGA | 271 | 102 | 0 | 0 | 4446 | MME | Ref:MME | |
| CLIC5 | Chloride intracellular channel 5 | AATCTGAACCAATTACC | CAGTCATCTG | 1,038 | 3,593 | 5 | 24 | 719 | 150 | CLIC5 | Ref:CLIC5 |
| NPHS1 | Nephrosis 1, congenital, Finnish type (nephrin) | TAAACATAAGTATGCTC | 1,041 | 2 | 0 | 549 | NPHS1 | Ref:NPHS1 | |||
| TCF21 | Transcription factor 21 | CGAGTGCTGAGCAAGGC | ATAGGATAGC | 451 | 817 | 2 | 0 | 514 | TCF21 | Ref:TCF21 | |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CCGCTGCGGGGCCCTGG | AGCGCCTGAG | 383 | 490 | 1 | 0 | 506 | CDKN1C | Ref:CDKN1C | |
| DDN | Dendrin | TGAACTTGGCCACATCA | 306 | 1 | 0 | 373 | DDN | Ref:DDN | |||
| THYMU2010816 | CDNA FLJ36413 fis, clone THYMU2010816 | TATCACTGGGGAGGGAA | ATTAGTCAAT | 203 | 449 | 2 | 0 | 285 | THYMU2010816 | ||
| FLJ22271 | CDNA: FLJ22271 fis, clone HRC03191 | GCTTTGTCGCAACGCTC | 82 | 0 | 0 | 263 | FLJ22271 | ||||
| PTPRO | Protein tyrosine phosphatase, receptor type, O | GATATACAACAGAAAAC | AACTGTGTAA | 226 | 755 | 4 | 0 | 215 | PTPRO | Ref:PTPRO | |
| FGF1 | Fibroblast growth factor 1 (acidic) | TAAAGGCCTTTAATAAG | TTCTTAGAAG | 474 | 102 | 0 | 24 | 210 | 4 | FGF1 | Ref:FGF1 |
| PTHR1 | Parathyroid hormone receptor 1 | TGACCAGGCGCTGGGGG | 1,143 | 8 | 96 | 141 | 12 | PTHR1 | Ref:PTHR1 | ||
| CLDN5 | Claudin 5 | GTAGGCGGCTGCCTCTT | 82 | 1 | 0 | 127 | CLDN5 | ||||
| LRRC2 | Leucine rich repeat containing 2 | GGACTGCACTCCAGCCT | 82 | 1 | 0 | 108 | LRRC2 | ||||
| AL049990 | MRNA; cDNA DKFZp564G112 (from clone DKFZp564G112) | ACTTGGAAATAAACAAA | 143 | 1 | 0 | 105 | AL049990 | ||||
| HTRA1 | HtrA serine peptidase 1 | ACCGACAGGCCAAAGGA | CGCAGGCAGA | 1,398 | 143 | 1 | 0 | 105 | HTRA1 | ||
| FAM65A | Family with sequence similarity 65, member A | GCTGCTGTCAGCACCCA | AGGCTGTTGT | 316 | 204 | 2 | 0 | 99 | FAM65A | ||
| SEMA3G | Semaphorin 3G | ACTGCCCCTGAGCTCTG | 674 | 7 | 24 | 94 | 28 | SEMA3G | |||
| SPOCK1 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 | AGAATACCTTAATACTG | TTTAATACTT | 226 | 122 | 1 | 0 | 92 | SPOCK1 | ||
| CRB2 | Crumbs homolog 2 (Drosophila) | TGCAGCAGTGGCAGCCT | 367 | 4 | 0 | 89 | CRB2 | ||||
| ENPEP | Glutamyl aminopeptidase (aminopeptidase A) | GCCTGGAATTGGATACA | ATATGAATTA | 632 | 143 | 2 | 96 | 79 | 1 | ENPEP | Ref:ENPEP |
| SLC45A1 | Solute carrier family 45, member 1 | GGCGTGGACATCTCTCT | 143 | 2 | 0 | 77 | SLC45A1 | ||||
| TNNT2 | Troponin T type 2 (cardiac) | ATGCATTTTGGGGGTTA | TGCTCCTCGC | 338 | 184 | 2 | 24 | 76 | 8 | TNNT2 | |
| CD34 | CD34 molecule | GGACCAGGTCTTGGAGC | 102 | 1 | 24 | 75 | 4 | CD34 | Ref;CD34 | ||
| EFNB1 | Ephrin-B1 | AGGGAAGAGGAAAGTGC | 102 | 2 | 0 | 68 | EFNB1 | Ref:EFNB1 | |||
| IL1RL1 | Interleukin 1 receptor-like 1 (ST2 Protein) | AGGGCAGGGACATCATC | TTTGTAGACT | 158 | 82 | 1 | 0 | 64 | IL1RL1 | ||
| ST6GALNAC3 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N- | TGATGCCCTTGAACACC | 286 | 5 | 0 | 63 | ST6GALNAC3 | Ref:ST6GALNAC3 | |||
| acetylgalactosaminide alpha-2,6-sialyltransferase 3 | |||||||||||
| NTNG1 | Netrin G1 | TGTAACAGCCCCCTCTA | 102 | 2 | 0 | 63 | NTNG1 | ||||
| SPOCK2 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 | CATAAAGGAAATCAAAT | TGGTAGGTTG | 248 | 1,123 | 18 | 0 | 61 | SPOCK2 | ||
| ABLIM2 | Actin binding LIM protein family, member 2 | ACACGCCAAGTCCCGTT | 122 | 2 | 0 | 60 | ABLIM2 | Ref:ABLIM2 | |||
| MGC16291 | Hypothetical protein MGC16291 | TGGGTCAAACTCTGAAA | 122 | 2 | 24 | 58 | 5 | MGC16291 | |||
| BCAM | Basal cell adhesion molecule (Lutheran blood group) | CCCGCCCCCGCCTTCCC | ACGTGGTATC | 632 | 3,205 | 55 | 788 | 58 | 4 | BCAM | Ref:BCAM |
| TPPP3 | Tubulin polymerization-promoting protein family member 3 | GTGACCCCAAGGCCAGT | 82 | 1 | 0 | 57 | TPPP3 | ||||
| C4orf31 | Chromosome 4 open reading frame 31 | TACATAAAATTAAAGAG | TAATCTAAGT | 226 | 429 | 8 | 0 | 57 | C4orf31 | ||
| GJA5 | Gap junction protein, alpha 5, 40kDa | GACCATTCCTCGGAGTA | AATCTTTGAT | 203 | 122 | 2 | 24 | 55 | 5 | GJA5 | RefGJA5 |
| ITGB8 | Integrin, beta 8 | TCTTGTATCAATGGCAG | 633 | 12 | 0 | 52 | ITGB8 | Ref:ITGB8 | |||
| TSPAN2 | Tetraspanin 2 | CCAAGGCACTGAATTAA | 143 | 3 | 0 | 49 | TSPAN2 | ||||
| WT1 | Wilms tumor 1 | CTGGTATATGGCTTCAA | TTACAAGATA | 406 | 143 | 3 | 0 | 49 | WT1 | Ref:WT1 | |
| COL4A3 | Collagen, type IV, alpha 3 (Goodpasture antigen) | TGCATTATTTTCCAGAT | 122 | 3 | 0 | 48 | COL4A3 | REF:COL4A3 | |||
| ALS2CL | ALS2 C-terminal like | CGATGCTGACGGGACCC | 796 | 17 | 0 | 47 | ALS2CL | ||||
| PLCE1 | Phospholipase C, epsilon 1 | AACGAACGTGGCTGTAT | 163 | 4 | 0 | 46 | PLCE1 | Ref:PLCE1 | |||
| WFS1 | Wolfram syndrome 1 (wolframin) | ACCCTCCTGTCCAGCAG | 204 | 4 | 0 | 46 | WFS1 | ||||
| PCOLCE2 | Procollagen C-endopeptidase enhancer 2 | ATGGAGGTATGAGGCCT | TATGTTCTCT | 271 | 408 | 9 | 24 | 45 | 17 | PCOLCE2 | |
| MRGPRF | MAS-related GPR, member F | AGGACCCACTGGGCAGC | CTCTTAAGGC | 316 | 327 | 8 | 0 | 43 | MRGPRF | ||
| SPTB | Spectrin, beta, erythrocytic | CAATCTGGGGCTGGCCC | 82 | 2 | 0 | 42 | SPTB | Ref:SPTB | |||
| ATP10A | ATPase, class V, type 10A | TCCTCTGCGCCAGGGGA | 204 | 5 | 0 | 41 | ATP10A | ||||
| USHBP1 | Usher syndrome 1C binding protein 1 | TCATAAACTGTCCTGGA | 122 | 3 | 0 | 40 | USHBP1 | ||||
| MAP6 | Microtubule-associated protein 6 | TACAGTAGTCTTGCTGG | 327 | 8 | 0 | 39 | MAP6 | ||||
| NFASC | Neurofascin homolog (chicken) | AGCAATGAAAAGGCCAA | 184 | 5 | 0 | 39 | NFASC | ||||
| PLA2R1 | Phospholipase A2 receptor 1, 180kDa | AATTTTGCAAAAAGGAA | 82 | 2 | 0 | 39 | PLA2R1 | ||||
| C1QL1 | Complement component 1, q subcomponent-like 1 | CGCGGCGGCGACGGCAC | 184 | 5 | 24 | 39 | 8 | C1QL1 | |||
| TMEM178 | Transmembrane protein 178 | CTTGTTAAATTTTAATG | 102 | 3 | 24 | 39 | 4 | TMEM178 | |||
| TP53I11 | Tumor protein p53 inducible protein 11 | TACCCCAAGGCCTGATG | 102 | 3 | 0 | 37 | TP53I11 | ||||
| SYNPO | Synaptopodin | ATATTAGGAAGTCGGGG | CATTTCTACC | 519 | 592 | 16 | 0 | 37 | SYNPO | Ref:SYNPO | |
| DACH1 | Dachshund homolog 1 (Drosophila) | TAGGACCTATGAAAATT | 82 | 2 | 0 | 37 | DACH1 | ||||
| TTMA | Two transmembrane domain family member A | CTTTATTGAGTGTTATC | 225 | 6 | 0 | 37 | TTMA | ||||
| FABP1 | Fatty acid binding protein 1, liver | ACATTGGGTGACATTGT | 102 | 3 | 24 | 35 | 4 | FABP1 | |||
| RAMP3 | Receptor (G protein-coupled) activity modifying protein 3 | AGCTTGTGGCCTCTATC | 1,000 | 29 | 0 | 34 | RAMP3 | ||||
| EMCN | Endomucin | CTACTTTGTACATATAA | TTTTCTTTAA | 361 | 449 | 13 | 0 | 34 | EMCN | ||
| RAPGEF3 | Rap guanine nucleotide exchange factor (GEF) 3 | AGGAGGGGCTGGGACTG | 184 | 6 | 72 | 33 | 3 | RAPGEF3 | |||
| FAM20B | Major histocompatibility complex, class I, B | TCCCGGCCCGGCCGCGG | 82 | 2 | 0 | 33 | FAM20B | ||||
| NPNT | Nephronectin | GTAAAGGTATAAGCCTT | CATTTTTAAT | 383 | 286 | 9 | 0 | 32 | NPNT | Ref:NPNT | |
| UACA | Uveal autoantigen with coiled-coil domains and ankyrin repeats | AGTTCTGTTTCACAAGT | 82 | 3 | 0 | 32 | UACA | ||||
| KLK7 | Kallikrein-related peptidase 7 | AGCCACCACGGCCAGCC | 592 | 19 | 96 | 31 | 6 | KLK7 | |||
| LIMS2 | LIM and senescent cell antigen-like domains 2 | CAGATGGAGGCCTCTGG | 776 | 25 | 24 | 31 | 32 | LIMS2 | |||
| TYRO3 | TYRO3 protein tyrosine kinase | GGGCGGGTCCTAGCTGT | 1,143 | 37 | 72 | 31 | 16 | TYRO3 |
Transcripts represented by SAGE tags enriched 30 fold or more in glomeruli compared to pooled SAGE libraries from diverse tissues and cells. The gene symbol and gene name are shown for each tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each tag. This table with embedded links to SAGE Genie and links to references showing expression and/or function in glomeruli is provided as additional file 3
Table 2.
Transcripts represented by SAGE tags enriched 10 – 30 fold in glomeruli
| Symbol | Gene Name | NlaIII long Tag | Sau3A Tag | G:TPM Sau3A |
G:TPM NlaIII |
P:TPM NlaIII |
K:TPM NlaIII |
Ratio G: P |
Ratio G: K |
Link SAGE Genie |
Ref Link |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL4A4 | Collagen, type IV, alpha 4 | TAACTTTTGCAAGATGC | 122 | 4 | 0 | 29 | COL4A4 | Ref:COL4A4 | |||
| AQP1 | Aquaporin 1 | AGCTCCTGATCAGAGGC | 102 | 4 | 0 | 27 | AQP1 | AQP1 | |||
| NPR1 | Natriuretic peptide receptor A/guanylate cyclase A | AGCAGAGACAATTAAAA | 163 | 6 | 24 | 27 | 7 | NPR1 | NPR1 | ||
| PTGDS | Prostaglandin D2 synthase 21kDa | ACGGAACAATAGGACTC | CCGGCCAGCC | 5,323 | 2,368 | 88 | 24 | 27 | 99 | PTGDS | |
| CRHBP | Corticotropin releasing hormone binding protein | AATAAATACATTCAGAA | ATAGTTCTAA | 654 | 286 | 11 | 27 | CRHBP | |||
| TNNI1 | Troponin I type 1 (skeletal, slow) | AGGCACCTGGGGCTTCT | TGCGGGCCAA | 158 | 204 | 8 | 24 | 26 | 9 | TNNI1 | |
| ODZ2 | Odz, odd Oz/ten-m homolog 2 | ACAGTCACCACGAGGAG | 122 | 5 | 26 | ODZ2 | |||||
| EPAS1 | Endothelial PAS domain protein 1 | GAACTTTTCTGTAATGG | 82 | 3 | 48 | 24 | 2 | EPAS1 | |||
| PODXL | Podocalyxin-like | GAGGACACAGATGACTC | ATATATGTCT | 2,323 | 3,369 | 141 | 48 | 24 | 70 | PODXL | Ref:PODXL |
| TMEM204 | Transmembrane protein 204 | CGTGCGAGACACGTGTG | 204 | 9 | 23 | TMEM204 | |||||
| SMAD6 | SMAD family member 6 | TCTCCGGACGCCACCAA | 163 | 7 | 23 | SMAD6 | Ref:Samd6 | ||||
| CLEC3B | C-type lectin domain family 3, member B | ACCGGCGCCCGCATCGC | GTGTAGCCGG | 361 | 367 | 16 | 72 | 23 | 5 | CLEC3B | |
| IFITM3 | Interferon induced transmembrane protein 3 | AACCCCTGCTGCCTGGG | 102 | 4 | 23 | IFITM3 | |||||
| HOXA7 | Homeobox A7 | GTATGTTGTCTTGAGTT | 82 | 4 | 21 | HOXA7 | |||||
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | GTATGGGCCCTGGACCT | CCCAAGCCTG | 564 | 1,021 | 48 | 21 | CHI3L1 | |||
| FRY | Furry homolog (Drosophila) | TGAACTTGTTGCACTGC | 408 | 19 | 21 | FRY | |||||
| PEA15 | Phosphoprotein enriched in astrocytes 15 | TCTGCCCTTTTTTGTGG | 125 | 6 | 24 | 21 | 5 | PEA15 | |||
| ZNF250 | Zinc finger protein 250 | TGGAACCACAAGCAGCC | 143 | 7 | 20 | ZNF250 | |||||
| TGFBR3 | Transforming growth factor, beta receptor III | GCAAATCCTGTCGGTCT | CTCCTGTCTA | 180 | 122 | 6 | 20 | TGFBR3 | |||
| IGFBP5 | Insulin-like growth factor binding protein 5 | GAGTACGTTGACGGGGA | TTTGTCTTTT | 3,496 | 367 | 18 | 20 | IGFBP5 | Ref:IGFBP5 | ||
| FCN3 | Ficolin (collagen/fibrinogen domain containing) 3 | GACACCGAGGGGGGCGG | GTCAGCCACC | 180 | 715 | 36 | 48 | 20 | 15 | FCN3 | |
| LRRC32 | Leucine rich repeat containing 32 (GARP gene) | TTGCATACCCTGACCCC | TTTGAAAACA | 180 | 20 | 1 | 19 | LRRC32 | |||
| NOSTRIN | Nitric oxide synthase trafficker | CCACACGCAGATTCACT | TTTGAATGGG | 158 | 61 | 3 | 19 | NOSTRIN | Ref:Nostrin | ||
| HTRA1 | HtrA serine peptidase 1 | TTTCCCTCAAAGACTCT | 1,409 | 74 | 24 | 19 | 59 | HTRA1 | |||
| ITGA1 | Pelota homolog (Drosophila) | TGCCAGGTGCAGTCACA | 122 | 6 | 24 | 19 | 5 | ITGA1 | |||
| TNNC1 | Troponin C type 1 (slow) | TCCTCAACCCCAAATCC | 184 | 11 | 17 | TNNC1 | |||||
| DOPEY2 | Dopey family member 2 | AGAATTGCTTGAACCCA | 1,347 | 79 | 263 | 17 | 5 | DOPEY2 | |||
| FOXC1 | Forkhead box C1 | AGCCTGTACGCGGCCGG | ATTGTTAAAG | 519 | 245 | 15 | 24 | 17 | 10 | FOXC1 | |
| CEP3 | CDC42 effector protein (Rho GTPase binding) 3 | ATGCTTCTGCAGAGACT | TTGGGCCCTC | 226 | 163 | 10 | 17 | CEP3 | |||
| BGN | Biglycan | GCCTGTCCCTCCAAGAC | GAGAACGGGA | 158 | 776 | 48 | 72 | 16 | 11 | BGN | Ref:BGN |
| MAGI2 | Membrane associated guanylate kinase | TATTAATAGTCACAGAA | 102 | 7 | 16 | MAGI2 | |||||
| AIF1L | Allograft inflammatory factor 1-like | GGAGTGTGCGTGGACTG | 1,572 | 102 | 334 | 15 | 5 | C9orf58 | |||
| KCNH3 | Potassium voltage-gated channel, subfamily H, member 3 | TGCCCCTGCCTCTACCT | 163 | 11 | 15 | KCNH3 | |||||
| PARD6G | Par-6 partitioning defective 6 homolog gamma (C. elegans) | TCGTTCAGTGCCCCAGC | 82 | 5 | 15 | PARD6G | |||||
| TM4SF18 | Transmembrane 4 L six family member 18 | CAAGTATACCACCCTTC | 82 | 5 | 15 | TM4SF18 | |||||
| TENC1 | Tensin like C1 domain containing phosphatase (tensin 2) | AATAGGGGAAAAAAGAG | ACATGAATAG | 519 | 531 | 37 | 15 | TENC1 | Ref:TENC1 | ||
| DPP6 | Dipeptidyl-peptidase 6 | ACATTTGGTTAAAAAAA | 82 | 6 | 14 | DPP6 | |||||
| NES | Nestin | TGCTGACTCCCCCCATC | 2,001 | 138 | 72 | 14 | 28 | NES | Ref:NES | ||
| ADORA1 | Adenosine A1 receptor | TGACTAATAAAAAACTG | 82 | 6 | 14 | ADORA1 | Ref:ADORA1 | ||||
| VEGFA | Vascular endothelial growth factor A | TTTCCAATCTCTCTCTC | CCCTGGCTCC | 248 | 1,450 | 109 | 24 | 13 | 61 | VEGFA | Ref:VEGFA |
| ZNF135 | Zinc finger protein 135 | GGGAAACTCCATCTCTA | 102 | 8 | 13 | ZNF135 | |||||
| SLC48A1 | Solute carrier family 48 (heme transporter), member 1 | GTGCATCAGAGCGGGAA | 82 | 6 | 13 | SLC48A1 | |||||
| FARP1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 | GGGTAGTGTCAGTCGGA | 122 | 10 | 72 | 13 | 2 | FARP1 | |||
| C1orf115 | Chromosome 1 open reading frame 115 | TCGAAGATTCACTGGGA | 102 | 8 | 24 | 12 | 4 | C1orf115 | |||
| DAG1 | Dystroglycan 1 (dystrophin-associated glycoprotein 1) | CAGAGACGTGGCTGGCC | 1,286 | 104 | 48 | 12 | 27 | DAG1 | Ref:DAG1 | ||
| PPAP2A | Phosphatidic acid phosphatase type 2A | AAACACCAACAACTGGG | CAGATTGGTC | 248 | 286 | 23 | 12 | PPAP2A | Ref:PPA2A | ||
| SEMA3B | Semaphorin 3B | TGCCGCCCGCAGCCTGC | 612 | 50 | 72 | 12 | 9 | SEMA3B | |||
| CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | TATTTTGTTATGAATAG | 143 | 12 | 24 | 12 | 6 | CDC14A | |||
| CIRBP | Cold inducible RNA binding protein | TGCCCGGGGAATGTTCC | 82 | 7 | 12 | CIRBP | |||||
| CSRP1 | Cysteine and glycine-rich protein 1 | CAGGCGGGGTCCTAGGA | 82 | 7 | 12 | CSRP1 | |||||
| FAM20C | Family with sequence similarity 20, member C | CGCCCGTCGTGAATTCA | 367 | 31 | 119 | 12 | 3 | FAM20C | |||
| CLEC16A | C-type lectin domain family 16, member A | CTTCGTGGGTACTGAAC | 122 | 10 | 24 | 12 | 5 | CLEC16A | |||
| INF2 | Inverted formin, FH2 and WH2 domain containing | TCCAGCCCCTGAAGTTG | 184 | 16 | 48 | 12 | 4 | INF2 | |||
| BMP7 | Bone morphogenetic protein 7 | TGGAACCCGGTCTTGTG | 204 | 18 | 12 | BMP7 | Ref:BPM7 | ||||
| ZDHHC6 | Zinc finger, DHHC-type containing 6, transmembrane protein 4 | TGGTACTTCTCTTTTCC | AATGGATGTT | 1,128 | 592 | 51 | 12 | ZDHHC6 | |||
| PPAP2B | Phosphatidic acid phosphatase type 2B | ATGTAGGTGCCACCCAC | AACCACATGC | 654 | 184 | 16 | 119 | 11 | 2 | PPAP2B | Ref:PPA2B |
| TXNIP | Thioredoxin interacting protein | AGAAACTAGAGGGCAGG | 102 | 9 | 11 | TXNIP | |||||
| ITGA3 | Integrin, alpha 3 | GTACTGTAGCAGGGGAA | CTCCACAGAG | 180 | 817 | 72 | 24 | 11 | 34 | ITGA3 | Ref:ITGA3 |
| C19orf63 | Chromosome 19 open reading frame 63 | AAAGAGTCGGGGCTGGA | 82 | 7 | 48 | 11 | 2 | C19orf63 | |||
| IGFBP2 | Insulin-like growth factor binding protein 2, 36kDa | GCCTGTACAACCTCAAA | CAGGGAGCCC | 880 | 1,797 | 161 | 96 | 11 | 19 | IGFBP2 | Ref:IGFBP2 |
| PTGER4 | Prostaglandin E receptor 4 (subtype EP4) | TTTTGTTGCTCAGTGTT | 306 | 28 | 11 | PTGER4 | |||||
| FLRT3 | Fibronectin leucine rich transmembrane protein 3 | TATTTTTCTAGGCATAA | 82 | 8 | 24 | 11 | 3 | FLRT3 | |||
| VWA1 | Von Willebrand factor A domain containing 1 | CCCAGGACACCAGCTGG | 531 | 49 | 24 | 11 | 22 | VWA1 | |||
| LARGE | Like-glycosyltransferase | AAAGCCCAGTTCTGAAG | 82 | 8 | 24 | 11 | 3 | LARGE | |||
| LINGO1 | Leucine rich repeat and Ig domain containing 1 | AAGATGATATGAGGCCG | 122 | 12 | 10 | LINGO1 | |||||
| MYO1E | Myosin IE | TATGAATGTACTAAGTA | ATATACTGTA | 248 | 245 | 26 | 9 | MYO1E | Ref:MYO1E |
Transcripts represented by SAGE tags enriched 10 – 30 fold in glomeruli compared to pooled SAGE libraries. The gene symbol and gene name are shown for each unique NlaIII-anchored long SAGE tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each tag. This table with embedded links to SAGE Genie and links to references showing expression and/or function in glomeruli is provided as additional file 4
A previously published Sau3A-anchored SAGE library [9] prepared from microdissected human glomeruli contained 184 SAGE tags that were enriched in glomeruli relative to other micro-dissected nephron segments. These represented 156 well-characterized transcripts. As expected, the corresponding NlaIII SAGE tag for 143 of these was also observed in the current glomerular SAGE library (Tables 1 and 2 and Additional files 3 and 4 and additional file 2). For 47 transcripts represented in both libraries a 10-fold or greater enrichment of the NlaIII tag relative to non-glomerular cells and tissues was observed and is shown in Tables 1 and 2 and Additional files 3 and 4. The NlaIII tag corresponding to the remaining 96 transcripts identified in the Sau3A library was enriched relative to whole kidney, in keeping with the previous report [9], but less than 10 fold relative to non-renal tissues (additional file 2).
Many of the highly expressed and highly enriched transcripts observed in this library are encoded by genes already known to be unique or highly enriched in glomerular podocytes, for instance Podocin (NPHS2), Nephrin (NPHS1), transcription factor 21 (Pod1, FLJ35700), Protein Tyrosine Phosphatase Glepp 1 (PTPRO), Synaptopodin (SYNPO), indicating that this SAGE database appropriately represents glomerular transcripts and that it identifies transcripts enriched in glomeruli. Some of the SAGE tags enriched in glomeruli represent known endothelial cell-predominant genes, for instance Endomucin (EMCN), claudin 5 (CLDN5), NOSTRIN and CD34, consistent with abundant EC in glomeruli.
To independently demonstrate the utility of this database in defining enrichment of transcripts in glomeruli, RT-PCR comparing the level of expression of 19 transcripts enriched in the glomerular SAGE library with that in lung, spleen and liver was performed. Lung, liver and spleen were not represented in the pooled SAGE libraries used here. For each, glomeruli microdissected from the kidneys of three distinct donors were used. The source mRNA used for RT-PCR was distinct from that used for generation of the SAGE library. Transcripts were chosen to represent a spectrum of glomerular enrichment, and some well-known podocyte-predominant transcripts (TCF21, VEGFA) were included as internal controls. Overall, the degree of glomerular transcript enrichment observed by RT-PCR compared to lung, liver and spleen was similar to that observed by SAGE, though there was variation between lung, spleen and liver (Table 3). The wide range of expression observed in the three non-glomerular tissues was expected, as the pooled SAGE-based comparison does not take into account tissue-to-tissue variation in gene expression.
Table 3.
Ratio of mRNA abundance in glomeruli compared to lung, spleen and liver.
| SAGE | RT-PCR | |||||||
|---|---|---|---|---|---|---|---|---|
| Symbol | Name | LongTag | G: TPM | P: TPM | Ratio G: P | G: Lu | G: Sp | G: Li |
| SOST | Sclerosteosis | ACATATGAAAGCCTGCA | 82 | 0.00 | Infinity | 130 ± 48 | 13007 ± 4827 | 7969 ± 2957 |
| TCF21 | Transcription factor 21 | CGAGTGCTGAGCAAGGC | 817 | 1.59 | 514 | 49 ± 16 | 27 ± 9 | 384 ± 123 |
| FGF1 | Fibroblast growth factor 1 (acidic) | TAAAGGCCTTTAATAAG | 102 | 0.49 | 210 | 490 ± 70 | 166 ± 23 | 2565 ± 367 |
| SPOCK2 | sparc/osteonectin, cwcv and kazal-like domains | TGTGGAGTGTACTTGTT | 245 | 1.44 | 170 | 45 ± 13 | 39 ± 12 | 258 ± 75 |
| PTHR1 | Parathyroid hormone receptor 1 | TGACCAGGCGCTGGGGG | 1143 | 8 | 141 | 196 ± 55 | 51 ± 14 | 30 ± 8 |
| EFNB1 | Ephrin-B1 | AGGGAAGAGGAAAGTGC | 102 | 1.50 | 68 | 20 ± 4 | 17 ± 3 | 30 ± 6 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | TAGCAGCAACCGGCGGC | 1021 | 46 | 22 | 51 ± 17 | 45 ± 15 | 198 ± 66 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | GAGTACGTTGACGGGGA | 367 | 18 | 20 | 10 ± 4 | 22 ± 9 | 43 ± 17 |
| CLDN5 | Claudin 5 | GACCGCGGCTTCCGCCG | 715 | 38 | 19 | 5 ± 1 | 27 ± 7 | 24 ± 6 |
| FAM65A | Family with sequence similarity 65, member A | GGTTCCTGGTGCCCCTT | 755 | 43 | 17 | 18 ± 5 | 11 ± 3 | 51 ± 15 |
| FOXC1 | Forkhead box C1 | AGCCTGTACGCGGCCGG | 245 | 15 | 17 | 25 ± 6 | 302 ± 74 | 452 ± 110 |
| C9orf58 | Chromosome 9 open reading frame 58 | GGAGTGTGCGTGGACTG | 1572 | 102 | 15 | 36 ± 3 | 2 ± 0 | 171 ± 16 |
| NES | Nestin | TGCTGACTCCCCCCATC | 2001 | 138 | 14 | 22 ± 6 | 23 ± 6 | 339 ± 84 |
| VEGFA | Vascular endothelial growth factor A | TTTCCAATCTCTCTCTC | 1450 | 109 | 13 | 16 ± 4 | 53 ± 12 | 22 ± 5 |
| ZDHHC6 | Zinc finger, DHHC-type containing 6 | TGGTACTTCTCTTTTCC | 592 | 51 | 12 | Infinity | 60 ± 18 | Infinity |
| MYO1E | Myosin IE | TATGAATGTACTAAGTA | 245 | 26 | 9 | 11 ± 3 | 21 ± 6 | 13 ± 3 |
| MYL9 | Myosin, light chain 9, regulatory | GGAGTGTGCTCAGGAGT | 3287 | 454 | 7 | 4 ± 1 | 15 ± 4 | 45 ± 11 |
| ITM2B | Integral membrane protein 2B | TCACCTTAGGTAGTAGG | 3328 | 508 | 7 | 4 ± 1 | 4 ± 1 | 5 ± 1 |
| MYO1D | Myosin ID | ATTGTAGACAATGAGGG | 327 | 82 | 4 | 3 ± 0 | 7 ± 1 | 7 ± 1 |
The SAGE tag frequency in glomeruli (G: TPM) and SAGE library pool (P: TPM) as well as the relative SAGE tag enrichment in glomeruli (Ratio G: P) is shown. The transcript abundance relative to lung (G: Lu), spleen (G: Sp) and liver (G: Li) was determined by RT-PCR using distinct sets of microdissected glomeruli from kidneys of three different donors. G: glomeruli, Lu: lung, Sp: spleen, Li: liver. Mean ± SEM.
Finally, it is of note that 117 transcript tags observed 2 or more times and enriched > 500 fold in this glomerular library remain unidentified or poorly characterized (additional file 1). At least some of these will likely prove to be currently unknown glomerulus-predominant transcripts.
In Silico Interrogation of the Glomerular SAGE Database
The current database can be retrieved directly or interrogated in silico. It may be used to determine whether any specific gene is highly expressed in glomeruli, and to define transcripts that are highly enriched relative to other tissues for which SAGE libraries are available.
To assess whether a specific transcript is expressed in glomeruli, the SAGE tags uniquely identifying the transcript can be found at http://cgap.nci.nih.gov/SAGE/ using the "SAGE Anatomic Viewer" [21]. The "Digital Northern" tool is then used to evaluate the level of expression in the SAGE libraries of the collection, which includes the current library. The collection can also be interrogated using specific NlaIII SAGE tags of cDNA sequences for which a gene symbol may not yet have been assigned. The tag can be retrieved from any cDNA sequence by identifying the 17-nt sequence immediately 3' of the last NlaIII site (CATG) prior to the poly(A+) tail. Its frequency in the glomerular database is an indicator of the level of expression in human glomeruli. The 95% confidence interval for observing any tag with a true count of 4 is ± 3.96. Hence, any transcript producing a tag frequency of 4 per 48,905 (81.8 TPM) or greater has a 95% probability of being represented in this library. Failure to find the SAGE tag representing any specific transcript in this library indicates that its expression level is lower than the limit of detection, or that the transcript does not contain an NlaIII restriction site from which a SAGE tag could be generated.
The "LSAGE_Kidney_Glomeruli_Normal_B_bjballer1" database can also be compared directly to a single, or sets of other SAGE databases in the SAGE Genie collection using the "SAGE Digital Gene Expression Displayer (DGED)" tool at http://cgap.nci.nih.gov/SAGE/. This type of analysis will return data similar to those in additional file 1, though comparison can also be restricted to specific libraries rather than the pool of libraries evaluated here.
Finally, this SAGE library with matching transcript identification, glomerulus to pool ratio and glomerulus to kidney ratio is supplied as additional file 1, where the order is based on tag abundance. This data set contains only 18,152 SAGE tags, as any tag found only once and not in any other library was removed. The table can be retrieved without restriction and, if desired, sorted based on the degree of tag enrichment.
Discussion
This study established a human glomerular SAGE library that can be used for data mining by investigators with an interest in glomerular cell biology and pathophysiology. The library was appropriately enriched in SAGE tags representing transcripts known to be restricted to glomerular podocytes, including nephrin [24], podocin [25], synaptopodin [26], podocalyxin [27], transcription factor 21 [28], the protein tyrosine phosphatase receptor type O GLEPP1 [29], the cyclin dependent kinase inhibitor C1 [30] and nestin [31]. It is therefore likely that other transcripts whose SAGE tags are much more highly represented in this library compared to SAGE libraries from other tissues and cells are also expressed predominantly in glomeruli.
A SAGE library that used Sau3A as the anchoring restriction enzyme was previously produced from human glomerular mRNA [9]. It identified 155 highly expressed transcripts in glomeruli that were enriched in glomeruli when compared to microdissected non-glomerular nephron segments. Since the previously published glomerular SAGE library is based on the Sau3A anchoring restriction site, it does not allow in silico comparison of tag frequencies with the much greater collection of NlaIII-based SAGE libraries. All except 12 transcripts reported to be enriched in glomeruli by Chabardes-Garonne [9] were observed in the current glomerular SAGE library. The corresponding NlaIII tag for a subset of these (47 tags) was enriched > 10 fold when compared to non-renal tissues and cells (Tables 1 and 2 and Additional files 3 &4), providing independent evidence that these represent glomerulus-predominant transcripts.
The current study also identified 86 transcript tags that were enriched more than 10 fold in glomeruli, but which were not represented in the previous Sau3A anchored library (Tables 1 and 2 and Additional files 3 &4). Failure to find a Sau3A SAGE tag for known glomerulus-restricted genes like nephrin, or an NlaIII SAGE tag for endoglin and VCAM1, suggests either that the tag frequency was too low to be detected or that the required restriction site was absent from the transcript. The current study also shows that that several transcripts more highly expressed in glomeruli compared to other nephron segments [9] are not restricted to glomeruli when compared to non-renal tissues or cells (additional file 2). This is not surprising since some transcripts that are not shared between nephron epithelium and glomerular capillary tuft nevertheless may be highly expressed in other tissues.
Several transcripts not previously shown to have a specific function in glomeruli were highly expressed and enriched in glomeruli when compared to non-glomerular tissues. Among these, the tag for the chloride intracellular channel 5 (CLIC5) is very abundant in the glomerular transcript pool, and its frequency in glomeruli was more than 800 fold greater than in other tissues. The transcript "DKFZp564B076" whose SAGE tag was previously shown to be enriched in microdissected glomeruli [9] and later in cultured glomerular EC in this laboratory [22] is identical to the 3' end of CLIC5. CLIC5 is an ezrin-binding protein involved in maintaining actin-based microvilli in the placenta and actin-based stereocilia in the inner ear [32]. Its role in glomerular cell function is as yet undefined. The transcript for the basal cell adhesion molecule (BCAM) is also very abundant in glomeruli and enriched approximately 58 fold. BCAM is a glycoprotein that functions as a receptor for alpha5 laminin. BCAM immunoreactivity is observed in both, glomerular podocytes and glomerular EC, and mice deficient in BCAM have significant structural abnormalities of glomeruli [33]. Glomerular expression of the parathyroid hormone receptor 1 (PTHR1) was not expected. PTHR1 is very abundant in renal proximal tubule cells and could therefore represent proximal tubule contamination. However, since the PTHR1 SAGE tag was less abundant in renal cortex than in glomeruli (Table 1 and Additional file 3), its enrichment in this library cannot be due to proximal tubule contamination. Indeed, mesangial cells express PTHR1 [34]. More work is required to define the function of PTHR1 in mesangial cells. In this regard, it is of great interest that Sclerostin, an inhibitor of bone matrix formation whose expression is regulated by PTH, is also expressed at much higher levels in glomeruli than in most non-renal tissues and cells (Table 1 and Additional file 3) or in other nephron segments [9]. While we have no comparison with a bone SAGE library where sclerostin is likely expressed at high levels, the finding nonetheless suggests that it could be involved in regulating extracellular matrix depositon in glomeruli. Nephronectin, a ligand for integrin alpha8beta1 is known to be essential for renal development, and is expressed in renal epithelium. Enrichment of the nephronectin SAGE tag in the glomerular library relative to kidney cortex is in keeping with the observation by Brandenberger et al [35], who observed very strong nephronectin immunoreactivity in differentiating glomeruli. The secreted glycoprotein testican 2 SPARC (SPOCK2) belongs to in the osteonectin/SPARC family [36] is also highly expressed and enriched in glomeruli. Members of this family of proteins regulate cell-cell and cell-matrix interactions, and SPOCK2 is induced after glomerular injury [37]. The other protein in this family is connective tissue growth factor (CTGF). The SAGE tag for CTGF was observed at a high frequency in glomeruli (additional file 1) but it was not highly enriched relative to other tissues. Nonetheless, both SPOCK2 and CTGF likely play a critical role in regulating glomerular remodeling. In 2006 Lakhe-Reddy and coworkers [38] described the localization of beta 8 integrin to glomerular mesangial cells and observed that its expression may suppress mesangial cell dedifferentiation via Rac1 activation. The SAGE tag for integrin beta 8 was highly expressed and enriched in this glomerular library.
While several semaphorins are expressed in renal glomeruli, so far a role for semaphorin 3G, whose SAGE tag is abundant and enriched in this database, has not been described. Still, semphorin 3G, which has repulsive function via neuropilin 2 binding in the CSN neuronal guidance, is also highly expressed in kidney [39], begging the question whether it serves an important function is in glomeruli. Based on this study many other transcripts are highly enriched in glomeruli. It is hoped that other investigators will use this database as a tool to further define the transcriptome of glomerular cells in health and disease.
We did not observe the NlaIII SAGE tag for EHD3, a transcript previously shown to be unique for glomerular endothelial cells [12], in this library. A SAGE tag for EHD3 also is not observed in the previously published Sau3A-anchored library [9]. Failure to observe this tag does not detract from the previous observations but only suggests that the EHD3 transcript abundance was too low to generate a SAGE tag in the two glomerular SAGE libraries.
Finally, not all tags observed in this SAGE library have as yet been matched to a specific gene. For some of these unidentified SAGE tags, matching sequences within the human genome are observed, but whether they represent specific transcripts is currently not known.
Conclusion
We have constructed a new human glomerular SAGE library, based on the NlaIII anchoring restriction site. The database can be searched to determine whether specific transcripts are highly expressed and/or enriched in glomeruli and it can be used a resource to further study transcripts that appear to be glomerulus-enriched but whose function in glomeruli has not been investigated so far.
Availability and requirements
The SAGE database (GEO Accession #GSM199994) described here is available for download from http://www.ncbi.nlm.nih.gov/geo/. It can also be downloaded from, or interrogated in silico at http://cgap.nci.nih.gov/SAGE/ without restriction. The annotated database containing Tag sequences, glomerular frequencies, gene identification, as well as frequency ratios to pooled and kidney libraries is available as additional file 1.
Abbreviations
SAGE: Serial Analysis of Gene Expression; TPM: Tags per million; RT-PCR: Reverse Transcriptase Polymerase Chain Reaction; WT-1: Wilms Tumor 1; EC: Endothelial Cell(s); EST: Expressed Sequence Tag; cDNA: Complementary DNA; bp: base pair.
Authors' contributions
WF, JN and BJB developed the conceptual design of the study. Contributions by WF and JN were equivalent. WF isolated human glomeruli and prepared RNA from glomeruli for construction of the SAGE library, and performed work shown in Figure 1. AG microdissected human glomeruli for RT-PCR analysis. JN performed RT-PCR on microdissected human glomeruli. SK performed immunofluorescence analysis. BJB performed the bioinformatics work and wrote the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Annotated NlaIII-anchored human glomerular long SAGE library. Additional file 1 contains all (18,152) NlaIII-anchored long SAGE tags observed in the current glomerular library and at least twice in this + currently reported long SAGE libraries. The SAGE tags are sorted by frequency (a measure of expression of the corresponding transcript). Genes matched to each tag, chromosomal localization, absolute SAGE tag count in the glomerular library, frequency (TPM) in the glomerular and the pooled libraries and the ratio of glomerular: pooled frequencies are shown. Absolute counts and TPM for the corresponding short NlaIII-based SAGE tag in whole kidney and the ratio of SAGE tag TPM in glomeruli: kidney are shown. The dataset can be downloaded without restriction.
SAGE tags observed NlaIII and Sau3A glomerular libraries but enriched less than 10 fold. This dataset represents all Sau3A tags reported by Chabardes-Garonne (9) to be enriched in glomeruli when compared to microdissected nephron segments that were also observed in the current NlaIII anchored library, but for which the corresponding NlaIII tags were enriched less than 10 fold when compared to pooled non-renal tissues and cells.
Transcripts identfied for SAGE tags enriched 30 fold or more in Glomeruli compared to pooled SAGE libraries from diverse tissues and cells. The gene symbol and gene name are shown for each tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each tag.
Transcripts name for SAGE tags enriched 10 - 25 fold in Glomeruli compared to pooled SAGE libraries. The gene symbol and gene name are shown for each unique NlaIII-anchored long SAGE tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each. tag.
Contributor Information
Jenny Nyström, Email: Jenny.Nystroem@wlab.gu.se.
Wolfgang Fierlbeck, Email: wfierlbeck@web.de.
Anna Granqvist, Email: anna.granqvist@wlab.gu.se.
Stephen C Kulak, Email: skulak@ualberta.ca.
Barbara J Ballermann, Email: barbara.ballermann@ualberta.ca.
Acknowledgements
The work was supported by Establishment Grant CEG63108 from the Canadian Institutes of Health Research (CIHR) and by CIHR operating grant MOP641814. B.J. Ballermann holds the Tier 1 Canada Research Chair in Endothelial Cell Biology. J. Sorensson-Nystrom's work was supported by Grant #14764 from the Swedish Research Council. W. Fierlbeck was supported by a fellowship grant from the Deutsche Forschungsgemeinschaft (Fi 829/1-1)
References
- Patari-Sampo A, Ihalmo P, Holthofer H. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann Med. 2006;38(7):483–492. doi: 10.1080/07853890600978149. [DOI] [PubMed] [Google Scholar]
- Herrera GA. Plasticity of mesangial cells: a basis for understanding pathological alterations. Ultrastruct Pathol. 2006;30(6):471–479. doi: 10.1080/01913120600932594. [DOI] [PubMed] [Google Scholar]
- Ballermann BJ. Glomerular endothelial cell differentiation. Kidney Int. 2005;67(5):1668–1671. doi: 10.1111/j.1523-1755.2005.00260.x. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yokomori H, Yoshimura K, Fujimaki K, Nomura M, Hibi T, Oda M. Actin filaments around endothelial fenestrae in rat hepatic sinusoidal endothelial cells. Med Electron Microsc. 2004;37(4):252–255. doi: 10.1007/s00795-004-0262-3. [DOI] [PubMed] [Google Scholar]
- Deen WM, Lazzara MJ, Myers BD. Structural determinants of glomerular permeability. Am J Physiol Renal Physiol. 2001;281(4):F579–596. doi: 10.1152/ajprenal.2001.281.4.F579. [DOI] [PubMed] [Google Scholar]
- Tryggvason K, Wartiovaara J. How does the kidney filter plasma? Physiology (Bethesda) 2005;20:96–101. doi: 10.1152/physiol.00045.2004. [DOI] [PubMed] [Google Scholar]
- Ballermann BJ. Contribution of the endothelium to the glomerular permselectivity barrier in health and disease. Nephron Physiol. 2007;106(2):19–25. doi: 10.1159/000101796. [DOI] [PubMed] [Google Scholar]
- Jeansson M, Haraldsson B. Morphological and functional evidence for an important role of the endothelial cell glycocalyx in the glomerular barrier. Am J Physiol Renal Physiol. 2006;290(1):F111–116. doi: 10.1152/ajprenal.00173.2005. [DOI] [PubMed] [Google Scholar]
- Chabardes-Garonne D, Mejean A, Aude JC, Cheval L, Di Stefano A, Gaillard MC, Imbert-Teboul M, Wittner M, Balian C, Anthouard V. A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci USA. 2003;100(23):13710–13715. doi: 10.1073/pnas.2234604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Sun Y, Patrakka J, Mostad P, Norlin J, Xiao Z, Andrae J, Tryggvason K, Samuelsson T, Betsholtz C. Glomerulus-specific mRNA transcripts and proteins identified through kidney expressed sequence tag database analysis. Kidney Int. 2007;71(9):889–900. doi: 10.1038/sj.ki.5002158. [DOI] [PubMed] [Google Scholar]
- Betsholtz C, He L, Takemoto M, Norlin J, Sun Y, Patrakka J, Tryggvason K. The glomerular transcriptome and proteome. Nephron Exp Nephrol. 2007;106(2):e32–36. doi: 10.1159/000101790. [DOI] [PubMed] [Google Scholar]
- Patrakka J, Xiao Z, Nukui M, Takemoto M, He L, Oddsson A, Perisic L, Kaukinen A, Szigyarto CA, Uhlen M. Expression and subcellular distribution of novel glomerulus-associated proteins dendrin, ehd3, sh2d4a, plekhh2, and 2310066E14Rik. J Am Soc Nephrol. 2007;18(3):689–697. doi: 10.1681/ASN.2006060675. [DOI] [PubMed] [Google Scholar]
- Tryggvason K, Patrakka J, Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354(13):1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- el-Meanawy MA, Barathan S, Hayden PS, Iyengar SK, Schelling JR, Sedor JR. Serial analysis of gene expression. Methods Mol Med. 2003;86:257–274. doi: 10.1385/1-59259-392-5:257. [DOI] [PubMed] [Google Scholar]
- Misra RP. Isolation of glomeruli from mammalian kidneys by graded sieving. Am J Clin Pathol. 1972;58(2):135–139. doi: 10.1093/ajcp/58.2.135. [DOI] [PubMed] [Google Scholar]
- Ballermann BJ, Skorecki KL, Brenner BM. Reduced glomerular angiotensin II receptor density in early untreated diabetes mellitus in the rat. Am J Physiol. 1984;247(1 Pt 2):F110–116. doi: 10.1152/ajprenal.1984.247.1.F110. [DOI] [PubMed] [Google Scholar]
- Akmaev VR, Wang CJ. Correction of sequence-based artifacts in serial analysis of gene expression. Bioinformatics. 2004;20(8):1254–1263. doi: 10.1093/bioinformatics/bth077. [DOI] [PubMed] [Google Scholar]
- Emmersen J, Heidenblut AM, Hogh AL, Hahn SA, Welinder KG, Nielsen KL. Discarding duplicate ditags in LongSAGE analysis may introduce significant error. BMC Bioinformatics. 2007;8:92. doi: 10.1186/1471-2105-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16(11):953–959. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
- Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA. 2002;99(17):11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoelge G, Luo W, Fine D, Perschl AM, Fierlbeck W, Haririan A, Sorensson J, Rehman TU, Hauser P, Trevick JS. A SAGE-based comparison between glomerular and aortic endothelial cells. Am J Physiol Renal Physiol. 2005;288(6):F1290–1300. doi: 10.1152/ajprenal.00076.2004. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Ma D, Armugam A. Real-time detection of gene promoter activity: quantitation of toxin gene transcription. Nucleic Acids Res. 2001;29(12):E58–58. doi: 10.1093/nar/29.12.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R. Positionally cloned gene for a novel glomerular protein – nephrin – is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1(4):575–582. doi: 10.1016/S1097-2765(00)80057-X. [DOI] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24(4):349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- Mundel P, Heid HW, Mundel TM, Kruger M, Reiser J, Kriz W. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139(1):193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D, Sharkey DJ, Farquhar MG. Identification and characterization of podocalyxin – the major sialoprotein of the renal glomerular epithelial cell. J Cell Biol. 1984;98(4):1591–1596. doi: 10.1083/jcb.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126(24):5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- Thomas PE, Wharram BL, Goyal M, Wiggins JE, Holzman LB, Wiggins RC. GLEPP1, a renal glomerular epithelial cell (podocyte) membrane protein-tyrosine phosphatase. Identification, molecular cloning, and characterization in rabbit. J Biol Chem. 1994;269(31):19953–19962. [PubMed] [Google Scholar]
- Hiromura K, Haseley LA, Zhang P, Monkawa T, Durvasula R, Petermann AT, Alpers CE, Mundel P, Shankland SJ. Podocyte expression of the CDK-inhibitor p57 during development and disease. Kidney Int. 2001;60(6):2235–2246. doi: 10.1046/j.1523-1755.2001.00057.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Boyle S, Zhao M, Su W, Takahashi K, Davis L, Decaestecker M, Takahashi T, Breyer MD, Hao CM. Differential expression of the intermediate filament protein nestin during renal development and its localization in adult podocytes. J Am Soc Nephrol. 2006;17(5):1283–1291. doi: 10.1681/ASN.2005101032. [DOI] [PubMed] [Google Scholar]
- Gagnon LH, Longo-Guess CM, Berryman M, Shin JB, Saylor KW, Yu H, Gillespie PG, Johnson KR. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci. 2006;26(40):10188–10198. doi: 10.1523/JNEUROSCI.2166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahuel C, Filipe A, Ritie L, El Nemer W, Patey-Mariaud N, Eladari D, Cartron JP, Simon-Assmann P, Le Van Kim C, Colin Y. Genetic inactivation of the laminin alpha5 chain receptor Lu/BCAM leads to kidney and intestinal abnormalities in the mouse. Am J Physiol Renal Physiol. 2008;294(2):F393–406. doi: 10.1152/ajprenal.00315.2007. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Ruiz-Ortega M, Esbrit P, Ruperez M, Ortega A, Santos S, Blanco J, Ortega L, Egido J. Angiotensin II increases parathyroid hormone-related protein (PTHrP) and the type 1 PTH/PTHrP receptor in the kidney. J Am Soc Nephrol. 2002;13(6):1595–1607. doi: 10.1097/01.ASN.0000015622.33198.BF. [DOI] [PubMed] [Google Scholar]
- Brandenberger R, Schmidt A, Linton J, Wang D, Backus C, Denda S, Muller U, Reichardt LF. Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J Cell Biol. 2001;154(2):447–458. doi: 10.1083/jcb.200103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CJ, Sage EH. A prototypic matricellular protein in the tumor microenvironment – where there's SPARC, there's fire. J Cell Biochem. 2008;104(3):721–732. doi: 10.1002/jcb.21688. [DOI] [PubMed] [Google Scholar]
- Francki A, Sage EH. SPARC and the kidney glomerulus: matricellular proteins exhibit diverse functions under normal and pathological conditions. Trends Cardiovasc Med. 2001;11(1):32–37. doi: 10.1016/S1050-1738(01)00081-0. [DOI] [PubMed] [Google Scholar]
- Lakhe-Reddy S, Khan S, Konieczkowski M, Jarad G, Wu KL, Reichardt LF, Takai Y, Bruggeman LA, Wang B, Sedor JR. Beta8 integrin binds Rho GDP dissociation inhibitor-1 and activates Rac1 to inhibit mesangial cell myofibroblast differentiation. J Biol Chem. 2006;281(28):19688–19699. doi: 10.1074/jbc.M601110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Masuda T, Fukaya M, Kataoka H, Mishina M, Yaginuma H, Watanabe M, Shimizu T. Identification and characterization of a novel member of murine semaphorin family. Genes Cells. 2005;10(8):785–792. doi: 10.1111/j.1365-2443.2005.00877.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotated NlaIII-anchored human glomerular long SAGE library. Additional file 1 contains all (18,152) NlaIII-anchored long SAGE tags observed in the current glomerular library and at least twice in this + currently reported long SAGE libraries. The SAGE tags are sorted by frequency (a measure of expression of the corresponding transcript). Genes matched to each tag, chromosomal localization, absolute SAGE tag count in the glomerular library, frequency (TPM) in the glomerular and the pooled libraries and the ratio of glomerular: pooled frequencies are shown. Absolute counts and TPM for the corresponding short NlaIII-based SAGE tag in whole kidney and the ratio of SAGE tag TPM in glomeruli: kidney are shown. The dataset can be downloaded without restriction.
SAGE tags observed NlaIII and Sau3A glomerular libraries but enriched less than 10 fold. This dataset represents all Sau3A tags reported by Chabardes-Garonne (9) to be enriched in glomeruli when compared to microdissected nephron segments that were also observed in the current NlaIII anchored library, but for which the corresponding NlaIII tags were enriched less than 10 fold when compared to pooled non-renal tissues and cells.
Transcripts identfied for SAGE tags enriched 30 fold or more in Glomeruli compared to pooled SAGE libraries from diverse tissues and cells. The gene symbol and gene name are shown for each tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each tag.
Transcripts name for SAGE tags enriched 10 - 25 fold in Glomeruli compared to pooled SAGE libraries. The gene symbol and gene name are shown for each unique NlaIII-anchored long SAGE tag sequence. Where available, the corresponding short Sau3A SAGE tag (Ref 9) is shown. SAGE tag frequencies are shown as Tags per Million (TPM). Sau3A TPM is derived from Ref 9. Enrichment in glomeruli relative to pooled libraries (G:P) and relative to whole kidney (G:K) is shown for each. tag.