Abstract
Pandemic influenza viruses cause significant mortality in humans. In the 20th century, 3 influenza viruses caused major pandemics: the 1918 H1N1 virus, the 1957 H2N2 virus, and the 1968 H3N2 virus. These pandemics were initiated by the introduction and successful adaptation of a novel hemagglutinin subtype to humans from an animal source, resulting in antigenic shift. Despite global concern regarding a new pandemic influenza, the emergence pathway of pandemic strains remains unknown. Here we estimated the evolutionary history and inferred date of introduction to humans of each of the genes for all 20th century pandemic influenza strains. Our results indicate that genetic components of the 1918 H1N1 pandemic virus circulated in mammalian hosts, i.e., swine and humans, as early as 1911 and was not likely to be a recently introduced avian virus. Phylogenetic relationships suggest that the A/Brevig Mission/1/1918 virus (BM/1918) was generated by reassortment between mammalian viruses and a previously circulating human strain, either in swine or, possibly, in humans. Furthermore, seasonal and classic swine H1N1 viruses were not derived directly from BM/1918, but their precursors co-circulated during the pandemic. Mean estimates of the time of most recent common ancestor also suggest that the H2N2 and H3N2 pandemic strains may have been generated through reassortment events in unknown mammalian hosts and involved multiple avian viruses preceding pandemic recognition. The possible generation of pandemic strains through a series of reassortment events in mammals over a period of years before pandemic recognition suggests that appropriate surveillance strategies for detection of precursor viruses may abort future pandemics.
Keywords: H1N1, influenza A, swine, virus evolution, molecular clock
Pandemic influenza outbreaks pose a significant threat to public health worldwide as highlighted by the recent introduction of swine-derived H1N1 virus into humans (1). In the 20th century, 3 influenza viruses caused major pandemics: the 1918 H1N1 virus, the 1957 H2N2 virus (H2N2/1957), and the 1968 H3N2 virus (H3N2/1968) (2, 3). These pandemics were initiated by the introduction and successful adaptation of a novel hemagglutinin subtype to humans from an animal source, resulting in antigenic shift (4, 5). A number of hypotheses have been proposed for the development of pandemicity of the influenza virus, including direct introduction into humans from an avian origin and reassortment between avian and previously circulating human viruses, either directly in humans or through an intermediate mammalian host (6–9).
Based on studies of amino acid similarities of all 8 gene segments of A/Brevig Mission/1/1918 virus (BM/1918), it was concluded that this virus most likely was derived directly from an avian precursor that was introduced to humans shortly before the pandemic (10, 11). This interpretation is controversial because of variant gene phylogenies that either conflict with this theory or remain ambiguous because of a lack of contemporaneous viruses (12–14). Analysis of sequences generated from the H2N2/1957 and H3N2/1968 strains showed that these pandemics were caused by genetic reassortment between avian and pre-existing human viruses (8). The H2N2/1957 pandemic strain contained introduced hemagglutinin, neuraminidase, and PB1 genes, whereas the H3N2/1968 pandemic strain incorporated avian HA and PB1 genes (2).
However, the evolutionary history of these 3 pandemic viruses remains unclear, and that lack of understanding hinders the recognition of and preparedness for future influenza pandemics. We therefore investigated evolutionary mechanisms of pandemic emergence by conducting comparative genetic analyses of all available viruses associated with the emergence of the 1918, 1957, and 1968 pandemics.
Bayesian relaxed molecular clock phylogenetic methods, as implemented in BEAST, use flexible evolutionary models to infer the timing of evolutionary events, so that the evolutionary rate can vary among branches on the tree and uncertainty caused by missing data and unknown evolutionary rates can be incorporated (15). In the case of influenza, the times of most recent common ancestor (TMRCA) provide an estimate of when virus genes emerged in a given host that allows the time of interspecies transmission to be inferred.
Here we estimated the evolutionary history to investigate the possible date of introduction to humans of each of the genes for all 20th century pandemic influenza strains. Mean TMRCA estimates of each gene segment of H1N1 viruses shows that the components of the 1918 pandemic strain were circulating in mammalian hosts, i.e., swine and humans, at least 2 to 15 years before pandemic occurrence. Phylogenetic analyses suggest that the 1918 H1N1 pandemic virus most likely was generated by reassortment between mammalian viruses and a previous human strain and was not a pure avian virus. We also show that seasonal and classic swine H1N1 viruses were not derived directly from BM/1918; rather, their precursors co-circulated during the pandemic. Mean TMRCA estimates also suggest that the avian-derived genes of the H2N2 and H3N2 pandemic strains may have been introduced to humans on multiple occasions over a number of years.
Results and Discussion
Evolutionary Inferences on the Origin of BM/1918, Human and Swine H1N1 Viruses.
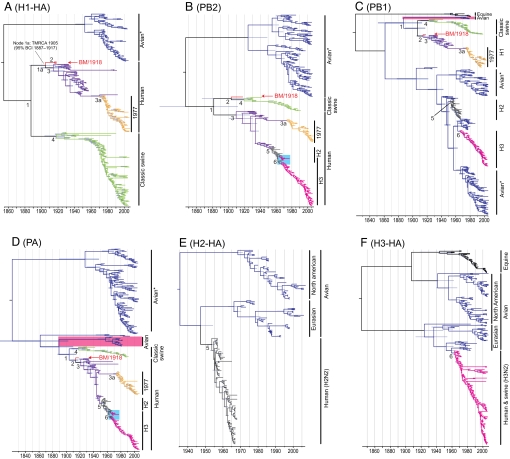
To establish when H1N1 virus genes were introduced to mammals, we co-estimated phylogenies and TMRCA for all known mammalian, i.e., swine and human, H1N1 virus genes (supporting information (SI) Table S1). For each of the 8 genes, mammalian H1N1 viruses (BM/1918, seasonal H1N1, and classic swine H1N1) formed monophyletic clades (node 1 in Fig. 1 and Figs. S1–S8 and Table 1). For 6 genes (H1, N1, PB2, NP, M, and NS), avian viruses formed distant monophyletic groups to mammalian H1N1 genes (Fig. 1 A and B and Figs. S1–S3 and S6–S8). In the PB1 and PA genes, a small clade of avian viruses formed a group more closely related to mammalian H1N1 viruses, providing direct phylogenetic evidence of a more recent avian source (Fig. 1 C and D and Figs. S4 and S5). The TMRCA estimates for node 1 ranged from 1881 [95% Bayesian credible interval (BCI) 1813–1912] for the PB2 gene to 1907 (BCI 1892–1918) for the N1 gene (Table 1). Ages of the H1 and NP genes at node 1 could not be calculated because of uncertainty in the phylogenies (Fig. 1A and Figs. S1 and S6). Importantly, TMRCAs at node 1 indicate that the PB2 and M gene precursors of all human H1N1 viruses were present in mammalian hosts (e.g., swine) at least 6 years before the 1918 pandemic (Table 1).
Fig. 1.
Dated phylogenies of influenza A virus genes. (A) H1-HA, (B) PB2, (C) PB1, (D) PA, (E) H2-HA, and (F) H3-HA gene trees scaled to time (Horizontal Axis) generated using the SRD06 codon model and uncorrelated relaxed clock model. Nodes correspond to mean TMRCAs, and blue horizontal bars at nodes represent the 95% BCIs of TMRCAs. Colored branches represent major influenza A virus lineages. The blue boxes in B and D indicate a period of co-circulation of H2 and H3 viruses. Identical phylogenetic trees with virus names are available online in the supporting information. TMRCAs and BCIs for each of the major influenza A virus lineages are given in Table 1.
Table 1.
Times of most recent common ancestors of human pandemic influenza viruses and related lineages
| Gene | Swine/Human H1N1 (node 1) | BM/1918 H1N1 (node 2) | Seasonal H1N1 (node 3) | H1N1 Re-introduction (node 3a) | H2N2 (node 5) | H3N2 (node 6) |
|---|---|---|---|---|---|---|
| PB2 | 1881 (1813, 1912) | 1903 (1867, 1918) | 1910 (1888, 1928) | 1974 (1967, 1977) | H1N1† | H2N2† |
| PB1 | 1906 (1890, 1918) | 1914 (1906, 1918) | BM/1918† | 1974 (1970, 1977) | 1954 (1951, 1957) | 1967 (1965, 1968) |
| PA | 1904 (1888, 1915) | 1914 (1906, 1918) | BM/1918 | 1974 (1969, 1977) | H1N1 | H2N2 |
| HA | 1885* | 1916 (1910, 1918) | 1913 (1895, 1925) | 1974 (1971, 1976) | 1955 (1952, 1957) | 1963 (1959, 1966) |
| NP | 1884* | 1909 (1884, 1918) | BM/1918 | 1974 (1969, 1977) | H1N1 | H2N2 |
| NA | 1907 (1892, 1918) | 1913 (1905, 1918) | BM/1918 | 1975 (1972, 1977) | 1950 (1945, 1955) | H2N2 |
| M | 1884 (1841, 1911) | 1896* | 1911 (1893, 1926) | 1972 (1965, 1977) | H1N1 | H2N2 |
| NS | 1899 (1876, 1917) | 1908 (1891, 1918) | 1915 (1900, 1926) | 1975 (1971, 1977) | H1N1 | H2N2 |
*Node not supported; therefore BCIs could not be estimated.
†These gene segments were derived from BM/1918, seasonal H1N1, and H2N2 viruses, respectively.
Preliminary phylogenetic analysis showed that the BM/1918 virus H1, N1, PB1, PA, and NP genes clustered with human H1N1 influenza A viruses, whereas its PB2, M, and NS genes clustered with swine. These relationships have high statistical support (bootstrap support >80%) except for the placement of BM/1918 virus in the M gene phylogeny. Estimates for BM/1918 virus TMRCAs (node 2) ranged from 1903 (BCI 1867–1918) for the PB2 gene to 1916 (BCI 1910–1918) for the HA gene (Table 1). These mean TMRCA estimates suggest that the BM/1918 virus genes were present in swine or human hosts 2 to 15 years before the pandemic. The TMRCA of the M gene at node 2 could not be estimated, but the TMRCA at node 1 indicated that the BM/1918 M gene precursor probably was present in mammalian hosts before 1911 (Fig. S7 and Table 1).
Interestingly, the TMRCA distributions of the PB2, NP, and NS genes of the BM/1918 virus (Table 1) suggest those genes have circulated in humans since the 1889 H3 influenza pandemic (16, 17). Earlier estimates of mutation rates of the NP gene suggested that human H1N1 and classic swine influenza viruses emerged from the avian source around 1912 or 1913 (18). However, our results suggest that the same NP gene lineage has been circulating in human influenza A viruses since the 19th century, consistent with the report by Gammelin et al. (19).
Extensive arguments, based primarily on similarity between consensus amino acid sequences, have been made that the BM/1918 virus was derived directly from an avian progenitor, contrary to phylogenetic evidence (10–13, 19, 20). These residue similarities may help explain the avian-like phenotype of the BM/1918 virus, particularly its high virulence in mammals (21).
Taken together, our results indicate that it is unlikely that the BM/1918 virus could have resulted from adaptation of an entire avian virus introduced directly into humans shortly before the pandemic. More likely, it was generated by reassortment between previously circulating swine and human strains and introduced avian viruses over a period of years.
It generally has been assumed that after the pandemic the BM/1918 virus established in humans to form the seasonal H1N1 influenza lineage (e.g., 2, 3, 9, 20). However, our phylogenetic analysis shows that only the PB1, PA, NP, and N1 genes of seasonal H1N1 were derived from BM/1918 (Fig. 1 and Figs. S1–S8). Comparisons of TMRCA estimates of the HA for the BM/1918 virus (node 2, TMRCA 1916, BCI 1910–1918) and seasonal H1N1 lineage (node 3, TMRCA 1913, BCI 1895–1925) indicate that these H1 lineages diverged (node 1a, TMRCA 1905, BCI 1887–1917) and co-circulated during the 1918 pandemic (Fig. 1A).
Phylogenetic relationships between BM/1918 and classic swine H1N1 virus PB2, M, and NS genes also indicate that classic swine H1N1 is a reassortant between BM/1918 and an unknown virus. As such, classic swine H1N1 is derived partially from BM/1918 and is not a precursor of the 1918 pandemic virus (Fig. 1 and Figs. S1–S8) (9).
It therefore appears that at least 3 reassortant H1N1 variants co-circulated: BM/1918 and the precursors of seasonal and classic swine H1N1 viruses. The co-circulation of the BM/1918 and seasonal H1N1 viruses may explain reports of influenza outbreaks of varying severity during the 1918 pandemic (22, 23). Here we provide the first evidence that seasonal H1N1 viruses were not derived directly from BM/1918 but co-circulated during the pandemic. This evidence may be relevant to the current emergence and potential pandemicity of swine-derived H1N1 viruses in humans (1).
Phylogenetic analyses of the re-emergent H1N1/1977 virus confirmed that each of 8 genes was directly derived from those H1N1 viruses circulating in the 1950s (Fig. 1 and Figs. S1–S8). Dating the time of emergence of each gene segment showed similar TMRCAs with a mean of ≈ 2 to 3 years before the detection of the viruses (Table 1 and Table S2). These results support the hypothesis that the re-emergence of H1N1/1977 most likely resulted from accidental laboratory re-introduction (2).
Emergence of H2N2 and H3N2 Pandemic Viruses.
Phylogenies confirmed that the H2N2/1957 was a genetic reassortant between previously circulating human and avian viruses, with the novel H2, N2, and PB1 genes derived from Eurasian avian sources (Fig. 1 A, C, and D, Figs. S2–S6 and S8–S11). The mean TMRCA estimates of the introduced genes of the H2N2 pandemic suggest that the introduction of these 3 genes into human populations occurred 2 to 6 years before the pandemic.
Ages of the novel H3 (TMRCA 1968, BCI 1967–1968) and PB1 (TMRCA 1967, BCI 1966–1968) genes indicated that the introduction occurred between 1966 and 1968 (Table 1). The remaining genes of the H3N2/1968 virus came from the previous human H2N2 virus. The upper BCI estimates of the human H3N2 PB1 and HA TMRCAs indicate that this virus may have circulated in humans as early as 1966; the last record of H2N2 in the human population was from 1968, indicating that H2N2 and H3N2 viruses co-circulated in humans for approximately 1 to 3 years (Table 1). This observation is consistent with the phylogenies of the shared genes, with the exception of the NS gene, in which late H2N2 and early H3N2 do not form separate monophyletic lineages (e.g., blue boxes in Fig. 1 B and D). Differences in the TMRCA estimates raise the possibility that the introduced genes of the H2N2 and H3N2 pandemic strains may have been introduced sequentially from multiple sources over a number of years. Because of a lack of sequence data for swine influenza from these periods, the involvement of swine in the generation of these pandemic strains cannot be precluded.
Conclusions
The results of our study have provided fresh insights into pandemic emergence by raising the possibility that all 3 pandemic influenza strains of the 20th century may have been generated through a series of multiple reassortment events and emerged over a period of years before pandemic recognition. Furthermore, results indicate that each of these strains was produced by reassortment between the previously circulating human virus and at least 1 virus of animal origin. The novel gene segments for the H2N2/1957 and H3N2/1968 pandemics seem to have originated from avian hosts, but the zoonotic sources of the introduced viral gene segments for the 1918 pandemic remain ambiguous. However, evidence suggests that, over a number of years, avian gene virus segments have entered mammalian populations where the viruses may have undergone reassortment with the prevailing human virus. Given the frequent interspecies transmission of influenza viruses between swine and humans, it is most likely that such reassortment events occurred in swine before pandemic emergence.
Interestingly, our analyses suggest that in the 1918 and 1957 pandemics novel NA and internal genes may have been introduced into the prevailing human virus strains before the acquisition of the novel pandemic HA. Frequent detection of seasonal human influenza strains in swine indicates that pandemic precursor viruses probably have circulated in either swine or human populations. The hypothetical precursors to the H2N2 and H3N2 pandemics have not been detected, probably because they originated in Asia where little or no surveillance was conducted at that time (2).
If future pandemics arise in this manner, this interval may provide the best opportunity for health authorities to intervene to mitigate the effects of a pandemic or even to abort its emergence. However, our findings argue the need for high-throughput characterization of all 8 gene segments of human virus isolates, even those that have unremarkable HA antigens, particularly of human viruses isolated in hotspots for zoonotic infections with avian influenza viruses. At present, global influenza surveillance in humans focuses attention primarily on hemagglutinin. Although this focus will continue to be required for strain selection for seasonal influenza vaccines, our findings argue that this surveillance will not suffice for early warning of an incipient pandemic.
Methods
Preliminary Phylogenetic Analyses and Data Preparation.
Provisional phylogenetic analyses were carried out for all available influenza gene sequences using the neighbor-joining method in PAUP* 4b10 (24) with a best-fit nucleotide substitution model (25) and an appropriate outgroup (Table S1). The purpose of these large-scale phylogenetic analyses was to identify relationships between pandemic strains and all other sequences. These lineages (in particular, avian, swine, and human) were identified in each tree as monophyletic clades with bootstrap support of 80% or higher.
Based on these preliminary analyses, 11 datasets were compiled for human influenza viruses: the hemagglutinin (HA: H1, H2, H3), neuraminidase (NA: N1, N2), and the 6 internal gene segments (PB2, PB1, PA, NP, M, and NS allele A), together with genes from representative influenza viruses isolated from other hosts (birds, swine, horses, and other mammals). Full details of the final datasets that were used for all subsequent analyses are given in Table S1.
Phylogenetic Inference, Estimation of Nucleotide Substitution Rates and Times of Divergence.
To estimate divergence times and rates of nucleotide substitutions in influenza A viruses, we applied a relaxed-clock Bayesian Markov chain Monte Carlo method as implemented in BEAST v1.4.8 (26). This method allows variable nucleotide substitution rates among lineages and also incorporates phylogenetic uncertainty by sampling phylogenies and parameter estimates in proportion to their posterior probability (26). The marginal likelihoods of 3 different clock models, strict clock, uncorrelated exponential clock (uced), and uncorrelated log-normal clock (ucld), were compared using a Bayes factor test for best fit (15, 27, 28). This test revealed that for all genes the uced model, which allows evolutionary substitution rates to vary within an exponential distribution along branches, was the best fit for the sequence data (Table S2). The outgroup sequences were not included in the BEAST analyses; rather, the relationships based on the tree topologies from the preliminary analyses described earlier were enforced as prior assumptions for the Bayesian analyses. Trees generated from the BEAST analyses were rooted by fixing basal node relationships of the major lineages (avian, swine, and human) in all phylogenies (Table S1).
In analyzing protein-coding sequences, we used the SRD06 codon position model to partition the data (29). The first partition unifies the first plus second codon positions, and the second partition describes the third codon position. Because each dataset included multiple non-mixing populations, a constant population coalescent tree prior over the unknown tree space and relatively uninformative priors over the remaining model parameter space were assumed for each dataset (15). We carried out 3 independent analyses for 20–60 million generations sampled to produce at least 10,000 trees for each data set to ensure adequate sample size of all analysis parameters including the posterior, prior, nucleotide substitution rates, and likelihoods (effective sample size > 200). The mean substitution rates, mean TMRCAs, and maximum clade credibility phylogenetic trees then were calculated after the removal of an appropriate burn-in (10%–15% of the samples in most cases, with 1 exception, in which ≈20% was removed for analyses of the PB1 gene) following visual inspection in TRACER version 1.4 (30).
TMRCA estimates of introduced genes of pandemic viruses were used to infer the time of incorporation of these genes to form the pandemic virus particle. For example, the H2N2/1957 and the H3N2/1968 pandemic strains were generated by known reassortment events between previously circulating human strains and introduced avian genes. The TMRCA estimates with BCIs of these introduced genes provide an estimated timeline for the generation of the pandemic strain.
Furthermore, because we are dealing with interspecies transmission events from a natural (avian) gene pool to other species, with very limited subtypes of influenza virus present, we believe it is reasonable to interpret TMRCAs as providing an estimate of time-bounds of interspecies transmission events. It has been well described that avian viruses rarely transmit to mammalian hosts, and in talking about initial transmission events, we have been very careful to indicate the uncertainty as to which mammalian host is involved. Likewise, transmission of influenza virus from swine to humans also is a rare event. The high level of host restriction between avian and mammalian hosts and host-adapted influenza viruses also supports our interpretation.
Supplementary Material
Acknowledgments.
This study was supported by the Area of Excellence Scheme of the University Grants Committee (Grant AoE/M-12/06) of the Hong Kong SAR Government, the National Institutes of Health [National Institute of Allergy and Infectious Disease (NIAID) contract HHSN266200700005C], and the Li Ka Shing Foundation. G.J.D.S. is supported by a career development award under NIAID contract HHSN266200700005C.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904991106/DCSupplemental.
References
- 1.Anonymous. Update: Infections with a swine-origin influenza A (H1N1) virus - United States and other countries, April 28, 2009. Morb Mortal Wkly Rep. 2009;58:431–433. [PubMed] [Google Scholar]
- 2.Kilbourne ED. Influenza pandemics of the 20th century. Emerging Infectious Diseases. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antiviral Therapy. 2007;12:581–591. [PMC free article] [PubMed] [Google Scholar]
- 4.Gething MJ, Bye J, Skehel J, Waterfield M . Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980;287:301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- 5.Fang R, Jou WM, Huylebroeck D, Devos R, Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: Animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981;25:315–323. doi: 10.1016/0092-8674(81)90049-0. [DOI] [PubMed] [Google Scholar]
- 6.Scholtissek C, Rohde W, von Hoyningen V, Rott R. On the origin of the human influenza virus subtype H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 7.Scholtissek C, Bürger H, Kistner O, Shortridge KF. The nucleoprotein as a possible factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 8.Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:192–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taubenberger JK, et al. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Morens DM. 1918 influenza: The mother of all pandemics. Emerging Infectious Diseases. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs MJ, Gibbs AJ. Molecular virology: Was the 1918 pandemic caused by a bird flu? Nature. 2006;440:E8. doi: 10.1038/nature04823. [DOI] [PubMed] [Google Scholar]
- 13.Antonovics JH, Hood ME, Baker CH. Molecular virology: Was the 1918 flu avian in origin? Nature. 2006;440:E9. doi: 10.1038/nature04824. [DOI] [PubMed] [Google Scholar]
- 14.Taubenberger JK, et al. Molecular virology: Was the 1918 pandemic caused by a bird flu? (Reply) Nature. 2006;440:E9–E10. doi: 10.1038/nature04823. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedson DS, Huber MA, Kasel JA, Webster RG. Presence of A-Equi-2 hemagglutinin and neuraminidase antibodies in man. Proc Soc Exp Biol Med. 1972;139:825–826. doi: 10.3181/00379727-139-36245. [DOI] [PubMed] [Google Scholar]
- 17.Masurel N, Marine WM. Recycling of Asian and Hong Kong influenza A virus hemagglutinins in man. Am J Epidemiol. 1973;97:44–49. doi: 10.1093/oxfordjournals.aje.a121483. [DOI] [PubMed] [Google Scholar]
- 18.Gorman OT, et al. Evolution of influenza A virus nucleoprotein genes: Implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gammelin M, et al. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990;7:194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- 20.Reid AH, Fannin TG, Janczewski TA, Lourens RM, Taubenberger JK. Novel origin of the 1918 pandemic influenza virus nucleoprotein gene. J Virol. 2004;78:12462–12470. doi: 10.1128/JVI.78.22.12462-12470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 22.Andreasen V, Viboud C, Simonsen L. Epidemiologic characterization of the 1918 influenza pandemic summer wave in Copenhagen: Implications for pandemic control strategies. J Infect Dis. 2008;197:270–278. doi: 10.1086/524065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry JM, Viboud C, Simonsen L. Cross-protection between successive waves of the 1918–1919 influenza pandemic: Epidemiological evidence from US Army camps and from Britain. J Infect Dis. 2008;198:1427–1434. doi: 10.1086/592454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swofford DL. Sunderland, MA: Sinauer Associates; 2001. PAUP*: Phylogenetic analysis using parsimony (and other methods) 4.0 Beta. [Google Scholar]
- 25.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 26.Drummond AJ, Rambaut A BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 28.Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 30.Rambaut A, Drummond AJ. Tracer v1.4: MCMC trace analyses tool. [Accessed June 20, 2008];2007 Available at: http://beast.bio.ed.ac.uk/Tracer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.