Abstract
Primary vesicoureteral reflux (pVUR) is one of the most common causes of pediatric kidney failure. Linkage scans suggest that pVUR is genetically heterogeneous with two loci on chromosomes 1p13 and 2q37 under autosomal dominant inheritance. Absence of pVUR in parents of affected individuals raises the possibility of a recessive contribution to pVUR. We performed a genome-wide linkage scan in 12 large families segregating pVUR, comprising 72 affected individuals. To avoid potential misspecification of the trait locus, we performed a parametric linkage analysis using both dominant and recessive models. Analysis under the dominant model yielded no signals across the entire genome. In contrast, we identified a unique linkage peak under the recessive model on chromosome 12p11-q13 (D12S1048), which we confirmed by fine mapping. This interval achieved a peak heterogeneity LOD score of 3.6 with 60% of families linked. This heterogeneity LOD score improved to 4.5 with exclusion of two high-density pedigrees that failed to link across the entire genome. The linkage signal on chromosome 12p11-q13 originated from pedigrees of varying ethnicity, suggesting that recessive inheritance of a high frequency risk allele occurs in pVUR kindreds from many different populations. In conclusion, this study identifies a major new locus for pVUR and suggests that in addition to genetic heterogeneity, recessive contributions should be considered in all pVUR genome scans.
Vesicoureteral reflux (VUR; OMIM no. 193000) is the retrograde flow of urine from the bladder to the ureters and the kidneys during micturation. Uncorrected, VUR can lead to repeated urinary tract infections, renal scarring and reflux nephropathy, accounting for up to 25% of pediatric end stage renal disease.1,2 VUR is commonly seen as an isolated disorder (primary VUR; pVUR), but it can also present in association with complex congenital abnormalities of the kidney and urinary tract or with specific syndromic disorders, such as renal-coloboma and branchio-oto-renal syndromes.3–8
pVUR has a strong hereditary component, with monozygotic twin concordance rates of 80%.9–12 Sibling recurrence rates of 30% to 65% have suggested segregation of a single gene or oligogenes with large effects.9,12–14 Interestingly however, the three published genome-wide linkage scans of pVUR have strongly suggested multifactorial determination.15–17 Two pVUR loci have been identified with genome-wide significance on chromosomes 1p13 and 2q37 under an autosomal dominant transmission with locus heterogeneity.15,16 Multiple suggestive signals have also been reported, but remarkably, these studies show little overlap.15–17 These data suggest that pVUR may be extremely heterogeneous, with mutations in different genes each accounting for a fraction of cases. The genes underlying pVUR loci have not yet been identified, but two recent studies have reported segregating mutations in the ROBO2 gene in up to 5% of pVUR families.18,19
Despite evidence for genetic heterogeneity and different subtypes of disease, genetic studies have all modeled pVUR as an autosomal dominant trait.15–17,20 Recessive inheritance has generally not been considered because the absence of affected parents can be explained by spontaneous resolution of pVUR with older age. However, many pVUR cohorts are composed of affected sibships or pedigrees compatible with autosomal recessive transmission, suggesting the potential for alternative modes of inheritance.9–12,16,17,20–22 Systematic family screening to clarify the mode of inheritance is not feasible for pVUR because the standard diagnostic tool, the voiding cystourethrogram (VCUG), is invasive and would expose participants to radiation. Formal assessment of a recessive contribution in sporadic pVUR has also been difficult because studies have been conducted in populations with low consanguinity rates.9–12,16,17,20–22 However, recent studies have identified an unexpected recessive contribution to several complex traits such as ductus arteriosus or autism.23,24 Thus, in addition to genetic heterogeneity, genes with alternative modes of transmission may segregate among pVUR families, and misspecification of the inheritance model may complicate mapping studies of this trait.
Several approaches can be considered to address the difficulties imposed by complex inheritance, variable penetrance, and genetic heterogeneity. Studying large, well characterized cohorts with newer single-nucleotide polymorphism (SNP)-based technologies can maximize inheritance information across the genome and increase the power of linkage studies.25 In addition, in the setting of locus heterogeneity and uncertainty about the mode of transmission, analysis under a dominant and a recessive model has greater power compared with nonparametric methods and more often results in detection of the correct mode of transmission without incurring a significant penalty for multiple testing.26–29 We combined these approaches in this study and successfully localized a major gene for VUR, which unexpectedly demonstrates autosomal recessive transmission.
RESULTS
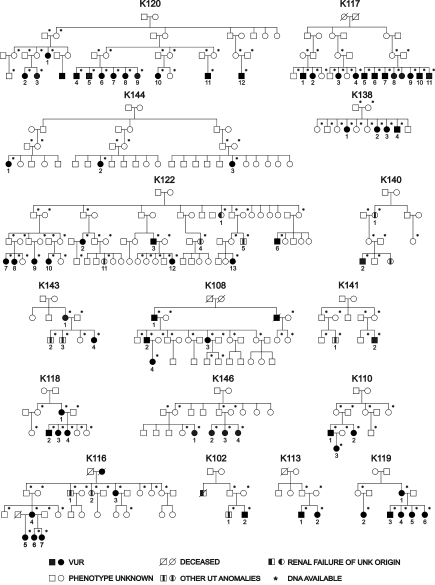
We identified 16 large Caucasian pedigrees from the United States (n = 8) and Italy (n = 8) ascertained through an index case with pVUR documented by positive VCUG and obtained DNA from 200 individuals (Figure 1 and Supplemental Table S1). Of the United States kindreds, six were of Hasidic Jewish origin (K117, K120, K122, K138, K144, K146), and two were of Irish American origin (K118, K119). Among the 184 relatives of the probands, 56 individuals (30%) had pVUR based on a positive VCUG without other renal or urologic defects, and 11 individuals (6%) had urinary tract abnormalities other than pVUR (Supplemental Table S1). The prevalence of pVUR and other urinary tract abnormalities among relatives was approximately 30-fold and 60-fold higher, respectively, than the reported prevalence in the general population.14,30 These data are consistent with the known familial aggregation of pVUR and other urinary tract defects, indicating a strong genetic effect on these traits. The remaining 117 individuals were considered as phenotype unknown. Based on our primary phenotype of pVUR without other urologic abnormalities, 12 families were informative for linkage (72 affected individuals, 22 males and 50 females). If individuals with non pVUR urinary tract defects were also considered as affected, all 16 families were informative (broad pVUR phenotype, 83 affected individuals, 28 males and 55 females).
Figure 1.
Pedigree structure of the 16 families studied. Asterisks (*) mark the individuals from whom DNA was available for the study. Patients with other urinary tract (UT) anomalies are indicated by a blackened rectangle within the symbol.
Interpretation of modes of inheritance in pVUR is complicated as a result of incomplete penetrance of the trait as well as spontaneous resolution of disease with increasing age. Examination of the pedigree structure revealed that seven families (44%) demonstrated parent-child transmission, suggestive of autosomal dominant inheritance. Absence of parent-child transmission in the remaining families was compatible either with dominant transmission with incomplete penetrance or recessive transmission of a high frequency gene. In the setting of genetic heterogeneity and uncertain mode of inheritance, parametric, LOD-based analysis under two simple models (dominant and recessive) is more powerful than allele sharing linkage methods.26–28 Therefore, our primary analysis was performed by computing heterogeneity LOD scores under both dominant and recessive models, using pVUR as our primary phenotype (12 informative families: K108, K110, K113, K116, K117, K118, K119, K120, K122, K138, K144, K146).
As with previous pVUR genome scans, analysis under genetic homogeneity did not identify any significant linkages under the dominant or recessive models.15–17 Under genetic heterogeneity and dominant transmission, the highest genome-wide heterogeneity LOD (HLOD) was on chromosome 8 (multipoint HLOD = 1.7, α = 0.6, nonparametric linkage [NPL] = 1.3, P = 0.05). However, after saturating this locus with 16 microsatellite markers, the HLOD decreased to zero. Combining microsatellite with SNP data has been shown to increase information content and improve the resolution of genome scans.25 Therefore, to exclude low marker informativeness as a cause of false negative results, we performed genome-wide SNP genotyping (Affymetrix 10K arrays) in 95 individuals in the most informative families. This analysis did not reveal novel loci across the genome, indicating that the absence of linkage under the dominant model is not due to lack of marker informativeness. Post hoc analysis of the seven pedigrees with parent-child transmission also did not reveal any promising signals. Hence, we found no significant or suggestive loci across the genome under the dominant model (supplemental Figure S1).
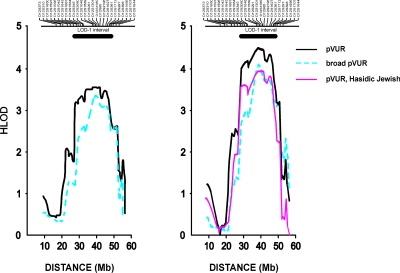
In contrast, heterogeneity analysis under the recessive model identified a single promising signal on chromosome 12 with an HLOD of 1.4 on pairwise analysis (D12S297, α = 0.64). Multipoint analysis augmented the HLOD to 2.7 across this interval (α = 0.65, peak at D12S1301). Importantly, this was the only peak with multipoint HLOD ≥2 across the entire genome under either the dominant or recessive model (Supplemental Figures S1 and S2). To confirm this finding, we performed high resolution mapping with 22 additional microsatellite markers and combined these with the SNP data, resulting in a mean intermarker distance of 0.77 Mb across the chromosome 12p11-q13 region. We observed positive pairwise HLODs across all markers within this interval, including one marker with genome-wide significance (D12S398, HLOD = 3.8, α = 0.82, Table 1).31 Next, multipoint analysis of linkage confirmed these results (multipoint HLOD 3.6 at marker D12S1048, α = 0.6, Figure 2A). This result exceeds traditional genome-wide significance thresholds.31,32 Furthermore, based on 1000 simulated genome-scans of the pedigrees under the assumption of no linkage, the empiric genome-wide significance thresholds for testing two models under genetic heterogeneity were 2.7 and 3.3 at P = 0.05 and 0.01, respectively. The HLOD of 3.6 therefore corresponds to an empiric genome-wide P value of 0.005, confirming genome-wide significance of findings by empiric criteria. The LOD-1 interval spans 22.7 Mb between markers rs1388659 and D12S361 and includes 161 genes. We next performed comparison of haplotypes across the chromosome 12 linkage regions to explore the possibility of a founder mutation, but found no evidence of shared segments among Hasidic or among Italian pedigrees (Supplemental Table S2). However, since these pedigrees were not closely related, the shared segments may be below the resolution of our typed markers.
Table 1.
Pairwise HLOD scores for Chr12 p11-q13
| Marker | Mb | pVUR Phenotype | Broad pVUR Phenotype |
|---|---|---|---|
| rs2054436 | 21.5 | 0.9 | 0.7 |
| rs725124 | 23.7 | 0.7 | 0.4 |
| D12S1591 | 24.0 | 2.2 | 1.2 |
| TSC54265 | 25.1 | 0.5 | 0.8 |
| D12S1596 | 25.8 | 1.5 | 1.2 |
| rs2343866 | 26.2 | 0.2 | 0.0 |
| rs1388659 | 27.1 | 0.4 | 0.3 |
| TA27A06P | 27.5 | 0.6 | 0.2 |
| D12S1643 | 29.2 | 1.4 | 0.7 |
| rs958478 | 29.3 | 0.1 | 0.0 |
| D12S1681 | 30.4 | 0.9 | 0.3 |
| D12S1584 | 31.6 | 0.7 | 0.1 |
| D12S345 | 33.0 | 2.6 | 1.9 |
| D12S2080 | 33.3 | 0.9 | 0.4 |
| rs1352123 | 37.4 | 0.6 | 0.9 |
| rs721709 | 39.0 | 1.6 | 1.4 |
| D12S1048 | 39.3 | 1.8 | 1.5 |
| rs2215456 | 40.6 | 1.6 | 0.9 |
| D12S1589 | 40.7 | 1.6 | 1.5 |
| D12S291 | 41.7 | 2.8 | 2.8 |
| rs721483 | 42.0 | 0.2 | 0.3 |
| TA91H06M | 42.4 | 0.8 | 0.5 |
| D12S1687 | 43.0 | 1.3 | 0.7 |
| rs1377002 | 43.2 | 0.3 | 0.2 |
| D12S85 | 45.6 | 0.3 | 0.2 |
| rs215389 | 46.1 | 1.3 | 1.4 |
| rs2254210 | 46.6 | 0.3 | 0.3 |
| D12S2196 | 47.0 | 1.3 | 1.2 |
| rs953673 | 47.4 | 1.4 | 1.1 |
| D12S1627 | 48.1 | 1.9 | 1.7 |
| rs4133070 | 48.4 | 1.6 | 1.8 |
| rs1316607 | 49.3 | 0.9 | 1.2 |
| D12S361 | 49.8 | 1.6 | 2.2 |
| D12S1712 | 50.7 | 2.0 | 1.9 |
| UT5029 | 50.9 | 1.3 | 1.4 |
| rs686339 | 51.3 | 0.1 | 0.1 |
| D12S398 | 51.5 | 3.8 | 3.1 |
| D12S1586 | 52.4 | 1.6 | 1.6 |
| D12S1707 | 53.3 | 1.0 | 1.2 |
| rs2371455 | 55.1 | 0.5 | 0.6 |
| D12S1644 | 55.8 | 1.0 | 0.4 |
Bolded numbers indicate HLOD scores >2. Broad phenotype includes individuals with pVUR or other urinary tract abnormalities.
Figure 2.
(A) HLOD plot of chromosome 12p11-q13 locus in the full cohort. The multipoint HLOD scores for the pVUR and broad pVUR phenotypes are shown on the y-axis. The x-axis denotes Mb distance based on the NCBI human physical map build 36.3. The location of the microsatellite markers genotyped is shown above the graph. The LOD-1 interval is indicated by the horizontal bar above the HLOD curve. (B) HLOD plot after post hoc exclusion of K117 and K122.
Alternative analyses were also performed to determine whether varying model parameters, analytic algorithm, or phenotype assignment impacted the chromosome 12 linkage results. Examination of pedigree LOD scores revealed that both Hasidic and nonHasidic pedigrees contributed to the linkage signal on chromosome 12. In addition, multipoint analysis in the six Hasidic Jewish pedigrees using marker allele frequencies from ethnicity matched controls revealed a peak HLOD of 2.6 (marker D12S291, α = 0.6). This further confirmed that most, but not all of the linkage signal originated from this subgroup (Table 2). Varying gene frequency (0.01 to 0.1) had negligible effects on overall linkage findings, with the best LOD scores achieved by modeling a high frequency risk allele (Table 3). Nonparametric analyses were also conducted and yielded a NPL score of 4.0 and P = 1 × 10−4 (at D12S1301), which falls just below the genome-wide significance threshold. Finally, to determine the impact of a broader phenotype assignment, we repeated the linkage analysis after including as affected all individuals with other renal and urologic clinical disorders. This expanded the cohort to 16 pedigrees and 83 affected individuals. Despite the broader phenotype, linkage to chromosome 12p11-q13 was confirmed, with a peak HLOD score of 3.4 (α = 0.5, Figure 2A). This locus remained the only suggestive or significant signal across the genome under either phenotype assignment scheme, with the next best multipoint HLOD score being 1.0 on chromosome 8. Altogether, these data demonstrate that the chromosome 12 linkage results were robust to varying analytic parameters and phenotype assignment criteria.
Table 2.
Peak multipoint LOD scores for each individual pedigree at the chromosome 12p11-q13
| Pedigree | Ethnicity | LODpVUR Phenotype | LODBroad Phenotype |
|---|---|---|---|
| 120 | Ashkenazi Jewish | 1.9 | 1.9 |
| 138 | Ashkenazi Jewish | 1.5 | 1.5 |
| 119 | Irish-American | 1.0 | 1.0 |
| 108 | Italian | 0.8 | 0.8 |
| 116 | Italian | 0.5 | -1.0 |
| 146 | Ashkenazi Jewish | 0.5 | 0.5 |
| 144 | Ashkenazi Jewish | 0.1 | 0.1 |
| 110 | Italian | -0.5 | -0.5 |
| 113 | Italian | -0.6 | -0.6 |
| 118 | Irish-American | -0.7 | -0.7 |
| 117 | Ashkenazi Jewish | -1.2 | -1.2 |
| 122 | Ashkenazi Jewish | -1.4 | -1.6 |
| 143 | Italian | N.I. | 0.6 |
| 140 | Italian | N.I. | 0.1 |
| 141 | Italian | N.I. | -0.1 |
| 102 | Italian | N.I. | -0.4 |
Broad phenotype includes individuals with pVUR or other urinary tract abnormalities. N.I., pedigree is not informative for pVUR phenotype.
Table 3.
Peak multipoint HLOD scores at chromosome 12p11-q13 with varying disease gene frequency
| Disease Gene Frequency | 0.01 | 0.05 | 0.10 |
|---|---|---|---|
| HLOD (α), pVUR phenotype | 3.4 (0.7) | 3.6 (0.6) | 3.4 (0.65) |
| HLOD (α), broad pVUR phenotype | 3.2 (0.6) | 3.4 (0.5) | 3.0 (0.5) |
Broad pVUR phenotype includes individuals with pVUR or other urinary tract abnormalities. The α value indicates the percent of families linked.
Finally, we scrutinized the genome in two large families (K117 and K122) that did not demonstrate linkage to chromosome 12 and were each large enough to achieve genome-wide significance (LOD >3). Remarkably, there was no evidence for linkage across the entire genome in either kindred under both dominant and recessive inheritance, even after incorporation of genome-wide SNPs into microsatellite data. The best signals were multipoint LOD scores of 1.3 with the dominant model on chromosome 8 (D8S592) for K117 and 1.2 with the recessive model for K122 on chromosomes 2 (SraP) and 10 (D10S1412), which are all below the suggestive significance threshold. Given the large size of these pedigrees and the comprehensive analyses performed across the genome, the absence of any linkage signals cannot be attributed to genetic heterogeneity or low power. These data suggest that problems such as complex structure or bilineal inheritance likely confound the analysis of linkage in these pedigrees. Because these two pedigrees did not map anywhere across the genome and consequently could only obscure linkage signals in our study, we performed post hoc analysis of the chromosome 12 locus after exclusion of these two complex kindreds (Figure 2B). This resulted in HLOD scores of 4.5 (α = 0.8) and 4.1 (α = 0.7) on chromosome 12p11-q13 for the pVUR and broad pVUR phenotypes, respectively (Figure 2B). Significantly, the remaining four Hasidic Jewish pedigrees demonstrate a LOD score of 3.9 with genetic homogeneity across the same region, indicating that most of the linkage signal originates from this subgroup (Figure 2B).
DISCUSSION
In the present study, we combined multiple approaches to overcome problems such as complex inheritance, incomplete penetrance, and genetic heterogeneity to localize pVUR susceptibility loci. Instead of studying sib pairs with nonparametric methods, we ascertained uniquely large families and analyzed the genome under both dominant and recessive transmission, because this approach avoids potential misspecification of the genetic model and maximizes power in the analysis of complex traits.26,29 The genome scan under the dominant model provided no signals across the entire genome. On the other hand, analysis under a recessive model localized a major susceptibility gene to an approximately 22-Mb interval on chromosome 12p11-q13, with a peak HLOD score of 3.6 (α = 0.6, NPL = 4.0, P = 1 × 10−4). This linkage peak was the only positive signal across the entire genome, exceeding genome-wide and empiric significance thresholds, and was not significantly changed by alternative analytic models or incorporation of a broader phenotype. These data emphasize the utility of parametric models for detection of major genes underlying complex traits.
The localization of a major gene with the recessive model may seem surprising because the literature has primarily focused on VUR segregating as a dominant trait.15,16,19,33–35 The absence of disease transmission across multiple generations has usually been attributed to incomplete penetrance or undetectable VUR due to spontaneous resolution in older individuals. However, given that many pVUR families consist of affected sib pairs, and considering the high degree of genetic heterogeneity of this trait, it is likely that genes with different modes of inheritance segregate among different pVUR pedigrees. The lack of parent-offspring transmission may therefore represent true recessive inheritance in some kindreds. Low penetrance recessive alleles imparting large effects have been implicated in several other complex traits such as patent ductus arteriosus, Hirschsprung disease, or autism.23,24,36 For these traits, gene localization was achieved by homozygosity mapping in consanguineous populations that were enriched for recessive disorders.23,24,36,37 Although we did not set out to study consanguineous populations, our cohort included six Hasidic Jewish kindreds, which contributed a significant proportion (but not all) of the linkage signal on chromosome 12 (Figure 2B). This population has many characteristics of a classic isolate, such as a limited set of founders, high endogamy, and recent expansion.38,39 In such a population, a common predisposing allele can achieve a high frequency due to founder effects, selection, or drift, enhancing the probability of recessive disorders.39,40 However, because several other pedigrees also contributed to the chromosome 12 linkage signal, these data suggest that recessive transmission applies to pVUR families of varying ethnicity.
The complexity of pVUR is further demonstrated by the lack of compelling signals across the genome in two high-density Hasidic Jewish kindreds (K117 and K122). These kindreds contained a total of 21 pVUR cases and were each predicted to be large enough to exceed LOD >3 under dominant (K117 and K122) or recessive (K117) models. Thus, the absence of linkage cannot be attributed to low power and genetic heterogeneity. Moreover, phenotyping error is unlikely, since all affecteds had VCUG-documented VUR. Such high-density pedigrees are commonly enriched for intrafamilial heterogeneity (the situation where affected individuals in a family have different risk alleles in the same gene or different genetic forms of disease).41 Therefore, the most likely alternative explanation is that these pedigrees have a complex hidden genealogical structure, such that risk alleles segregate across multiple lines of descent. This is a common phenomenon in population isolates, and the presence of all affected sibships in K117 would further support this possibility. Although intrafamilial heterogeneity reduces power in linkage analysis of complex traits, it does not produce false positive signals nor necessarily lead to false exclusion of linkage.41 The effects of such confounders can be mitigated by studying large cohorts and applying systematic analytic approaches, as demonstrated by the successful localization of a pVUR gene in our study.
There are 161 genes in the conservative LOD-1 interval on the chromosome 12 p11-q13 locus (National Center for Biotechnology Information [NCBI] Map Viewer). Among these, 19 genes have been implicated in human traits (OMIM). Based on a recently published study, 83 positional candidates have murine homologs and detectable expression in the murine metanephric mesenchyme and ureteric bud tip and stalk (Supplemental Table S3).42 These positional candidates may be pursued by systematic sequence analysis; however, it would be preferable to achieve further reduction of this locus through an interval-specific association study.43–45 Since the post hoc analysis indicated a strong linkage signal with homogeneity among the Hasidic Jewish families, association mapping may be especially suitable in this population because of its limited number of founders.38,39,46,47 Furthermore, one would expect enrichment for homozygous segments surrounding susceptibility genes among pVUR cases from this population. We did not detect regions of homozygosity within the chromosome 12 locus in the Hasidic Jewish patients, but this may be due to the lack of close consanguinity among families studied. Therefore, a higher resolution analysis may be required to detect autozygous segments in these kindreds.
These findings have many important implications for future genetic studies of pVUR. Because the chromosome 12 signal originated from pedigrees of varying ethnicity, recessive transmission may be applicable to pVUR kindreds from many different populations. Thus, in addition to genetic heterogeneity, variable modes of transmission should be considered in all pVUR linkage scans, and analysis under recessive transmission is henceforth warranted. It is also worth noting that sporadic disease cannot be differentiated from recessive transmission in the absence of an affected family member or consanguinity. Consequently, the recessive contribution to pVUR may have been underestimated in nonfamilial cases as well. If recessive transmission accounts for a significant fraction of sporadic pVUR, this offers a potentially powerful setting for a genome-wide association study. As demonstrated by recent studies of age-related macular degeneration, modest sized case/control cohorts may be quite successful in such situations.48 Thus, association scans in sporadic pVUR may offer an additional promising approach for resolving the genetic basis of this trait.
CONCISE METHODS
Patients and Phenotypes
All families were ascertained through an index case diagnosed with primary VUR by a VCUG. Index cases and family members had no evidence of secondary causes of VUR or syndromic abnormalities. In addition, we conducted extensive family history interviews and searched medical records to identify family members diagnosed with pVUR documented by VCUG or any clinical or urologic abnormalities that may be pathogenically related to pVUR (e.g., other urologic anomalies or a diagnosis of ESRD in absence of any obvious cause such as glomerulonephritis and/or diabetes mellitus). This led to the identification of 56 relatives with pVUR and 11 relatives with other renal or urologic abnormalities (Supplemental Table S1). For linkage analysis, we considered all individuals with pVUR diagnosed with VCUG as affected, leaving all others as phenotype unknown. Subsequently, we performed linkage analysis by also including the 11 additional family members with clinically related abnormalities in the affected cohort. Because VUR is known to resolve with age, we classified individuals that did not undergo a VCUG and those with a negative VCUG as phenotype unknown in all analyses (affected only analysis). All individuals gave informed consent, and the study protocol adhered to the Declaration of Helsinki and was approved by the Western Institutional Review Board for Columbia University and the ethics committees at the University of Brescia and at the Gaslini Institute.
Genotyping
Total genomic DNA was isolated from peripheral white blood cells of the patients and relatives using standard procedures. We performed genome-wide scans using both microsatellites and SNPs. The microsatellite scan was performed with 393 microsatellites (intermarker distance approximately 10 cM) genotyped across the genome in all 200 individuals (Marshfield Mammalian Genotyping Service). To maximize inheritance information across the genome, we also typed 95 individuals (65 affecteds) in the most complex families with 10,204 SNPs using the GeneChips Mapping 10K 2.0 Arrays (Affymetrix, Santa Clara, California). DNA processing and gene-chip hybridization were performed as suggested by the manufacturer. We fine-mapped two loci suggestive of linkage on chromosome 8 and chromosome 12 (with 16 and 22 polymorphic microsatellite markers, respectively). Integration of the most informative SNPs from 10K GeneChip data (minor allele frequency [MAF] ≥0.2) with the fine mapping microsatellites on the chromosome 12 locus yielded an average marker spacing of 0.77 Mb and average information content of 0.9 (standard deviation = 0.1).
Analysis of Linkage
We performed pairwise and multipoint analyses of linkage, using FASTLINK4.1,49 and SimWalk2 2.90,50 respectively. Since pVUR is known to be genetically heterogeneous, we computed parametric LOD scores under a dominant and a recessive model, with disease gene frequencies of 0.01 and 0.05, respectively. We used penetrance of 75% and phenocopy of 0.01 for both models (since affected only analysis was performed, penetrance parameters did not affect LOD statistics). For comparison, we concurrently computed nonparametric statistics with the SimWalk2 program (NPLpairs score and associated exact P value). We calculated allele frequencies on the basis of the frequencies observed in the dataset for the genome-wide microsatellites, whereas for the SNP data, we based frequencies on Caucasian allele frequencies provided by Affymetrix. In the fine mapping experiments, we obtained control allele frequencies from 40 Ashkenazi Jewish individuals. We used published thresholds for significant linkage under heterogeneity (LOD = 3.3).31,32 Moreover, we estimated empiric thresholds of significance by performing 1000 genome scans with microsatellites spaced every 10 cM across the genome under the hypothesis of no linkage, using the same structure as our pedigrees, and performing pairwise genome scans under the dominant and recessive models described above.
DISCLOSURES
None.
Supplementary Material
Acknowledgments
We thank all of the patients and their families for participating in the study. This study was supported by National Institutes of Health 1R21 DK073903–01. Genome-wide STR genotyping was performed by the Mammalian Genotyping Service at the Marshfield clinic (NO1-HV-48141). P.L.W. is supported by a National Kidney Foundation Clinical Research Fellowship grant and National Institute of Child Health and Human Development HD052890. S.S.C. is supported by the Telethon Grant GFP05012. We are grateful to J.D. Terwilliger and David Greenberg for their insightful comments. We would also like to thank Jonathan Barasch and Kai Schmidt-Ott for the mouse gene expression data. This study was presented in part at the 2008 annual meeting of the American Society of Nephrology.
The URLs for data presented herein are as follows:
NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview (build 36.3).
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (vesicoureteral reflux).
NCBI HomoloGene, http://www.ncbi.nlm.nih.gov/homologene (release 63)
Published online ahead of print. Publication date available at www.jasn.org.
P.L.W. and S.S.C. contributed equally to this paper
Supplemental information for this article is avialable online at http://www.jasn.org/.
REFERENCES
- 1.Ardissino G, Dacco V, Testa S, Bonaudo R, Claris-Appiani A, Taioli E, Marra G, Edefonti A, Sereni F: Epidemiology of chronic renal failure in children: data from the ItalKid project. Pediatrics 111: e382–e387, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA: Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant 11: 366–373, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C: A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet 15: 157–164, 1997 [DOI] [PubMed] [Google Scholar]
- 4.Hoskins BE, Cramer CH, Silvius D, Zou D, Raymond RM, Orten DJ, Kimberling WJ, Smith RJ, Weil D, Petit C, Otto EA, Xu PX, Hildebrandt F: Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am J Hum Genet 80: 800–804, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F: SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A 101: 8090–8095, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont ME, Sullivan MJ, Dobyns WB, Eccles MR: Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet 9: 358–364, 1995 [DOI] [PubMed] [Google Scholar]
- 7.Hinchliffe SA, Chan YF, Jones H, Chan N, Kreczy A, van Velzen D: Renal hypoplasia and postnatally acquired cortical loss in children with vesicoureteral reflux. Pediatr Nephrol 6: 439–444, 1992 [DOI] [PubMed] [Google Scholar]
- 8.Karnak I, Woo LL, Shah SN, Sirajuddin A, Kay R, Ross JH: Prenatally detected ureteropelvic junction obstruction: clinical features and associated urologic abnormalities. Pediatr Surg Int 24: 395–402, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Kenda RB, Fettich JJ: Vesicoureteric reflux and renal scars in asymptomatic siblings of children with reflux. Arch Dis Child 67: 506–508, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noe HN: The long-term results of prospective sibling reflux screening. J Urol 148: 1739–1742, 1992 [DOI] [PubMed] [Google Scholar]
- 11.Tobenkin MI: Hereditary vesicoureteral reflux. South Med J 57: 139–147, 1964 [DOI] [PubMed] [Google Scholar]
- 12.Kaefer M, Curran M, Treves ST, Bauer S, Hendren WH, Peters CA, Atala A, Diamond D, Retik A: Sibling vesicoureteral reflux in multiple gestation births. Pediatrics 105: 800–804, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Noe HN, Wyatt RJ, Peeden JN, Jr, Rivas ML: The transmission of vesicoureteral reflux from parent to child. J Urol 148: 1869–1871, 1992 [DOI] [PubMed] [Google Scholar]
- 14.Scott JE, Swallow V, Coulthard MG, Lambert HJ, Lee RE: Screening of newborn babies for familial ureteric reflux. Lancet 350: 396–400, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Feather SA, Malcolm S, Woolf AS, Wright V, Blaydon D, Reid CJ, Flinter FA, Proesmans W, Devriendt K, Carter J, Warwicker P, Goodship TH, Goodship JA: Primary, nonsyndromic vesicoureteric reflux and its nephropathy is genetically heterogeneous, with a locus on chromosome 1. Am J Hum Genet 66: 1420–1425, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly H, Molony CM, Darlow JM, Pirker ME, Yoneda A, Green AJ, Puri P, Barton DE: A genome-wide scan for genes involved in primary vesicoureteric reflux. J Med Genet 44: 710–717, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte ML, Bertoli-Avella AM, de Graaf BM, Punzo F, Lama G, La Manna A, Grassia C, Rambaldi PF, Oostra BA, Perrotta S: A genome search for primary vesicoureteral reflux shows further evidence for genetic heterogeneity. Pediatr Nephrol 23: 587–595, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu W, van Eerde AM, Fan X, Quintero-Rivera F, Kulkarni S, Ferguson H, Kim HG, Fan Y, Xi Q, Li QG, Sanlaville D, Andrews W, Sundaresan V, Bi W, Yan J, Giltay JC, Wijmenga C, de Jong TP, Feather SA, Woolf AS, Rao Y, Lupski JR, Eccles MR, Quade BJ, Gusella JF, Morton CC, Maas RL: Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am J Hum Genet 80: 616–632, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertoli-Avella AM, Conte ML, Punzo F, de Graaf BM, Lama G, La Manna A, Polito C, Grassia C, Nobili B, Rambaldi PF, Oostra BA, Perrotta S: ROBO2 gene variants are associated with familial vesicoureteral reflux. J Am Soc Nephrol 19: 825–831, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman CJ, Bailey RR, Janus ED, Abbott, G.D, Lynn, K.L: Vesicoureteric reflux: segregation analysis. Am J Med Genet 20: 577–584, 1985 [DOI] [PubMed] [Google Scholar]
- 21.de Vargas A, Evans K, Ransley P, Rosenberg AR, Rothwell D, Sherwood T, Williams DI, Barratt TM, Carter CO: A family study of vesicoureteric reflux. J Med Genet 15: 85–96, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried K, Yuval E, Eidelman, A, Beer S: Familial primary vesicoureteral reflux. Clin Genet 7: 144–147, 1975 [DOI] [PubMed] [Google Scholar]
- 23.Mani A, Meraji SM, Houshyar R, Radhakrishnan J, Ahangar M, Rezaie TM, Taghavinejad MA, Broumand B, Zhao H, Nelson-Williams C, Lifton RP: Finding genetic contributions to sporadic disease: a recessive locus at 12q24 commonly contributes to patent ductus arteriosus. Proc Natl Acad Sci U S A 99: 15054–15059, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, Al-Saad S, Ware J, Joseph RM, Greenblatt R, Gleason D, Ertelt JA, Apse KA, Bodell A, Partlow JN, Barry B, Yao H, Markianos K, Ferland RJ, Greenberg ME, Walsh CA: Identifying autism loci and genes by tracing recent shared ancestry. Science 321: 218–223, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN: Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility Loci. Am J Hum Genet 75: 948–965, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodge SE, Abreu PC, Greenberg DA: Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60: 217–227, 1997 [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg DA, Hodge SE: Linkage analysis under ‘random’ and ‘genetic’ reduced penetrance. Genet Epidemiol 6: 259–264, 1989 [DOI] [PubMed] [Google Scholar]
- 28.Greenberg DA: Linkage analysis assuming a single-locus mode of inheritance for traits determined by two loci: inferring mode of inheritance and estimating penetrance. Genet Epidemiol 7: 467–479, 1990 [DOI] [PubMed] [Google Scholar]
- 29.Abreu PC, Greenberg DA, Hodge SE: Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet 65: 847–857, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida J, Tsuchiya M, Tatsuma N, Murakami M: Mass screening for early detection of congenital kidney and urinary tract abnormalities in infancy. Pediatr Int 45: 142–149, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247, 1995 [DOI] [PubMed] [Google Scholar]
- 32.Faraway JJ: Distribution of the admixture test for the detection of linkage under heterogeneity. Genet Epidemiol 10: 75–83, 1993 [DOI] [PubMed] [Google Scholar]
- 33.Devriendt K, Groenen P, Van Esch H, van Dijck M, Van de Ven W, Fryns JP, Proesmans W: Vesico-ureteral reflux: a genetic condition? Eur J Pediatr 157: 265–271, 1998 [DOI] [PubMed] [Google Scholar]
- 34.Lewy PR, Belman AB: Familial occurrence of nonobstructive, noninfectious vesicoureteral reflux with renal scarring. J Pediatr 86: 851–856, 1975 [DOI] [PubMed] [Google Scholar]
- 35.Sanna-Cherchi S, Reese A, Hensle T, Caridi G, Izzi C, Kim YY, Konka A, Murer L, Scolari F, Ravazzolo R, Ghiggeri GM, Gharavi AG: Familial vesicoureteral reflux: testing replication of linkage in seven new multigenerational kindreds. J Am Soc Nephrol 16: 1781–1787, 2005 [DOI] [PubMed] [Google Scholar]
- 36.Puffenberger EG, Kauffman ER, Bolk S, Matise TC, Washington SS, Angrist M, Weissenbach J, Garver KL, Mascari M, Ladda R, Slaugenhaupt SA, Chakravarti A: Identity-by-descent and association mapping of a recessive gene for Hirschsprung disease on human chromosome 13q22. Hum Mol Genet 3: 1217–1225, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Lander ES, Botstein D: Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236: 1567–1570, 1987 [DOI] [PubMed] [Google Scholar]
- 38.Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B: Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci U S A 97: 6769–6774, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostrer H: A genetic profile of contemporary Jewish populations. Nat Rev Genet 2: 891–898, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Risch N, Tang H, Katzenstein H, Ekstein J: Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet 72: 812–822, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durner M, Greenberg DA, Hodge SE: Inter- and intrafamilial heterogeneity: effective sampling strategies and comparison of analysis methods. Am J Hum Genet 51: 859–870, 1992 [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, Li JY, Lu B, Costantini F, Schiffer M, Bottinger E, Barasch J: Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol 16: 1993–2002, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA: Complement factor H polymorphism and age-related macular degeneration. Science 308: 421–424, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA: Complement factor H variant increases the risk of age-related macular degeneration. Science 308: 419–421, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006 [DOI] [PubMed]
- 46.Bronstein M, Pisante A, Yakir B, Darvasi A: Type 2 diabetes susceptibility loci in the Ashkenazi Jewish population. Hum Genet 124: 101–104, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Friedrichsen DM, Stanford JL, Isaacs SD, Janer M, Chang BL, Deutsch K, Gillanders E, Kolb S, Wiley KE, Badzioch MD, Zheng SL, Walsh PC, Jarvik GP, Hood L, Trent JM, Isaacs WB, Ostrander EA, Xu J: Identification of a prostate cancer susceptibility locus on chromosome 7q11-21 in Jewish families. Proc Natl Acad Sci U S A 101: 1939–1944, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J: Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cottingham RW, Jr, Idury RM, Schaffer AA: Faster sequential genetic linkage computations. Am J Hum Genet 53: 252–263, 1993 [PMC free article] [PubMed] [Google Scholar]
- 50.Sobel E, Lange K: Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58: 1323–1337, 1996 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.