Abstract
The α1- and β1-subunits of human soluble guanylate cyclase (sGC) were coexpressed in the Sf9 cells/baculovirus system. In addition to the native enzyme, constructs with hexahistidine tag at the amino and carboxyl termini of each subunit were coexpressed. This permitted the rapid and efficient purification of active recombinant enzyme on a nickel-affinity column. The enzyme has one heme per heterodimer and was readily activated with the NO donor sodium nitroprusside or 3-(5′-hydroxymethyl-2′furyl)-1-benzyl-indazole (YC-1). Sodium nitroprusside and YC-1 treatment potentiated each other in combination and demonstrated a remarkable 2,200-fold stimulation of the human recombinant sGC. The effects were inhibited with 1H-(1,2,4)oxadiazole(4,3-a)quinoxalin-1one (ODQ). The kinetics of the recombinant enzyme with respect to GTP was examined. The products of the reaction, cGMP and pyrophosphate, inhibited the enzyme. The extent of inhibition by cGMP depended on the activation state of the enzyme, whereas inhibition by pyrophosphate was not affected by the enzyme state. Both reaction products displayed independent binding and cooperativity with respect to enzyme inhibition. The expression of large quantities of active enzyme will facilitate structural characterization of the protein.
Keywords: nitric oxide, cGMP
The signaling molecule NO plays an important role in many biological processes (1, 2). To mediate these effects, NO binds to and regulates several cellular targets. The best-characterized target for the signal transduction of NO is the heme-containing enzyme soluble guanylyl cyclase (sGC) (3, 4). sGC catalyzes the conversion of GTP to cGMP and pyrophosphate (PPi) and functions as a receptor for NO. Synthesized cGMP, in turn, regulates various effector proteins, such as protein kinases, phosphodiesterases, and ion channels.
sGC is a heterodimer, composed of 73- to 82-kDa α- and 70-kDa β-subunits (5, 6), which contains one heme prosthetic group per heterodimer (7). At least two isoforms for each subunit have been cloned from mammals (8–12), and sGC homologues were identified in other species, including medaka fish (13), insects Drosophila melanogaster (14), Anopheles sp. (15), and the nematode Caenorhabditis elegans (GenBank accession no. AF235027). The basal activity of sGC enzyme is markedly stimulated (up to 400-fold) upon NO interaction with the heme prosthetic group (16, 17). Carbon monoxide has also been shown to weakly stimulate the enzyme (18, 19). Both NO- and CO-dependent stimulation of sGC can be enhanced by the allosteric activator 3-(5′-hydroxymethyl-2′furyl)-1-benzyl-indazole (YC-1) (20). YC-1 alone can also stimulate the enzyme in a NO-independent manner. The exact mechanism of YC-1-dependent stimulation is not completely understood and is currently a subject of intense investigation (21–23).
sGC has an enormous clinical significance. sGC plays a critical role in smooth muscle contractility, platelet reactivity, central and peripheral neurotransmission, as well as other effects (3, 4, 24). The enzyme also plays a key role in the generation of the NO-induced hypotension in septic shock (25, 26). Despite such clinical importance, the biochemical properties of the human sGC are not completely characterized. The current knowledge of sGC properties, with a few exceptions, is based on studies of bovine and rat sGC isoforms. This does not reflect the absence of interest in human sGC, but the technical difficulties associated with it. The widely used inhibitors 1H-(1,2,4)oxadiazole(4,3-a)quinoxalin-1one (ODQ), LY-83583, and methylene blue affect the heme prosthetic group not only of sGC but other hemoproteins. Unfortunately, no specific sGC inhibitors are known. A thorough characterization of human sGC isoforms will be beneficial for the development of specific sGC regulatory compounds (activators and inhibitors) and their further application to clinical medicine.
In this report, we adapted the insect Sf9 cells/baculovirus system for overexpression of relatively large quantities of an enzymatically active human sGC protein. Purification and characterization of human recombinant sGC and its inhibition by reaction products under various conditions are reported.
Materials and Methods
cDNA Isolation and Baculovirus Construction.
The human sGC α1 and β1 subunits coding sequences were PCR-amplified from a gene pool of cDNAs from human cerebral cortex (Invitrogen) by using primers corresponding to the first 18 and last 22 bp of the known coding sequences of α1- and β1-subunits (GenBank accession nos. Y15723 and X66534, respectively). The obtained PCR fragments were inserted into the EcoRI site of pAcHLT-B or pVL1392 baculovirus transfer vectors (PharMingen) and used to generate the recombinant virus by using BaculoGold DNA according to the manufacturer's protocol (PharMingen). The recombinant viruses named αN-sGC and βN-sGC are based on the pAcHLT-B vector and express sGC subunits carrying a 5-kDa N-terminal tag consisting of hexahistidine and a thrombin cleavage site. Recombinant baculoviruses expressing the untagged subunits were named α-sGC and β-sGC, respectively. To generate the αC-sGC virus expressing the α-subunit with a C-terminal tag, a hexahistidine tag was inserted in-frame by PCR in front of the stop codon of the α1-subunit coding sequenced cloned in pVL1392 vector. High-titer virus stocks were obtained from Sf9 cells cultured in monolayer and infected at a multiplicity of infection (moi) <1 and harvested 7 days postinfection.
Sf9 Cell Culture and Production of Recombinant sGC.
Sf9 cells were cultured in SF900II medium supplemented with 10% (vol/vol) FCS, 1% (vol/vol) Pluronic 68, and 1% of penicillin and streptomycin at 27°C. Spinner cultures (2 × 106/ml) were cotransfected with a moi ≈5. To generate human recombinant sGC enzymes carrying various tags, Sf9 cells were cotransfected with α1- and β1-expressing viruses. For example, to generate αNβ-sGC enzyme, Sf9 cells were cotransfected with αN-sGC and β-sGC viruses.
Purification of sGC.
sGC-expressing Sf9 cells were harvested 3 days postinfection, washed in PBS, and suspended in lysis buffer (50 mM of phosphate buffer at pH 7.2, 100 mM NaCl, 10% glycerol, 1 mM PMSF, and 5 μg/ml each of pepstatin A, leupeptin, aprotinin, and chymostatin). Cells were disrupted by sonication and centrifuged at 100,000 × g for 60 min at 4°C. The high-speed supernatant fraction was slowly passed over a 5-ml column of His-Bind resin (Novagen) and followed by 10 bed volume washes each with the lysis buffer and the lysis buffer with 50 mM imidazole. The enzyme was eluted with 5 bed volumes of 150 mM imidazole in lysis buffer. Fractions of 2 ml each were collected and 5 mM DTT and 1 mM EDTA were added to each fraction. The sGC-containing fractions were pooled and used for studies. The enzyme was 95–98% pure as determined by SDS/PAGE (see below). Protein concentration was determined by the Bradford method (27).
Western Blotting.
The proteins were resolved by 7.5% SDS/PAGE (28) and visualized by Coomassie blue R250 or silver staining of the gel. For Western blot analysis, proteins were transferred to nitrocellulose membranes and the 6His-tagged sGC subunits were detected by anti-6His antibody (Invitrogen). Hexahistidine-tagged and untagged subunits were also detected by antibodies against human α- or β-subunits obtained in these studies as follows: High-speed pellets from Sf9 cells expressing the subunits individually were washed three times by resuspension and centrifugation in PBS and resolved on a preparative 7.5% polyacrylamide gel. The α- and β-subunits were excised from the gel and were used to produce rabbit polyclonal antibodies.
sGC Activity Assay.
sGC activity was measured by the formation of [32P]cGMP from [α-32P]GTP as described (29). A total of 200 μM of GTP, Mg2+ as cofactor, and sodium nitroprusside (SNP) as NO-donor were used in all experiments unless otherwise stated. CO-dependent activation of sGC was measured with reaction mixtures saturated with CO or argon in gas-tight tubes with rubber stoppers. The concentration of GTP, cGMP, and PPi was varied as indicated in the reaction when the GTP-Km and Ki for the end products were determined.
Spectroscopic Studies.
UV/visible spectra between 360 and 750 nm were recorded by using a Shimadzu-2501 PC equipped with a TCC temperature controller. Heme concentration in the sample was measured by the pyridine-hemochromagen assay (30). Briefly, 15% (vol/vol) pyridine and 150 mM NaOH were added to the sample, and the spectrum was recorded between 350 and 650 nm. Heme content was determined based on the λ557 value of 33.4 mM−1·cm−1 for the reduced pyridine hemochromagen (30).
Results
Coexpression of α1 and β1 in Sf9 Cells.
Recombinant human sGC was generated by cotransfection of Sf9 cells with recombinant viruses expressing α- and β-subunits. An equal molar expression of the two subunits and a high basal/stimulating activity were achieved by adding 1:1 ratio of α- and β-containing virus at a moi of 5 (data not shown). Maximal protein expression was observed at 72 h after infection of the Sf9 cells with the recombinant viruses.
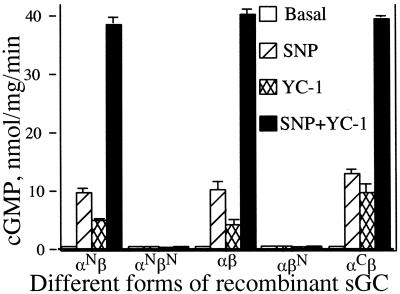
We tested the activity in crude lysates of expressed sGC containing the hexahistidine tag at amino or carboxyl termini of α- and β-subunits. Although the basal activity of sGC was similar for all recombinant proteins, the stimulated activity of these enzymes differed depending on the position of the hexahistidine tag (Fig. 1). The native (αβ) as well as the enzymes carrying the tag only on the α-subunit (αNβ, αCβ) were stimulated by NO and the YC-1 activator. However, the enzymes carrying the tag on the N terminus of the β-subunit (αβN and αNβN) displayed basal activity, but were not stimulated by NO or by YC-1 (Fig. 1). A similar pattern was observed when Mn2+ rather then Mg2+ was used as a cofactor in the reaction (data not shown). Because both αNβ and αCβ displayed properties similar to the native enzyme, they were chosen for the expression, purification, and characterization.
Figure 1.
sGC activity in the cytosol of Sf9 cells expressing different forms of sGC. Activities for the αβ, αNβ, αCβ, αβN, and αNβN recombinant enzymes were assayed with Mg2+ as a cofactor in the absence or presence of SNP (100 μM) and/or YC-1 (100 μM). Data are mean ± SD of three independent experiments.
Purification of Human Recombinant sGC.
Purification of the αNβ and αCβ enzymes by Ni2+ affinity chromatography was performed as described in Materials and Methods. The results of a representative purification of human αCβ sGC are summarized in Table 1. The difference in the recoveries of basal and SNP enzymes (Table 1) reflect the separation of heme-containing and non-heme enzyme with imidazole elution from the Ni2+-column (data not shown). Both αNβ and αCβ human recombinant sGC enzymes were purified to apparent homogeneity by Ni2+-affinity chromatography as judged by SDS/PAGE and silver staining (Fig. 2 Inset). The purified enzyme had the expected molecular mass of ≈155 kDa (native enzyme plus the tag) as determined by gel filtration (data not shown). The purified enzyme displayed a basal specific activity of 5.6 nmol/min per mg and a NO-stimulated specific activity of 3,245 nmol/min per mg (a 580-fold activation by SNP). These values are well in the 1–20 μmol/min per mg range of specific activities reported earlier for native activated enzyme from other species (see ref. 31 for review).
Table 1.
Purification of sGC expressed in a baculovirus/Sf9 system
| Purification Step | Protein, mg | Specific
activity, nmol/min per mg
|
Total activity,
nmol/min
|
Purification, -fold
|
Recovery,
%
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Basal | SNP | Basal | SNP | Basal | SNP | Basal | SNP | ||
| Lysate | 308 | 0.32 | 9.75 | 98.6 | 3003 | 1 | 1 | 100 | 100 |
| Ni2+ | 0.62 | 5.6 | 3245 | 3.5 | 2012 | 17.5 | 332 | 3.5 | 67 |
Sf9 insect cell lysate was prepared from 2 × 109 cells cotransfected with baculoviruses carrying αC- and β-subunits. Similar yields and specific activities were observed when αN- and β-baculoviruses were used for transfection or when other preparations were examined.
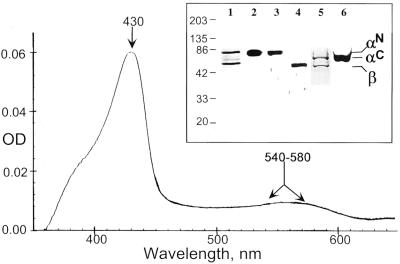
Figure 2.
Analysis of purified sGC protein. Spectral analysis of the purified human sGC enzyme displayed a typical Soret band (430 nm) and the broad α/β band (540–580 nm) indicated by arrows. (Inset) A total of 250 ng of purified recombinant αNβ (lanes 1–3) and αCβ (lanes 4–6) sGC was separated by 7.5% SDS/PAGE and analyzed by silver staining (lanes 1 and 5) or by Western blotting. The identity of the upper bands as α-subunit was confirmed by anti-hexahistidine antibody (lanes 2 and 6) and validated by antibodies to α-sGC subunit (lane 3). The identity of the lower band as β-subunit was confirmed by antibodies raised against human β-sGC subunit (lane 4). The different mobility of αN and αC subunits reflects the difference in the size of the tag. The sizes of the molecular mass markers are indicated in kDa.
Basal sGC activities of both the lysate and the purified enzyme were higher when assayed with Mn2+ as a cofactor than when Mg2+ was used, whereas the SNP and/or YC-1 stimulated activities were lower in the presence of Mn2+ (data not shown). These observations are in good agreement with the properties of a native functional sGC with a full complement of heme (32).
Characterization of Human Recombinant sGC.
Purified recombinant human sGC showed a Soret peak at 431 nm and a broad α/β band around 540–580 nm (Fig. 2). The Soret peak shifted to 399 nm upon stimulation with 1 mM of the NO-donor (2-(N,N-diethylamino)-diaznenolate-2-oxide (DEA/NO), and to 420 nm when the sample was saturated with CO gas. YC-1 did not have any effect on the spectra of basal or stimulated sGC. The heme/sGC ratio was estimated by measuring the heme amounts in the purified enzyme by the pyridine-hemochromagen assay. We found 0.9 equivalents of heme per sGC heterodimer. These observations confirm earlier findings (7, 19, 33) and demonstrate that the recombinant human sGC purified in these studies is a fully functional sGC.
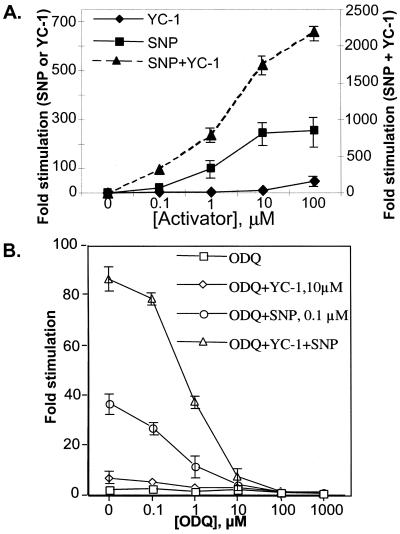
The effects of the NO-donor SNP, of the allosteric sGC activator YC-1, and of the inhibitor ODQ on αNβ and αCβ sGC enzymes were also tested (Fig. 3). SNP stimulated the enzyme in a dose-dependent manner with a maximal ≈300-fold stimulation at 100 μM and an EC50 of 2 μM (Fig. 3A). The allosteric stimulator YC-1 alone has a 50-fold stimulating effect at 100 μM (Fig. 3A). Moreover, the combined treatment of human sGC with various YC-1 amounts in the presence of 100 μM SNP demonstrated a synergistic activation over the whole range of YC-1 concentration (from 0.1 to 100 μM). The combination of 100 μM of SNP and 100 μM YC-1 resulted in a remarkable 2,200-fold stimulation of the enzyme. Under these activating conditions, the specific activity of purified recombinant human sGC is approximately 22 μmol/min per mg.
Figure 3.
Regulation of sGC activity by various compounds. (A) Synergistic effects of SNP and YC-1. The activity of the purified recombinant sGC was measured upon stimulation with various concentration of SNP (■) or YC-1 (⧫). The synergistic effect of various concentrations of YC-1 on 100 μM SNP-dependent activation of sGC was also analyzed (▴). Mg2+ was used as a cofactor. Data are expressed as -fold stimulation of activated over basal activity. Data are presented as mean of three independent experiments performed in triplicates ± SD for αNβ enzyme. Similar activation levels were obtained with αCβ enzyme. Note that the left scale corresponds to SNP or YC-1 stimulation, whereas the right scale corresponds to SNP + YC-1 stimulation. (B) ODQ-dependent inhibition of the human sGC. sGC activity was measured in the absence or presence of increasing concentration of ODQ. The enzyme was not treated (□) or stimulated by 0.1 μM SNP (○), 10 μM YC-1 (◊), or combined treatment of 0.1 μM SNP and 10 μM YC-1 (▵). Mg2+ was used as a cofactor. Data are presented as mean of three independent experiments performed in triplicates ± SD for αNβ enzyme. Identical inhibition was observed with αCβ enzyme. Some error bars fall within the symbols.
ODQ has been identified previously as an inhibitor of sGC (34, 35). We investigated the inhibitory effects of ODQ on SNP- and/or YC-1-stimulated human sGC. ODQ showed a concentration-dependent inhibition of sGC stimulated by either SNP (0.1 μM) or YC-1 (10 μM). Moreover, ODQ also inhibited the human enzyme activated by the combination of SNP (0.1 μM) and YC-1 (10 μM) with an IC50 of ≈0.8 μM (Fig. 3B). The combined SNP/YC-1 stimulation was completely inhibited by 100 μM of ODQ (Fig. 3B). A similar IC50 (1 μM) for ODQ was determined with rat and bovine sGC enzymes (32, 35).
Carbon monoxide has been shown to stimulate sGC activity, and this stimulation was markedly potentiated by YC-1 (20). The effects of CO and CO/YC-1 together on the activity of human sGC are summarized in Table 2. The samples were saturated either with CO gas or with argon as a control. CO was a poor activator of sGC and only stimulated the enzyme 4-fold over the basal activity. The addition of YC-1 to CO-treated enzyme markedly enhanced sGC activity up to 78-fold at 10 μM of YC-1 and 221-fold at 100 μM YC-1. These results are in good agreement with previous findings (20).
Table 2.
Stimulation of sGC with CO and YC-1
| Conditions | Specific activity, nmol/min per mg | Stimulation, -fold |
|---|---|---|
| Basal | 4.4 ± 0.5 | 1 |
| CO | 15.7 ± 0.7 | 4 |
| CO + YC-1 (10 μM) | 347 ± 32 | 78 (22.1) |
| CO + YC-1 (100 μM) | 979 ± 34 | 221 (62.3) |
All experiments were performed on samples saturated with CO (or argon for basal measurements) as described in Materials and Methods. Mg2+ was used as cofactor. The values for the specific activity are means (±SD) of three independent experiments performed for αNβ. αCβ enzyme showed a similar activity and stimulation (data not shown). Values in parentheses represent -fold activation by YC-1 over the CO stimulation.
Kinetic Comparison of sGC with Various Stimulatory Conditions.
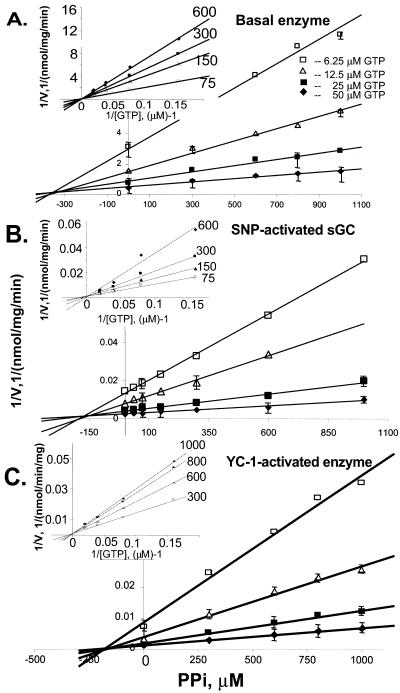
To probe the nature of the changes that occur in the active center of sGC after stimulation with various activators, we tested several kinetic characteristics of the enzyme. We initially measured the effect of sGC activators on the Km of Mg2+-GTP. We found the Km for GTP with purified basal enzyme was 65 ± 5 μM. Upon activation by SNP and YC-1, the Km for GTP slightly decreased to 47 ± 6 μM and 35 ± 4 μM, respectively. The values were similar for both αNβ and αCβ enzymes.
We next studied the inhibition of αNβ and αCβ enzymes by the reaction products PPi and cGMP under different stimulatory conditions. The product inhibition was measured for basal, SNP-, and YC-1-stimulated αNβ and αCβ enzymes under various concentrations of PPi and GTP. Both reaction products exhibited inhibitory properties, but their potency was different and depended on the state of the enzyme. PPi inhibited αNβ and αCβ sGC enzymes in the basal state, and SNP or YC-1 stimulated states with comparable efficiency. The Dixon plots of PPi inhibition for αCβ enzyme are presented in Fig. 4. The transformation of the same data (Fig. 4 Insets) revealed that the double-reciprocal plots (1/activity vs. 1/substrate concentration) at various concentrations of PPi indicated a competitive type of inhibition for PPi with basal, SNP-activated, or YC-1-activated enzyme. The calculated Ki value for PPi presented in Table 3 was in the range of 175 to 300 μM and is slightly higher for the basal enzyme and almost identical for the YC-1- and SNP-stimulated enzyme.
Figure 4.
Inhibition of sGC activity by PPi. The PPi inhibition was measured for the basal (A), SNP-stimulated (B), and YC-1-activated (C) human sGC. The activity was measured at various concentrations of PPi and GTP. The data are represented as Dixon plot (1/velocity vs. concentration of the inhibitor). The Ki for PPi was determined as the projection to the abscissa at the intersection point of the plots. The replotting of the data as 1/velocity vs. 1/[GTP]. (Insets) Indicated competitive type of inhibition by PPi. The concentrations of PPi are indicated for each plot (μM). The plots are representative of three independent measurements for both αNβ and αCβ enzymes with similar results. The plotted data are means of triplicate value ± SD. Some error bars fall within the symbols.
Table 3.
Inhibition of sGC by PPi and cGMP
| PPi
|
cGMP, IC50, mM
|
|||
|---|---|---|---|---|
| Enzyme state | Ki, μM | Inhibition type | αNβ | αCβ |
| Basal | 300 ± 25 | Competitive | >50 | >50 |
| YC-1 stimulated | 180 ± 20 | Competitive | 13 | >100 |
| SNP-stimulated | 175 ± 18 | Competitive | 6 | 12 |
Ki values are represented as the average (±SD) of three independent experiments for αNβ enzyme. αCβ enzymes demonstrated similar results. A total of 100 μM YC-1 or SNP was used to stimulate the enzymes.
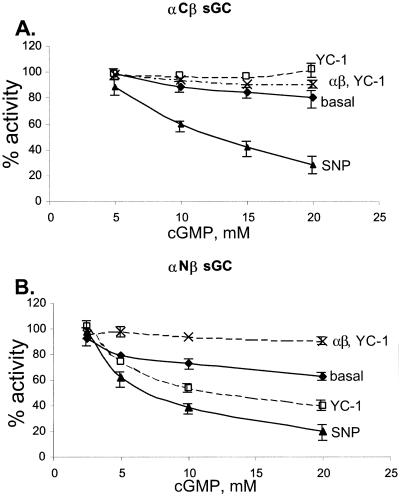
On the other hand, cGMP was a poor inhibitor of the enzyme. Measurable cGMP-dependent inhibition was observed only at millimolar concentrations of cGMP. The magnitude of inhibition depended on the enzyme's state. The basal αNβ and αCβ enzymes displayed a measurable inhibition only at cGMP concentrations higher than 5 mM (Fig. 5). At 20 mM cGMP concentration, both basal enzymes were inhibited by 30% and 20%, respectively. Both enzymes were inhibited to a similar extent by cGMP in the SNP-stimulated state, displaying only 20–30% activity in the presence of 20 mM cGMP. The IC50 values for cGMP are presented in Table 3. However, when cGMP inhibition of YC-1-stimulated enzyme was analyzed, we observed a significant difference between the enzymes. We found that the αCβ enzyme activated by YC-1 was not inhibited by cGMP even at 20 mM concentration, whereas the αNβ enzyme was more susceptible to cGMP inhibition and was inhibited by 60% at this concentration of cGMP. Despite this difference between the αNβ and αCβ variants of sGC, both enzymes were inhibited less by cGMP in the YC-1-induced conformation than in the SNP-induced conformation. When partially purified untagged αβ enzyme stimulated by YC-1 was tested, less than 10% inhibition was observed with 20 mM cGMP (Fig. 5), which is similar to the finding with the αCβ enzyme. These data suggested that YC-1, when compared with SNP, induced a conformational change in the enzyme with a decreased affinity for cGMP. SNP induced a change in the conformation of the basal enzyme that rendered it more susceptible to cGMP inhibition.
Figure 5.
Inhibition of sGC activity by cGMP. cGMP inhibition of αCβ (A) and αNβ (B) sGC was measured for basal (⧫), SNP-stimulated (▴), and YC-1-activated (□, dotted lines) enzymes. The inhibition of partially purified nontagged enzyme is also plotted for reference (crosses, dashed line). The plots are representative of three independent measurements for each of the recombinant enzymes. The plotted data are means of triplicate value ± SD. Some error bars fall within the symbols.
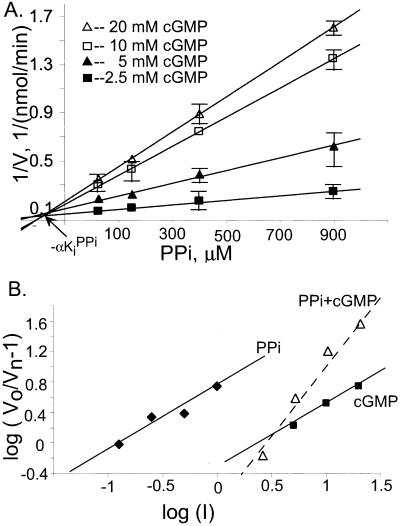
Next, we investigated the combined effect of PPi and cGMP as inhibitors. We monitored the sGC activity in the presence of various concentrations of PPi (0–1,000 μM) and cGMP (0–20 mM) at a constant concentration of GTP (50 μM). The plotting of the data according to Yonetani and Theorell (36) presented in Fig. 6A indicated that the apparent Ki for PPi (the intersection with the abscissa) decreases with the increased concentration of cGMP, suggesting cooperativity with respect to their inhibition. We also calculated the fractional velocities Vn/Vo, where Vo is the velocity of the uninhibited reaction and Vn are the velocities at various concentrations of inhibitors. These calculations were used to generate a median-effect plot for PPi (Fig. 6B, ♦), cGMP (Fig. 6B, ▪), and combined PPi/cGMP inhibition (Fig. 6B, ▵, dashed line), according to the method of Chou and Talalay (37). We found that, in this case, the PPi and cGMP plots were parallel with very similar slopes of m = 0.82 and m = 0.86 for PPi and cGMP, respectively. The slope values m are close to 1, indicating that both inhibitors follow similar first-order kinetics. In the case of combined inhibition, only data points with a constant PPi/GMP concentration ratio (1:20) were used (37). The plot of the combined PPi/cGMP inhibition is not parallel to the PPi or cGMP plots, and the m value of the slope increased to 1.9. These data indicate that cGMP and PPi are competitive inhibitors whose binding are not mutually exclusive.
Figure 6.
Combined inhibition by cGMP and PPi. (A) Data were plotted according to Yonetani-Theorell (36). The activity of SNP-stimulated human sGC was measured at 50 μM of GTP and various concentrations of PPi (abscissa) and cGMP (2.5 mM, 5 mM, 10 mM, and 20 mM, as indicated). The plots are representative of two independent experiments for recombinant enzymes. The plotted data are means of triplicate value ± SD. Some error bars fall within the symbols. (B) Data were plotted by the method of Chou-Talalay (37). The activity of SNP-stimulated human sGC was measured at 50 μM of GTP and various concentrations of PPi and cGMP. Inhibition by PPi alone (⧫) or cGMP alone (■) is compared with combined PPi/cGMP inhibition at PPi/GMP molar ratio of 1:20 (▵). The abscissa is log([PPi]) (⧫), log([cGMP]) (■), or log([PPi] + [cGMP]) (▵).
Discussion
The analysis of the primary sequence of the α- and β-subunits of sGC indicated that both NH2 and COOH termini of these subunits are not conserved and vary in length when compared with sGC from other species. This suggested that the additional 36 amino acids that contain the hexahistidine tag, the protein kinase A, and the thrombin cleavage sites at the amino termini of α- and β-subunits should not significantly affect the activity of the enzyme. However, we found that this tag completely abolished the sGC responsiveness to NO and YC-1, when the NH2 terminus of the β-subunit was tagged, whereas basal activity was not affected. A similar effect was observed by Wedel and coworkers (38) after 64 amino acids from the NH2 terminus of the β-subunit were deleted. Although in both cases the His-105 residue of the β-subunit responsible for the coordination of heme iron is intact, the deletion of 64 or the addition of 36 residues induces conformational changes in the heme binding pocket and modifies the enzyme's responsiveness to NO. Wedel and coworkers also deleted 131 amino acids from the N-terminal portion of the α-subunit and found that the sGC enzyme carrying that deletion also had an impaired ability to be activated (38). However, in our hands, the addition of the tag to the NH2 terminus of α-subunit did not affect the properties of the enzyme, suggesting that the amino terminus of the α-subunit has a greater structural flexibility than the β-subunit. Zabel and coworkers (39) fused a considerably larger glutathione S-transferase-tag on the amino terminus of the α-subunit. This resulted in a functional enzyme that was stimulated only 22-fold by NO vs. 300- to 600-fold stimulation observed in our studies. Despite the high specific activity and NO responsiveness of the αNβ enzyme, we decided to generate an additional enzyme carrying only a minimal hexahistidine tag on the COOH terminus of the α-subunit.
The analysis of both αNβ and αCβ demonstrated that these recombinant enzymes are fully functional and display the characteristics specific for a native enzyme. The recombinant enzymes contained a 0.9:1 heme/enzyme ratio, had a proper Soret band (Fig. 2), were activated by NO, CO, and/or YC-1 (Figs. 1 and 3, and Tables 1 and 2), and displayed the expected shifts in the heme spectra upon NO or CO treatments (data not shown). The YC-1 stimulator not only activated the enzymes alone, but also synergized with NO and CO for both αNβ and αCβ enzymes. Although no significant differences between αNβ and αCβ in these experiments were observed, we performed the kinetic analyses of the sGC under various conditions for both recombinant proteins.
The catalytic rate of sGC increases several hundred fold as a result of NO binding to the heme prosthetic group. The catalytic rate of the enzyme is also increased upon treatment with YC-1, which acts at a yet unknown allosteric site. Although YC-1 sensitizes the enzyme to either NO or CO, it also can activate alone. The catalytic properties of sGC are changed as a result of conformational changes in the enzyme's active center, which were induced by NO binding to heme or YC-1 binding to its allosteric site. We decided to test by means of kinetic analyses of human recombinant sGC whether the NO or YC-1 binding results in identical or different active conformations. First, we tested the Km for the natural substrate (Mg-GTP) of the enzyme in basal, YC-1-, and SNP- induced conformational states. We found that the Km for the substrate was slightly higher for the basal enzyme than for conformations induced by NO or YC-1. We observed modest changes in the Km value after the enzyme stimulation (65 μM for basal, 47 μM for SNP, and 35 μM for YC-1 enzyme), which is similar with earlier studies (7). Others, however, found 3- to 4-fold greater Km values for basal enzyme and similar Km values for NO- and YC-1-stimulated bovine enzyme (40). These differences are most probably because of species differences of the enzyme preparation used. The analysis of the PPi-dependent inhibition demonstrated that for all three conformations PPi exhibited a competitive type of inhibition with similar 200–300 μM Ki values for all three states, indicating a similar mode of interaction at the PPi-binding site. However cGMP, the other product of the reaction, was a poor inhibitor of sGC. Basal enzyme has a distinctly lower affinity to cGMP with an IC50 in the 30–40 mM range, as estimated by the extrapolation of the inhibitory curves (Fig. 5 and Table 3). Our measurements indicated that SNP-stimulated enzyme was most susceptible to cGMP inhibition, although even in this case the IC50 was in the 6–10 mM range (Fig. 5, ▴, and Table 3). At 20 mM cGMP, the enzyme still retained ≈20% of its activity. The most interesting finding was that YC-1-stimulated enzyme displayed a cGMP-dependent inhibition different from both basal and SNP-stimulated enzymes. Although the cGMP-inhibition was different for αNβ and αCβ, in both cases the enzymes had a lower affinity to cGMP in the YC-1-induced conformation than in the SNP-induced state (Fig. 5, □). YC-1 stimulated αCβ was not inhibited by cGMP in the tested range of cGMP concentrations, whereas the αNβ enzyme was inhibited by 60%. When partially purified untagged YC-1-stimulated αβ enzyme was tested for comparison (Fig. 5 A and B, crosses) we found an absence of inhibition similar to the αCβ enzyme. The differences between αCβ and αNβ most probably reflect the perturbation induced by the 36 residues of the N-terminal tag near a YC-1 binding site. Because the YC-1 stimulation requires for its function the presence of the prosthetic group assumed to be in the NH2-terminal half of the enzyme (41), we propose that it is the presence of the N-terminal tag on the α-subunit that affected the αNβ enzyme. These data indirectly suggest the involvement of the N-terminal portion of α-subunit in the YC-1 binding.
We also tested the mode of interaction of PPi and cGMP with the sGC by measuring the simultaneous inhibition of sGC by PPi and cGMP. The analysis of the data according to Yonetani and Theorell (36) indicated a cooperative inactivation of sGC when both reaction products are present (Fig. 6A). The Chou-Talalay method revealed that cGMP and PPi act as mutually nonexclusive inhibitors (Fig. 6B), suggesting that their binding sites are not overlapping. Both compounds followed first-order kinetics, indicating that only one molecule of PPi or cGMP is bound per sGC heterodimer. The cooperative inhibition of sGC by both reaction products suggests that a reverse reaction might occur, as was demonstrated in similar conditions for adenylyl cyclase (42).
Although sGC was identified more than 25 years ago (43) and its physiological and clinical importance is well established, no specific inhibitors of the enzyme have been developed. Although ODQ, methylene blue, and LY-83583 are used in many reports as sGC inhibitors, none of these agents are specific inhibitors. The compounds also affect other hemoproteins (44). With very few exceptions (45), no studies on sGC inhibition by substrate analogs have been performed. We believe that the present report will initiate the search for a specific sGC inhibitor that will have utility in many biological experiments and perhaps be useful clinically.
Acknowledgments
We thank Dr. Pei-Feng Chen, Dr. Ah-Lim Tsai, and Dr. Vladimir Berka for helpful discussions. Research support was provided by the John S. Dunn Foundation, the Harold and Leila Y. Mathers Foundation, the Welch Foundation, National Space Biomedical Research Institute, U.S. Army Medical Research, and the University of Texas.
Abbreviations
- sGC
soluble guanylyl cyclase
- SNP
sodium nitroprusside
- YC-1
3-(5′-hydroxymethyl-2′furyl)-1-benzyl-indazole
- ODQ
1H-(1,2,4)oxadiazole(4,3-a)quinoxalin-1one
- moi
multiplicity of infection
- PPi
pyrophosphate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190333697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190333697
References
- 1.Ignarro L J, Cirino G, Casini A, Napoli C. J Cardiovasc Pharmacol. 1999;34:879–886. doi: 10.1097/00005344-199912000-00016. [DOI] [PubMed] [Google Scholar]
- 2.Lane P, Gross S S. Semin Nephrol. 1999;19:215–229. [PubMed] [Google Scholar]
- 3.Murad F. Adv Pharmacol. 1994;26:1–335. [PubMed] [Google Scholar]
- 4.Martin E, Davis K, Bian K, Lee Y C, Murad F. Semin Perinatol. 2000;24:2–6. doi: 10.1016/s0146-0005(00)80045-2. [DOI] [PubMed] [Google Scholar]
- 5.Garbers D L. J Biol Chem. 1979;254:240–243. [PubMed] [Google Scholar]
- 6.Kamisaki Y, Saheki S, Nakane M, Palmieri J A, Kuno T, Chang B Y, Waldman S A, Murad F. J Biol Chem. 1986;261:7236–7241. [PubMed] [Google Scholar]
- 7.Gerzer R, Bohme E, Hofmann F, Schultz G. FEBS Lett. 1981;132:71–74. doi: 10.1016/0014-5793(81)80429-2. [DOI] [PubMed] [Google Scholar]
- 8.Giuili G, Scholl U, Bulle F, Guellaen G. FEBS Lett. 1992;304:83–88. doi: 10.1016/0014-5793(92)80594-7. [DOI] [PubMed] [Google Scholar]
- 9.Koesling D, Herz J, Gausepohl H, Niroomand F, Hinsch K D, Mulsch A, Bohme E, Schultz G, Frank R. FEBS Lett. 1988;239:29–34. doi: 10.1016/0014-5793(88)80539-8. [DOI] [PubMed] [Google Scholar]
- 10.Koesling D, Harteneck C, Humbert P, Bosserhoff A, Frank R, Schultz G, Bohme E. FEBS Lett. 1990;266:128–132. doi: 10.1016/0014-5793(90)81523-q. [DOI] [PubMed] [Google Scholar]
- 11.Nakane M, Saheki S, Kuno T, Ishii K, Murad F. Biochem Biophys Res Commun. 1988;157:1139–1147. doi: 10.1016/s0006-291x(88)80992-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakane M, Arai K, Saheki S, Kuno T, Buechler W, Murad F. J Biol Chem. 1990;265:16841–16845. [PubMed] [Google Scholar]
- 13.Mikami T, Kusakabe T, Suzuki N. Eur J Biochem. 1998;253:42–48. doi: 10.1046/j.1432-1327.1998.2530042.x. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa S, Miyamoto I, Aruga J, Furuichi T, Okano H, Mikoshiba K. J Neurochem. 1993;60:1570–1573. doi: 10.1111/j.1471-4159.1993.tb03324.x. [DOI] [PubMed] [Google Scholar]
- 15.Caccone A, Garcia B A, Mathiopoulos K D, Min G S, Moriyama E N, Powell J R. Insect Mol Biol. 1999;8:23–30. doi: 10.1046/j.1365-2583.1999.810023.x. [DOI] [PubMed] [Google Scholar]
- 16.Humbert P, Niroomand F, Fischer G, Mayer B, Koesling D, Hinsch K D, Gausepohl H, Frank R, Schultz G, Bohme E. Eur J Biochem. 1990;190:273–278. doi: 10.1111/j.1432-1033.1990.tb15572.x. [DOI] [PubMed] [Google Scholar]
- 17.Katsuki S, Arnold W, Mittal C, Murad F. J Cyclic Nucleotide Res. 1977;3:23–35. [PubMed] [Google Scholar]
- 18.Brune B, Schmidt K U, Ullrich V. Eur J Biochem. 1990;192:683–688. doi: 10.1111/j.1432-1033.1990.tb19276.x. [DOI] [PubMed] [Google Scholar]
- 19.Stone J R, Marletta M A. Biochemistry. 1994;33:5636–5640. doi: 10.1021/bi00184a036. [DOI] [PubMed] [Google Scholar]
- 20.Friebe A, Schultz G, Koesling D. EMBO J. 1996;15:6863–6868. [PMC free article] [PubMed] [Google Scholar]
- 21.Friebe A, Koesling D. Mol Pharmacol. 1998;53:123–127. doi: 10.1124/mol.53.1.123. [DOI] [PubMed] [Google Scholar]
- 22.Sharma V S, Magde D, Kharitonov V G, Koesling D. Biochem Biophys Res Commun. 1999;254:188–191. doi: 10.1006/bbrc.1998.9812. [DOI] [PubMed] [Google Scholar]
- 23.Stone J R, Marletta M A. Chem Biol. 1998;5:255–261. doi: 10.1016/s1074-5521(98)90618-4. [DOI] [PubMed] [Google Scholar]
- 24.Denninger J W, Marletta M A. Biochim Biophys Acta. 1999;1411:334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A, Brar R, Wang P, Dee L, Skorupa G, Khadour F, Schulz R, Parrillo J E. Am J Physiol. 1999;276:R265–R276. doi: 10.1152/ajpregu.1999.276.1.R265. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A, Haery C, Parrillo J E. Crit Care Clin. 2000;16:251–287. doi: 10.1016/s0749-0704(05)70110-x. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Schultz G, Bohme E. In: Methods of Enzymatic Analysis. Bergmeyer H U, Bergmeyer J, Grasl M, editors. Weinheim, Germany: Verlag Chemie; 1984. pp. 379–389. [Google Scholar]
- 30.de Duve C. Acta Chem Scand. 1948;2:264–289. doi: 10.3891/acta.chem.scand.02-0264. [DOI] [PubMed] [Google Scholar]
- 31.Koesling D. Methods. 1999;19:485–493. doi: 10.1006/meth.1999.0891. [DOI] [PubMed] [Google Scholar]
- 32.Hoenicka M, Becker E M, Apeler H, Sirichoke T, Schroder H, Gerzer R, Stasch J P. J Mol Med. 1999;77:14–23. doi: 10.1007/s001090050292. [DOI] [PubMed] [Google Scholar]
- 33.Stone J R, Marletta M A. Biochemistry. 1996;35:1093–1099. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 34.Garthwaite J, Southam E, Boulton C L, Nielsen E B, Schmidt K, Mayer B. Mol Pharmacol. 1995;48:184–188. [PubMed] [Google Scholar]
- 35.Schrammel A, Behrends S, Schmidt K, Koesling D, Mayer B. Mol Pharmacol. 1996;50:1–5. [PubMed] [Google Scholar]
- 36.Yonetani T, Theorell H. Arch Biochim Biophys. 1964;106:243–251. doi: 10.1016/0003-9861(64)90184-5. [DOI] [PubMed] [Google Scholar]
- 37.Chou T C, Talalay P. Eur J Biochem. 1981;115:207–216. doi: 10.1111/j.1432-1033.1981.tb06218.x. [DOI] [PubMed] [Google Scholar]
- 38.Wedel B, Harteneck C, Foerster J, Friebe A, Schultz G, Koesling D. J Biol Chem. 1995;270:24871–24875. doi: 10.1074/jbc.270.42.24871. [DOI] [PubMed] [Google Scholar]
- 39.Zabel U, Hausler C, Weeger M, Schmidt H H. J Biol Chem. 1999;274:18149–18152. doi: 10.1074/jbc.274.26.18149. [DOI] [PubMed] [Google Scholar]
- 40.Denninger J W, Schelvis J P, Brandish P E, Zhao Y, Babcock G T, Marletta M A. Biochemistry. 2000;39:4191–4198. doi: 10.1021/bi992332q. [DOI] [PubMed] [Google Scholar]
- 41.Hobbs A J. Trends Pharmacol Sci. 1997;18:484–491. doi: 10.1016/s0165-6147(97)01137-1. [DOI] [PubMed] [Google Scholar]
- 42.Dessauer C W, Gilman A G. J Biol Chem. 1997;272:27787–27795. doi: 10.1074/jbc.272.44.27787. [DOI] [PubMed] [Google Scholar]
- 43.Kimura H, Murad F. Metabolism. 1975;24:439–445. doi: 10.1016/0026-0495(75)90123-7. [DOI] [PubMed] [Google Scholar]
- 44.Feelisch M, Kotsonis P, Siebe J, Clement B, Schmidt H H. Mol Pharmacol. 1999;56:243–253. doi: 10.1124/mol.56.2.243. [DOI] [PubMed] [Google Scholar]
- 45.Brandwein H J, Lewicki J A, Waldman S A, Murad F. J Biol Chem. 1982;257:1309–1311. [PubMed] [Google Scholar]