Abstract
The MMACHC gene product of the cblC complementation group, referred to as the cblC protein, catalyzes the in vitro and in vivo decyanation of cyanocobalamin (vitamin B12). We hypothesized that the cblC protein would also catalyze the dealkylation of newly internalized methylcobalamin (MeCbl) and 5′-deoxyadenosylcobalamin (AdoCbl), the naturally occurring alkylcobalamins that are present in the diet. The hypothesis was tested in cultured endothelial cells using [57Co]-AdoCbl and MeCbl analogs consisting of [57Co]-labeled straight-chain alkylcobalamins ranging from C2 (ethylcobalamin) to C6 (hexylcobalamin). [57Co]-AdoCbl was converted to [57Co]-MeCbl by cultured bovine aortic endothelial cells, suggesting that a dealkylation process likely involving the cblC protein removed the 5′-deoxyadenosyl alkyl group. Surprisingly, all of the straight-chain alkylcobalamins served as substrates for the biosynthesis of both AdoCbl and MeCbl. Dealkylation was then assessed in normal skin fibroblasts and fibroblasts derived from 3 patients with mutations in the MMACHC gene. While normal skin fibroblasts readily converted [57Co]-propylcobalamin to [57Co]-AdoCbl and [57Co]-MeCbl, there was little or no conversion in cblC mutant fibroblasts. These studies suggest that the CblC protein is responsible for early processing of both CNCbl (decyanation) and alkylcobalamins (dealkylation) in mammalian cells.
Keywords: MMACHC gene product, cblC protein, vitamin B12, adenosylcobalamin, alkylcobalamin, dealkylation, cobalamin, endothelial cells, skin fibroblasts, cblC complementation group
Introduction
Cobalamin (Cbl) is an essential micronutrient required by all cells in the body. Adenosylcobalamin (AdoCbl) and methylcobalamin (MeCbl) serve as coenzymes for methylmalonyl-CoA mutase (EC 5.4.99.2) and methionine synthase (EC 2.1.1.13), respectively. Insufficient dietary intake of Cbl, malabsorption, defective transport, or impaired intracellular processing and coenzyme biosynthesis can lead to clinical cobalamin deficiency [1]. Accumulating evidence suggests that the intracellular processing of dietary cobalamins and cyanocobalamin (CNCbl, vitamin B12) precedes the biosynthesis of AdoCbl and MeCbl [2–6]. At least one early processing step is dependent on the MMACHC gene product (hereafter referred to as the cblC protein), which is defective in patients with cobalamin disorders belonging to the cblC complementation group.
The cblC complementation group, first described in a patient 4 decades ago by Harvey Mudd and colleagues [7], contains the largest number of inherited defects of cobalamin metabolism (OMIM 277400). Patients with the cblC defect usually present with combined homocystinuria and methylmalonic aciduria, suggesting impaired methionine synthase and methylmalonyl-CoA mutase activities, respectively. Lerner-Ellis et al. recently identified the gene in the cblC locus on chromosome region 1p using homozygosity mapping and haplotype analyses and named it MMACHC for “methylmalonic aciduria type C and homocystinuria” (Gene ID 25974) [5]. Cultured skin fibroblasts from patients with mutations in the MMACHC gene are unable to utilize CNCbl for the biosynthesis of AdoCbl and MeCbl [8, 9]. Recently, Kim et al. discovered that the cblC protein catalyzes the reductive decyanation of CNCbl [6]. Because the alkylcobalamins MeCbl and AdoCbl are major cobalamin forms found in mammalian tissues, plasma and milk [10–12], we hypothesized that the cblC protein would also catalyze the dealkylation of newly internalized (dietary) alkylcobalamins. Herein, we provide strong evidence that: 1) aortic endothelial cells possess the required machinery to synthesize AdoCbl and MeCbl from both natural and xenobiotic alkylcobalamins; 2) the processing machinery responsible for dealkylation reactions displays broad substrate specificity; and, 3) patient fibroblasts with mutations in the MMACHC gene are unable to perform dealkylation reactions. This is consistent with a role for the involvement of the cblC protein in dealkylation processing of newly absorbed alkylcobalamins.
Materials and Methods
Synthesis and purification of ethylcobalamin (EtCbl), propylcobalamin (PrCbl), butylcobalamin (BuCbl), pentylcobalamin (PnCbl) and hexylcobalamin (HxCbl)
Xenobiotic alkylcobalamins were synthesized by the reaction of cob(I)alamin with the corresponding alkylhalide [13, 14] and purified by HPLC as described in the Supplementary Material (see Fig. S1, Supplementary Material). [57Co]-Alkylcobalamins were synthesized as described above using [57Co]-CNCbl (MP Biomedicals, Solon, OH) as the starting material (specific activity: 379 μCi/μg). 5′-Chloro-5′-deoxyadenosine, synthesized as described by Jacobsen et al. [12], was used to synthesize [57Co]-AdoCbl.
Cell culture lines and [57Co]-cobalamin metabolic labeling
Bovine aortic endothelial cells (BAEC) were cultured in 162 cm3 flasks (Corning) and grown in vitamin B12-free, folic acid-free Ham’s F12/DME (1:1) medium supplemented with 5% FBS, 2.0 mM L-glutamine, penicillin (100 units/mL), streptomycin (0.1 mg/ml) and 50 nM (6S)-N5-methyltetrahydrofolic acid (Eprova AG). The amount of cobalamin present in the 5% FBS-supplemented culture medium (33 pM) was shown to be sufficient to support normal growth of BAEC. Normal and cblC mutant fibroblasts were grown in Advanced DMEM (Gibco) culture medium supplemented with 10% FBS (final cobalamin concentration, 66 pM). For [57Co]-Cbl metabolic labeling experiments, cells were passaged at a ratio of 1:2. [57Co]-CNCbl was added to achieve a final concentration of 0.2 nM, and cells were grown to 100% confluency (~48 h).
cblC patient cell lines
Dr. David Watkins, McGill University, kindly provided human cblC mutant skin fibroblasts from patients with severe disease (WG1801, WG2176 and WG3354). The Repository for Mutant Human Cell Strains, Montreal Children’s Hospital, Montreal, Canada (http://www.cellbank.mcgill.ca/) provided patient information on the cblC lines. Patient WG1801 was a 2 month-old male of Turkish ethnicity, son of first cousins, with two brothers who were possible carriers of the inborn error. Patient WG2176 was a 7-month old male of Hong Kong Chinese ethnicity with a healthy older sister and an affected fetal brother who was aborted. Patient WG3354 was a female of Pakistani ethnicity with both parents and younger siblings heterozygous for the cblC mutation.
Assessment of dealkylation in BAEC and human fibroblasts
Cells were passaged at a ratio of 1:2 in medium containing 0.125 nM (specific activity: 379 mCi/mg) of the desired [57Co]-cobalamin. After 48 h, cells were harvested, total cobalamins extracted with 80% aqueous ethanol and the intracellular cobalamin profile determined as recently described by Hannibal et al. [15]. The cell cultures were protected from light at all times to prevent photolysis of the alkylcobalamins.
Stability of [57Co]-alkylcobalamins in the culture medium
Conditioned medium (1 ml) from 48-h old cultures was extracted with a 1:1 mixture of phenol/chloroform, taken to dryness in a Speedvac, reconstituted with 0.4 ml of phosphate-buffered saline (PBS). Cobalamin standards were added to the sample, the mixture was filtered (0.22 μ filter) and analyzed by HPLC as previously described [15]. Workup of the conditioned culture medium was conducted under dim-red light.
Extraction and analysis of intracellular cobalamins
Confluent cells were harvested by trypsinization and washed three times with Dulbecco’s PBS. Extraction of cobalamins from cell pellets was performed as previously described [15]. Extracted [57Co]-cobalamins were mixed with unlabeled cobalamin standards and separated by gradient reverse-phase HPLC using an Agilent 1100 System equipped with a Zorbax SB C-18 column (4.6 × 250 mm, 5 μm particle size, Agilent) as previously described [15]. Elution was monitored with UV detection at 254 nm. In typical runs, 60 fractions were collected. The radioactivity associated with each fraction was counted using a gamma-counter (Gamma 4000 Beckman-Coulter).
Cobalamin uptake studies
Cells were seeded at an initial density of ~50% and allowed to grow for 24 h. After 24 h, half of the conditioned culture medium was replaced with fresh medium and [57Co]-CNCbl added to a final concentration of 0.2 nM (specific activity: 379 μCi/mg). Uptake was followed by counting the radioactivity in a γ-counter at 6, 12, 24, 48 and 72 h, both in spent medium and in washed cell pellets. Total cobalamin values were normalized to cellular protein concentration.
Cytotoxicity
To rule out cytotoxic effects of the xenobiotic alkylcobalamins under our culture conditions, BAEC were grown in the presence of 1 μM of each of the xenobiotic alkylcobalamins (supraphysiological concentration) as the major source of cobalamin for two weeks, with medium plus fresh alkylcobalamin changes every three days. Morphological changes were monitored by phase-contrast microscopy. To assess for cobalamin deficiency, total homocysteine and methylmalonic acid concentrations were determined in the conditioned culture medium. Cell number and cell viability were determined by hemocytometry and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [16], respectively. Cellular senescence was assessed by determining b-galactosidase activity using a commercial kit (Sigma).
Biochemical analyses
Total homocysteine in conditioned culture medium was determined by the method of Jacobsen et al. using monobromobimane and HPLC with fluorescence detection [17]. Values were normalized to cellular protein concentration. The concentration of methylmalonic acid in conditioned culture medium was determined by gas chromatography and mass spectrometry (GC/MS) in the Department of Clinical Pathology, Cleveland Clinic by a method modified from Hoffmann et al [18]. Total protein concentration was determined by the bicinchoninic acid assay (Thermo Scientific) using bovine serum albumin as a standard.
Results
Cobalamin uptake and cofactor biosynthesis by BAEC
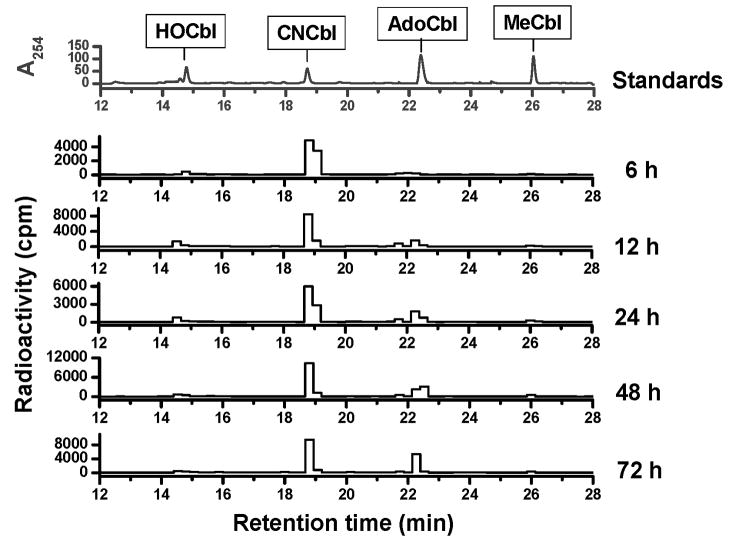
Because little is known about B12 metabolism in cardiovascular cells and tissues, the ability of BAEC to internalize [57Co]-CNCbl and synthesize [57Co]-AdoCbl and [57Co]-MeCbl was first determined. [57Co]-CNCbl was added to pre-confluent cells in culture and total cobalamin uptake was measured at 6, 12, 24, 48 and 72 h in washed cell pellets. Data for uptake of [57Co]-CNCbl by BAEC are shown in Table 1. Maximum uptake occurs at ~48 h of growth under the culture conditions described above. For each time point, the intracellular cobalamin profile was also examined and the results for cofactor biosynthesis are summarized in Table 1. The kinetics of biosynthesis of the active cofactors, AdoCbl and MeCbl is shown in Fig. 1. Although AdoCbl appeared to be the most abundant cobalamin form at all times, the ratio AdoCbl/MeCbl varied over time. BAEC also utilized [57Co]-hydroxocobalamin ([57Co]-HOCbl) and [57Co]-AdoCbl as substrates for cofactor biosynthesis (Fig. S2, Supplementary Material). These results are summarized in Table 2. Thus, endothelial cells possess the machinery for the efficient conversion of the natural cofactor AdoCbl into MeCbl as well as for utilizing HOCbl and CNCbl as substrates for cofactor biosynthesis (see Table 2).
Table 1.
Uptake and processing of [57Co]-CNCbl by BAEC.
| Time (b) | [57Co]-Cbl uptake1 (fmol/mg protein) | [57Co]-Cofactor biosynthesis2 (fmol/mg protein) | ||
|---|---|---|---|---|
| Conditioned medium | Intracellular | AdoCbl | MeCbl | |
| 6 | 10.5 | 0.36 | 0.006 ± 0.002 | 0.003 ± 0.001 |
| 12 | 8.8 | 0.44 | 0.035 ± 0.005 | 0.007 ± 0.002 |
| 24 | 8.4 | 0.54 | 0.044 ± 0.014 | 0.008 ± 0.001 |
| 48 | 8.3 | 0.59 | 0.090 ± 0.021 | 0.012 ± 0.003 |
| 72 | 8.5 | 0.60 | 0.113 ± 0.016 | 0.008 ± 0.002 |
Uptake results represent three pooled samples per time point.
Cofactor biosynthesis is expressed as mean ± standard deviation (n=3)
Fig. 1. Processing [57Co]-CNCbl in BAEC.
Kinetics of cobalamin biosynthesis in BAEC for 6, 12, 24, 48 and 72 h. BAEC were grown in the presence of [57Co]-CNCbl (0.2 nM final concentration; 0.1 μCi/ml culture medium) as the cobalamin source. Absorbance at 254 nm for cobalamin standards is shown in the upper chromatogram. Radioactivity for [57Co]-cobalamins at 6, 12, 24, 48 and 72 h is shown in the bottom chromatograms.
Table 2.
Processing of natural xenobiotic cobalamins by BAEC1.
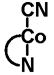
| Source | Intracellular cobalamin (%) | Ratio AdoCbl/MeCbl |
||||
|---|---|---|---|---|---|---|
| AdoCbl | MeCbl | HOCbl | CNCbl | Others2 | ||
|
|
58.1 | 4.5 | 20.4 | <1 | 17.0 | 12.9 |
|
33.6 | 7.5 | 3.7 | 47.1 | 8.1 | 4.5 |
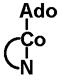
|
57.1 | 8.6 | 10.2 | <1 | 23.1 | 6.6 |
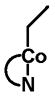
|
59.1 | 5.5 | 15.4 | <1 | 20.0 | 10.7 |
|
|
32.7 | 8.1 | 7.7 | <1 | 51.5 | 4.0 |
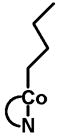
|
28.8 | 4.0 | 12.4 | <1 | 54.8 | 7.2 |
|
43.2 | 7.4 | 15.1 | <1 | 34.3 | 5.8 |
|
48.5 | 8.7 | 16.2 | <1 | 26.6 | 5.6 |
BAEC were cultured in the presence of [57Co]-cobalamins for 48 h. Intracellular [57Co]-cobalamins were extracted from washed cells and separated by HPLC as described in Experimental Procedures.
“Others” includes unprocessed Cbl source and unidentified corrinoids.
Cytotoxicity of xenobiotic alkylcobalamins
It has been reported that EtCbl and PrCbl form inactive complexes with apo-methionine synthase in vitro [19]. Therefore, examined the effects of high concentrations of each of the non-radioactive xenobiotic alkylcobalamins on cultured BAEC were examined. None of the xenobiotic alkylcobalamins appeared to be cytotoxic (Fig. S3, Supplementary Material). In addition, cell viability (MTT assay), senescence (β-galactosidase activity), and export of homocysteine and methylmalonic into the culture medium were also examined. None of these markers were altered in the presence of xenobiotic cobalamins compared to cells supplemented with CNCbl, AdoCbl or no cobalamin at all (data not shown). The lack of cytotoxicity of EtCbl, PrCbl and the other extended alkylcobalamins suggests that they are converted to non-toxic cobalamins, which serve as substrates for the biosynthesis of MeCbl and AdoCbl.
Dealkylation of xenobiotic alkylcobalamins by BAEC
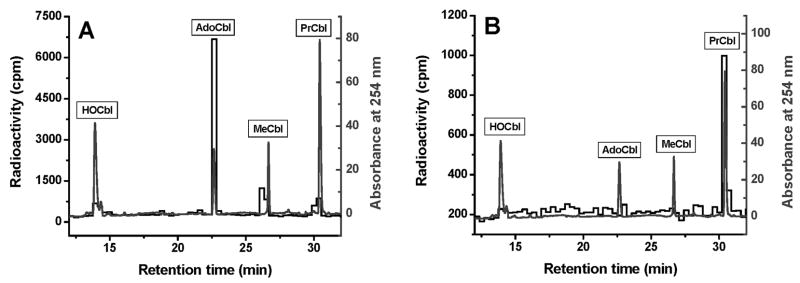
The ability of BAEC to convert a series of [57Co]-labeled xenobiotic alkylcobalamins with increasing β-axial ligand alkyl chain length into the active coenzyme forms was determined. Cells were grown for 48 h in the presence of 125 pM [57Co]-alkylcobalamin. Cobalamins present in cells and in the conditioned culture medium were extracted and analyzed as described under Materials and Methods. The intracellular cobalamin profile of cells grown in the presence of [57Co]-PrCbl is shown in Fig. 2A. Cells were able to dealkylate [57Co]-PrCbl and efficiently convert it to the two natural cofactors AdoCbl and MeCbl. Very little [57Co]-PrCbl was recovered from the cell extracts (Fig. 2A). Examination of the 48-h conditioned culture medium (Fig. 2B) revealed a prominent [57Co]-PrCbl peak but little or no other [57Co]-labeled cobalamins suggesting that the [57Co]-PrCbl substrate was stable and that there was little or no export of cellular cobalamins from BAEC into the conditioned medium. Similarly, all of the other [57Co]-labeled alkylcobalamins used in this study were stable and did not degrade in the culture medium (data not shown). [57Co]-AdoCbl was the major form of cobalamin found in BAEC after feeding with [57Co]-labeled alkylcobalamins. Table 2 summarizes the results obtained for the xenobiotic alkylcobalamin series (EtCbl, PrCbl, BuCbl, PnCbl, HxCbl), as well as results from the naturally occurring cobalamin forms (AdoCbl, CNCbl and HOCbl).
Fig. 2. Processing [57Co]-PrCbl in BAEC.
A. Typical cobalamin profile from BAEC grown in the presence of [57Co]-PrCbl (0.125 nM final concentration; 0.06 μCi/ml culture medium) as the cobalamin source for 48 h. Absorbance at 254 nm for cobalamin standards is shown in the lighter tracing; radioactivity for [57Co]-cobalamins is shown in the darker tracing. B. Cobalamins extracted from conditioned medium after 48 h. The darker tracing shows that [57Co]-PrCbl is largely intact after 48 h in the culture medium.
Genetic background, biochemical characterization and ability of the cblC cell lines to perform decyanation of [57Co]-CNCbl
A summary of the mutations present in the cblC cell lines used in this study and the age of onset of the disease are presented in Table 3. The three patient cblC cell lines present distinct mutations in the MMACHC gene, which led in all cases to a severe impairment of cobalamin metabolism. Total levels of homocysteine and methylmalonic acid were assessed in the conditioned culture medium of normal and cblC mutant fibroblasts grown for 7 days, and are shown in Table 3. All cblC cell lines excreted increased levels of both Hcy and MMA compared to normal fibroblasts. In addition, all cblC cell lines were unable to decyanate [57Co]-CNCbl and synthesize [57Co]-MeCbl and [57Co]-AdoCbl (Table 3). In contrast normal fibroblasts performed decyanation and subsequent cofactor biosynthesis efficiently. Patient cell line WG3354 performed decyanation to form HOCbl, however, there was no detectable cofactors biosynthesis (Table 3). Overall, this is the biochemical phenotype expected for combined methylmalonic aciduria and homocystinuria, hence, the cblC patient cell lines selected herein represent a suitable model to investigate the role of the cblC protein in the dealkylation process.
Table 3.
Genetic and biochemical background of the cblC patient cell lines used in this study.
| Cell line | Mutation 1 | Mutation 2 | Age of Onset | Metabolites in conditioned culture mediumb | Decyanation of [57Co]-CNCbl and cofactor biosynthesis (%)b | ||||
|---|---|---|---|---|---|---|---|---|---|
| Hcy (nmol/mg protein) | MMA (nmol/mg protein) | HOCbl | CNCbl | AdoCbl | MeCbl | ||||
| Normal | a | a | a | 11.2 ± 11.8 | 0.76 ± 0.07 | 19.5 ± 3.7 | 28.9 ± 2.6 | 25.3 ± 4.8 | 26.4 ± 1.1 |
| WG1801 | c.217C>T | c.217C>T | <2 months | 52.8 ± 25.5 | 6.67 ± 0.09 | NDc | 100 | ND | ND |
| WG2176 | c.1-234A>G | c.609G>A | Birth | 92.3 ± 23.2 | 3.57 ± 0.03 | ND | 100 | ND | ND |
| WG3354 | c.435_436dclAT | c.435_436dclAT | <2 months | 48.1 ± 8.1 | 2.46 ± 0.27 | 27.9 ± 5.9 | 72 ± 5.9 | ND | ND |
Values represent mean ± standard deviation (n=3). Total Hey and MMA were determined in the conditioned culture medium of cells grown for 7 days. Difference in Hey and MMA levels between the normal and the cblC mutant cell lines were statistically significant as determined by Student’s t-test at the 95% level of confidence (p<0.05)
Values represent mean ± standard deviation (n=3).
ND: Not detectable.
Dealkylation of xenobiotic alkylcobalamins by normal and cblC mutant fibroblasts
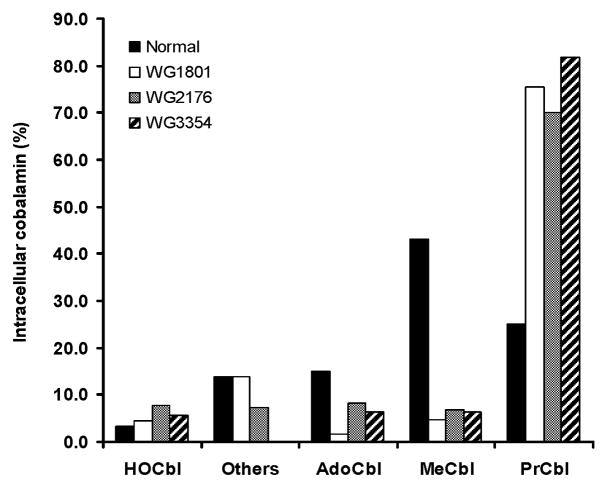
Because patients with a defective MMACHC gene are unable to utilize CNCbl as a substrate for cofactor biosynthesis [1, 4, 9] (Table 3), we hypothesized that cblC-derived skin fibroblasts would be incapable of dealkylating newly internalized alkylcobalamins. To test this hypothesis, normal and cblC mutant cell lines were incubated with [57Co]-PrCbl as described, and after 48 h, the intracellular cobalamin profiles were examined. As shown in Fig. 3, there was a much reduced capacity for the cblC mutant lines to convert [57Co]-PrCbl to [57Co]-AdoCbl and [57Co]-MeCbl, and most of the cobalamin in the mutant cells was unprocessed [57Co]-PrCbl. However, normal fibroblasts were very efficient at converting [57Co]-PrCbl into [57Co]-MeCbl, the predominate form, and, to a lesser extent, [57Co]-AdoCbl (Fig. 3). These results are consistent with a role for the cblC protein in removing alkyl groups from the β-axial ligand position of alkylcobalamins.
Fig. 3. Processing of [57Co]-PrCbl by human normal and cblC mutant fibroblasts (WG1801, WG2176 and WG3354).
Cells were cultured in the presence of 0.125 nM [57Co]-PrCbl (0.06 μCi/ml culture medium) for 48 h. [57Co]-labeled cobalamins were then extracted and analyzed by HPLC as described[15]. Results represent two pooled samples per cell line. “Others” refers to cobalamins not quantitatively determined in the present study, which include glutathionylcobalamin, sulphitocobalamin and nitrocobalamin (See ref [15]).
Discussion
The primary objective of the current work was to demonstrate that mammalian cells are capable of processing alkylcobalamins and to provide evidence that the processing is mediated by the cblC protein. Bovine aortic endothelial cells (BAEC) were used for assessing dealkylation processing and conversion to AdoCbl and MeCbl. The vascular endothelium appears to play important roles in cobalamin homeostasis. For example, endothelial cells synthesize and secrete considerable amounts of TC, the serum B12-binding protein that delivers the vitamin to cells throughout the body [20]. However, there is little information on cobalamin processing and coenzyme biosynthesis by vascular endothelial cells. We hypothesized that the vascular endothelium is able to utilize CNCbl and alkylcobalamins as substrates for the synthesis of AdoCbl and MeCbl and that the CblC protein recently shown to catalyze the in vitro decyanation of CNCbl [6], also catalyzes the dealkylation of alkylcobalamins in BAEC.
Our results demonstrate that cultured BAEC convert CNCbl to both AdoCbl and MeCbl. The amount of AdoCbl synthesized is always greater than the amount of MeCbl synthesized (Table 1). However, there was considerable variation in the AdoCbl/MeCbl product ratio depending on the substrate. When HOCbl and CNCbl were used as substrates, the AdoCbl/MeCbl product ratio was 12.9 and 4.5, respectively. Next, we assessed the ability of BAEC to utilize AdoCbl as a substrate for the biosynthesis of MeCbl as reported for cultured human lymphocytes several years ago [3]. Since AdoCbl and MeCbl are naturally occurring alkylcobalamins in circulation [10, 21], BAEC must have a system for dealkylating these endogenous alkylcobalamins that are delivered to the cell. We find that BAEC are indeed capable of converting AdoCbl to MeCbl.
We then determined whether the putative “dealkylase” activity would remove other alkyl groups from the β-axial position of cobalamins. Xenobiotic straight-chain alkylcobalamins were synthesized and purified, ranging from CH3CH2- (ethylcobalamin) to CH3(CH2)5-(hexylcobalamin). None of the straight-chain alkylcobalamins appeared to be cytotoxic to cultured BAEC. Surprisingly, all of the xenobiotic alkylcobalamins served as substrates for the synthesis of AdoCbl and MeCbl with AdoCbl/MeCbl product ratios ranging from 4.0 to 10.7 (Table 2). While our study demonstrates that the dealkylase system has broad substrate specificity for the ligand coordinating at the β-axial position of the cobalamin molecule, it does not address whether this activity is associated with one or more proteins.
The mystery of how decyanation of CNCbl occurs was recently solved by the in vitro studies of Kim et al. [6]. Decyanation is a process that is dependent on the activity of the CblC protein. The protein is a monomer of ~29 kDa, which catalyzes the reductive decyanation of CNCbl using a flavoprotein oxidoreductase for transferring reducing equivalents from NADPH [6]. Human fibroblasts that carry mutations in the MMACHC gene exhibit impaired cobalamin processing of CNCbl and little or no AdoCbl and MeCbl biosynthesis [4, 22]. Three cblC mutant fibroblasts isolated from severely ill and genetically unrelated patients were used in the present study to assess dealkylation in vivo. The biochemical profile of the cblC mutant cell lines WG1801, WG2176 and WG3354 resembled that reported for other cblC cell lines, i.e., substantial export of Hcy and MMA into culture medium (indicative of functional cobalamin deficiency) and poor or negligible utilization of CNCbl as a substrate for cofactor biosynthesis. The inability of the cblC mutant fibroblasts to utilize [57Co]-PrCbl as a substrate for AdoCbl and MeCbl biosynthesis is consistent with the hypothesis that the cblC protein catalyzes the dealkylation. In contrast, normal fibroblasts were able to use PrCbl efficiently to generate both AdoCbl and MeCbl. Based on the observations that 1) human recombinant MMACHC catalyzes decyanation of CNCbl [6]; 2) cblC mutant fibroblasts are unable to utilize CNCbl as a substrate for coenzyme biosynthesis [4, 23]; and, 3) cblC mutant fibroblasts are unable to perform dealkylation of [57Co]-PrCbl (this study), we propose that the cblC protein is responsible for catalyzing the removal of alkyl groups from the β-ligand position of alkylcobalamins.
The phenotypic expression of combined hyperhomocysteinemia and methylmalonicacidemia is associated with patients from the cblF, cblC and cblD complementation groups. Of these, cblF is unlikely to be a dealkylase since its impairment leads to accumulation of newly internalized B12 in lysosomes [24–26]. Recent work now shows that the cblF gene product is a B12 lysosomal membrane transporter [27]. The cblD locus is complex since it can lead to either isolated or combined defects in methionine synthase and methylmalonyl-CoA mutase [4, 28] and, for this reason, is also unlikely to encode a dealkylase that is shared by both AdoCbl and MeCbl synthesis pathways. Hence, the cblC locus appears to be the most likely candidate for encoding an alkylcobalamin dealkylase function.
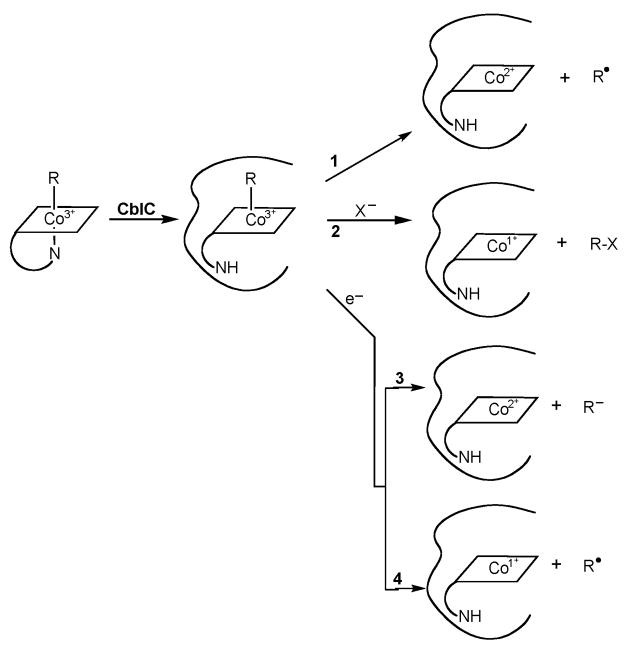
The current work shows that newly internalized alkylcobalamins undergo dealkylation processing, a likely prerequisite for generating the biologically active cobalamin forms AdoCbl and MeCbl, and that the dealkylase activity requires the cblC protein. In light of our results and previous findings, alternative mechanisms for cobalamin processing in vivo should be considered. Our current thoughts on the possible mechanisms by which the cblC protein could process newly internalized natural and xenobiotic alkylcobalamins are summarized in Fig. 4. Binding of the natural alkylcobalamins MeCbl and AdoCbl to the MMACHC chaperone has been shown to induce the “base-off” conformation in which the α-axial dimethylbenzimidazole ligand is not coordinated to the cobalt. This could be important in enhancing the reactivity of the β-axial ligand [6]. A variety of mechanistic alternatives can be considered for the removal of an alkyl group from the β-axial position of cobalamins (Fig. 4). First, homolysis of the cobalt-carbon bond would generate cob(II)alamin and an alkyl radical (Reaction 1, Fig. 4). Second, nucleophilic displacement of the alkyl group would result in the formation of cob(I)alamin and the transfer of the alkyl carbocation to the acceptor (Reaction 2, Fig. 4) [29, 30]. Third, reductive dealkylation could occur resulting in the formation of either cob(II)alamin or cob(I)alamin and the departure of the alkyl group as a carbanion or a radical, respectively (Reactions 3 and 4, Fig. 4). A number of in vitro studies with human recombinant cblC protein are currently underway in our laboratories to elucidate the mechanism of dealkylation catalyzed by the surprisingly versatile cblC protein.
Fig. 4. Possible mechanisms for the dealkylation of alkylcobalamins mediated by the cblC protein.
Formation of the base-off conformation of the cobalamin leads to an enhanced reactivity of the upper axial ligand. Reaction 1: homolysis of the cobalt-carbon bond would generate cob(II)alamin and an alkyl radical. Reaction 2: nucleophilic displacement of the alkyl group would result in the formation of cob(I)alamin and the transfer of the alkyl carbocation to the acceptor as described by model studies [29, 30]. Reactions 3 and 4: reductive dealkylation could occur resulting in the formation of either cob(II)alamin or cob(I)alamin and the departure of the alkyl group as a carbanion or a radical, respectively.
Supplementary Material
Acknowledgments
This research was funded by the National Heart, Lung and Blood Institute of the National Institutes of Health (HL71907 to DWJ).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosenblatt DS, Fenton WA. Inherited disorders of folate and cobalamin transport and metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic & Molecular Bases of Inherited Disease. Mc-Graw Hill; New York: 2001. pp. 3897–3933. [Google Scholar]
- 2.Gravel RA, Mahoney MJ, Ruddle FH, et al. Genetic complementation in heterokaryons of human fibroblasts defective in cobalamin metabolism. Proc Natl Acad Sci U S A. 1975;72:3181–3185. doi: 10.1073/pnas.72.8.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quadros EV, Jackson B, Hoffbrand AV, et al. Interconversion of cobalamins in human lymphocytes in vitro and the influence of nitrous oxide on sythesis of cobalamin coenzymes. In: Zagalak B, Friedrich W, editors. Vitamin B12. Walter de Gruyter; Berlin: 1979. pp. 1045–1054. [Google Scholar]
- 4.Suormala T, Baumgartner MR, Coelho D, et al. The cblD defect causes either isolated or combined deficiency of methylcobalamin and adenosylcobalamin synthesis. J Biol Chem. 2004;279:42742–42749. doi: 10.1074/jbc.M407733200. [DOI] [PubMed] [Google Scholar]
- 5.Lerner-Ellis JP, Tirone JC, Pawelek PD, et al. Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat Genet. 2006;38:93–100. doi: 10.1038/ng1683. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Gherasim C, Banerjee R. Decyanation of vitamin B12 by a trafficking chaperone. Proc Natl Acad Sci U S A. 2008;105:14551–14554. doi: 10.1073/pnas.0805989105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mudd SH, Levy HL, Abeles RH. A derangment in B12 metabolism leading to homocystinemia, cystathioninemia and methylmalonic aciduria. Biochem Biophys Res Commun. 1969;35:121–126. doi: 10.1016/0006-291x(69)90491-4. [DOI] [PubMed] [Google Scholar]
- 8.Mahoney MJ, Rosenberg LE. Defective metabolism of vitamin B12 in fibroblasts from children with methylmalonicaciduria. Biochem Biophys Res Commun. 1971;44:375–381. doi: 10.1016/0006-291x(71)90610-3. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg LE, Patel L, Lilljeqvist AC. Absence of an intracellular cobalamin-binding protein in cultured fibrblasts from patients with defective synthesis of 5′-deoxyadenosylcobalamin and methylcobalamin. Proc Natl Acad Sci U S A. 1975;72:4617–4621. doi: 10.1073/pnas.72.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimsing P, Nexo E, Hippe E. Determination of cobalamins in biological material. II. The cobalamins in human plasma and erythrocytes after desalting on nonpolar adsorbent material, and separation by one-dimensional thin-layer chromatography. Anal Biochem. 1983;129:296–304. doi: 10.1016/0003-2697(83)90553-5. [DOI] [PubMed] [Google Scholar]
- 11.Gimsing P, Nexo E. The forms of cobalamin in biological materials. In: Hall CA, editor. The Cobalamins. Vol. 10. Churchill Livingstone; New York: 1983. pp. 7–30. [Google Scholar]
- 12.Raaberg L, Nexo E, Poulsen SS, et al. Cobalamin and its binding protein in rat milk. Scand J Clin Lab Invest. 1989;49:529–535. doi: 10.3109/00365518909089132. [DOI] [PubMed] [Google Scholar]
- 13.Kim SH, Chen HL, Feilchenfeld N, et al. Thermal decomposition and cobalt-carbon bond dissociation energies of organocobalamins: neopentyl-, (cyclopentylmethyl)-, (cyclohexylmethyl)-, (tetrahydrofurfuryl)- and ((tetrahydro-2H-pyryl)methyl)cobalamin. J Am Chem Soc. 1988;110:3120–3126. [Google Scholar]
- 14.Pratt JM. Inorganic Chemistry of Vitamin B12. Academic Press; London: 1972. [Google Scholar]
- 15.Hannibal L, Axhemi A, Glushchenko AV, et al. Accurate assessment and identification of naturally occurring cellular cobalamins. Clin Chem Lab Med. 2008;46:1739–1746. doi: 10.1515/CCLM.2008.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferstion and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen DW, Gatautis VJ, Green R, et al. Rapid HPLC determination of total homocysteine and other thiols in serum and plasma: sex differences and correlation with cobalamin and folate levels in normal subjects. Clin Chem. 1994;40:873–881. [PubMed] [Google Scholar]
- 18.Hoffmann G, Aramaki S, Blum-Hoffmann E, et al. Quantitative analysis for organic acids in biological samples: batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin Chem. 1989;35:587–595. [PubMed] [Google Scholar]
- 19.Yamada K, Yamada S, Tobimatsu T, et al. Heterologous high level expression, purification, and enzymological properties of recombinant rat cobalamin-dependent methionine synthase. J Biol Chem. 1999;274:35571–35576. doi: 10.1074/jbc.274.50.35571. [DOI] [PubMed] [Google Scholar]
- 20.Quadros EV, Rothenberg SP, Jaffe EA. Endothelial cells from human umbilical vein secret functional transcobalamin II. Am J Physiol. 1989;256:C296–C303. doi: 10.1152/ajpcell.1989.256.2.C296. [DOI] [PubMed] [Google Scholar]
- 21.Gimsing P. Determination of cobalamins in biological materials. 1. Improvement in the unequal recovery of cobalamins by preincubation with cadium acetate. Anal Biochem. 1983;129:288–295. doi: 10.1016/0003-2697(83)90552-3. [DOI] [PubMed] [Google Scholar]
- 22.Rosenblatt DS, Aspler AL, Shevell MI, et al. Clinical heterogeneity and prognosis in combined methylmalonic aciduria and homocystinuria (cblC) J Inherit Metab Dis. 1997;20:528–538. doi: 10.1023/a:1005353530303. [DOI] [PubMed] [Google Scholar]
- 23.Rosenblatt DS. Inborn errors of folate and cobalamin metabolism. In: Carmel R, Jacobsen DW, editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge, UK: 2001. pp. 244–258. [Google Scholar]
- 24.Rosenblatt DS, Hosack A, Matiaszuk NV, et al. Defect in vitamin B12 release from lysosomes: Newly described inborn error of vitamin B12 metabolism. Science. 1985;228:1319–1321. doi: 10.1126/science.4001945. [DOI] [PubMed] [Google Scholar]
- 25.Watkins D, Rosenblatt DS. Failure of lysosomal release of vitamin B12: A new complementation group causing methylmalonic aciduria (cblF) Am J Hum Genet. 1986;39:404–408. [PMC free article] [PubMed] [Google Scholar]
- 26.Shih VE, Axel SM, Tewksbury JC, et al. Defective lysosomal release of vitamin B12 (cblF): a hereditary cobalamin metabolic disorder associated with sudden death. Am J Med Genet. 1989;33:555–563. doi: 10.1002/ajmg.1320330431. [DOI] [PubMed] [Google Scholar]
- 27.Rutsch F, Gailus S, Miousse IR, et al. Identification of a putative lysosomal cobalamin exporter altered in the cblF defect of vitamin B12 metabolism. Nat Genet. 2009;41:234–239. doi: 10.1038/ng.294. [DOI] [PubMed] [Google Scholar]
- 28.Coelho D, Suormala T, Stucki M, et al. Gene identification for the cblD defect of vitamin B12 metabolism. N Engl J Med. 2008;358:1454–1464. doi: 10.1056/NEJMoa072200. [DOI] [PubMed] [Google Scholar]
- 29.Hogenkamp HPC, Bratt GT, Sun SZ. Methyl transfer from methylcobalamin to thiols. A reinvestigation. Biochemistry. 1985;24:6428–6432. doi: 10.1021/bi00344a018. [DOI] [PubMed] [Google Scholar]
- 30.Hogenkamp HPC, Bratt GT, Kotchevar AT. Reaction of alkylcobalamins with thiols. Biochemistry. 1987;26:4723–4727. doi: 10.1021/bi00389a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.