Abstract
Synthetic small duplex RNAs that are complementary to gene promoters can activate or inhibit target gene expression. The potency and robustness of gene modulation by these RNAs suggests that natural mechanisms may exist to facilitate recognition of sequences within gene promoters by endogenous small RNAs. Here we describe computational methods for identifying potential miRNA target sites within gene promoters. These methods will facilitate investigations of whether miRNAs interact with sequences outside of 3′ untranslated regions and suggest new targets for the design of synthetic modulators of gene expression.
Synthetic small duplex RNAs complementary to gene promoters within chromosomal DNA have been reported to be potent inhibitors or activators of target gene expression in mammalian cells.1–5. We refer to these synthetic RNAs as antigene RNAs (agRNAs) to distinguish them from small duplex RNAs that target mRNA. agRNAs recruit members of the argonaute (AGO) protein family to RNA transcripts that originate from the target gene promoter in either the sense or antisense direction.6–9 Data suggests that recognition of the target RNA occurs in close proximity to the chromosome, resulting in transcriptional modulation of the target gene.
One remarkable feature of the synthetic agRNAs that we have examined is the potency and robustness of their activity when they are introduced into cells. This potency, coupled with the presence of protein machinery that facilitates their function, suggests that endogenous small RNAs may possess the ability to recognize gene promoters. If RNA could direct proteins to specific gene promoters, such RNA-mediated modulation of transcription might have evolutionary advantages relative to the development of gene-specific protein transcription factors.
Synthetic duplex RNAs that are complementary to mRNA (small interfering RNAs or siRNAs) are also potent and robust agents for modulating gene expression.10 siRNAs are known to have endogenous analogs that regulate gene expression called microRNAs (miRNAs).11 miRNAs are processed inside the cell from RNA precursors that contain stem-loop structures. These stem-loop structures are processed by the double-stranded nucleases Drosha and Dicer to produce mature miRNAs.
As of the current release of the miRNA repository (miRBase v12.0), 866 human miRNAs have been annotated, but this number continues to increase. Several miRNAs that recognize sequences within the 3′-untranslated regions (3′UTR) of mRNA transcripts have been characterized. Many miRNAs, however, have no known targets12,13 while some can recognize multiple mRNAs13, suggesting that the determinants of miRNA interactions are complex and poorly understood.
Two reports based on computational analyses have suggested that miRNAs can modulate gene expression through promoter recognition. Dahiya and co-workers used publically available software (RegRNA) to search for potential miRNA target sites within the promoter of the E-cadherin gene.14 They identified one potential binding site for miR-373 within the E-cadherin promoter and reported that introduction of a synthetic miR-373 mimic increased expression of the gene by 6 fold at the level of the mRNA. Rossi and co-workers searched for perfect complementarity between miRNAs and gene promoters.15 Their analysis suggested that miR-320 targets the genomic location from which it is transcribed and showed that expression of miR-320 and the adjacent gene, POLR3D, are anti-correlated.
The above-mentioned studies either analyzed a single gene promoter or used highly stringent sequence comparison criteria. These approaches were not intended to assess broader potential for miRNAs to recognize gene promoters, warranting a more thorough evaluation of the relationship between miRNAs and promoter sequences.
A practical justification for more comprehensive studies is that validating natural gene targets of miRNAs is a complex and difficult process. The development of systematic and efficient methods for identifying promoter sequences that may be miRNA targets is essential for prioritizing predictions and efficiently allocating experimental resources towards validating the most promising targets. Here we examine computational methods for predicting potential miRNA targets within gene promoters and demonstrate that promoters are strong candidates for miRNA regulation.
Sequence Acquisition
To identify putative promoter-targeting miRNAs we constructed a database comprised of miRNA and gene promoter sequences from public sequence repositories. Promoter sequences were acquired from the UCSC genome browser (hg 18) and consisted of the 200 nucleotides immediately 5′ to the annotated transcription start site for each gene.16,17 We chose 200 base sequences (−200 to −1) for initial evaluations but larger promoter regions can also be examined. Mature miRNA sequences were obtained from miRBase (Build 12.0), which contains sequences of experimentally determined precursor and mature miRNAs.18,19,20
Analysis of seed sequence matches
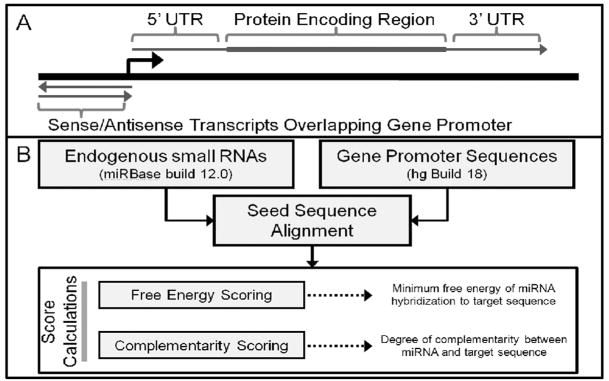
Synthetic promoter-targeting RNAs recognize non-coding (ncRNA) transcripts that overlap gene promoters. We used promoter DNA sequences to construct datasets representing potential ncRNA transcripts in both the sense and antisense direction for each gene promoter as we hypothesize that endogenous small RNAs would also recognize these ncRNA transcripts. For comparison we also obtained the sequences of the 5′UTR, coding sequences (CDS), and 3′UTR for each gene (Fig. 1A).
Figure 1.
(A) Diagram of sequences that are potential miRNA targets. (B) Schematic of algorithm used to predict miRNA targets within gene promoters.
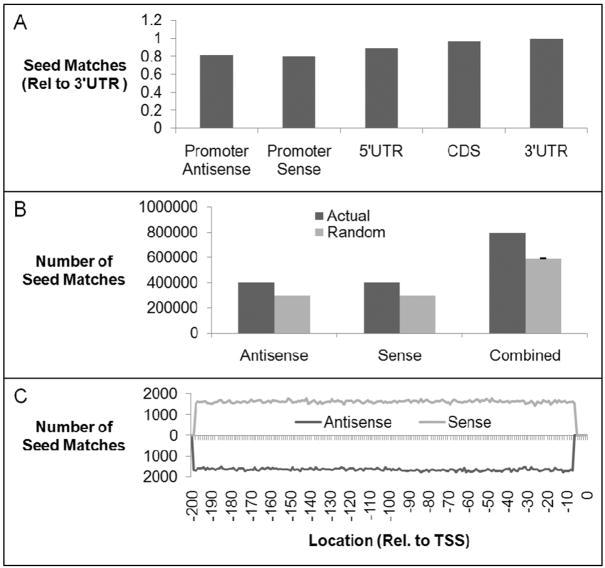
A basic requirement for target recognition by miRNAs is perfect complementarity between the target sequences and bases 2–8 of the mature miRNA sequence, called the seed sequence. We determined the number of seed matches within potential sense and antisense transcripts that overlap gene promoters and compared them to seed matches within the 3′UTR region of mRNAs (Fig. 1B). We found that seed matches within promoter-overlapping transcripts occur 80% as frequently as seed matches within 3′UTRs, indicating that gene promoter sequences have the potential to be miRNA targets (Fig. 2A). Our analysis detected the previously reported complementarity between miR-320 and the POLR3D promoter.15
Figure 2.
Gene promoters contain predicted miRNA target sites. (A) The frequency of seed sequence matches in promoter regions, 5′UTRs, coding regions, and 3′UTRs. (B) Comparison of seed matches within promoter-overlapping transcripts vs. randomized promoter sequences. (p < .01) (C) Distribution of seed match locations within sense and antisense transcripts that overlap gene promoters from −1 to −200 relative to the +1 transcription start site.
To evaluate the statistical significance of seed matches within gene promoter sequences we tabulated the frequency of occurrences of seed matches in 100 randomizations of each promoter sequence. We found that seed matches occur 75% as frequently within randomized as opposed to actual promoter sequences (Fig. 2B). The excess of observed to expected seed sequence matches within promoter sequences was similar for both putative sense and antisense transcripts. This result implies that promoter sequences are enriched for potential targets for recognition by miRNAs. Matches are equally distributed throughout the 200 base gene promoter segments surveyed, suggesting that no particular region of a gene promoter is more likely than another to contain a predicted miRNA target site (Fig. 2C).
Ranking matches
Our analysis identified nearly 800,000 miRNA seed matches within 27,345 gene promoter sequences (Fig. 2B). This large number required investigation of additional factors to prioritize target predictions. Although not necessarily a prerequisite for miRNA function, the minimum free energy (MFE) of hybridization between miRNAs and their predicted target sites have been successfully used to predict miRNA target sites within 3′UTRs21 We reasoned that MFE values may also be useful for prioritizing miRNA target predictions within gene promoters.
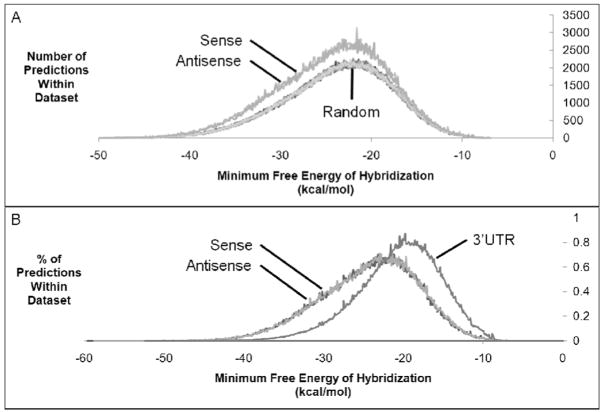
The MFE values were calculated for miRNA hybridization to predicted target sites (based on seed sequence matches, hereafter simply referred to as predictions) within putative promoter-overlapping transcripts and within 10 randomizations of promoter sequences. We found that predictions with lower MFE values occurred more frequently in actual promoter sequences than in randomized sequences (Fig. 3A). The difference between the distributions of MFE values demonstrates that predictions with low MFE values occur more often than would be expected at random, implying that these predictions are more likely to be biologically significant and that MFE values will be useful criteria for prioritizing target predictions.
Figure 3.
MFE properties of miRNA target predictions. (A) Distribution of MFE values for predictions within promoter-overlapping transcripts as compared to randomized promoter sequences. (B) Distribution of MFE values for predictions within promoter-overlapping transcripts as compared to 3′UTRs.
During the course of the MFE analysis we identified several miRNA target predictions within gene promoters that had notably low MFE values. These observations prompted us to compare the MFE values for target predictions within gene promoters to target predictions within 3′UTRs (Fig. 3B). We calculated the mean MFE value for all predictions within gene promoters to be −24.27 kcal/mol and −24.32 kcal/mol for putative sense and antisense promoter-overlapping transcripts, respectively. The mean MFE for all predictions within 3′UTRs was −20.57 kcal/mol, more than 3.5 kcal/mol higher than predictions within promoters. The difference in mean MFE values suggests that, on average, miRNA recognition of sequences at gene promoters would be more energetically favorable than recognition of 3′UTR sequences.
To further evaluate the differences between target predictions within gene promoters and 3′UTRs, we examined the distribution of MFE values for all predictions within the different sequence datasets. As previously indicated by the mean MFE values, roughly 50% of target predictions within gene promoters had MFE values below −24.3 kcal/mol. Interestingly, only 22% of predictions within 3′UTRs had MFE values below −24.3 kcal/mol (Fig. 3B). The difference in MFE value distributions demonstrates that gene promoters are enriched relative to 3′UTRs for predicted target sites with low free energies of hybridization and may actually represent more favorable miRNA targets than 3′UTRs.
Another criterion used in miRNA target prediction is sequence complementarity. Sequence complementarity alone has been used successfully to predict miRNA target sites within 3′UTRs.22 We used the Needleman-Wunsch algorithm23 to evaluate the degree of sequence complementarity between miRNAs and predicted target sites within gene promoters (Fig. 1B). We identified over 200 individual miRNAs with near perfect complementarity to their predicted target sites within gene promoters. A selected subset of these predictions is listed in Fig. 4. The high degree of complementarity between miRNAs and gene promoters further demonstrates that gene promoters are promising candidates for miRNA targets.
Figure 4.
Examples of predicted miRNA targets within sequences of promoter-overlapping transcripts.
Strong evidence that gene expression can be modulated using synthetic duplex RNAs that are complementary to gene promoters suggests that natural gene regulation may include recognition of gene promoters by miRNAs. Such recognition would have evolutionary advantages, given the large difference between protein transcription factors and miRNAs in their efficiency of generating new selectivity for gene promoters through mutation.
Our computational algorithm that can be used to identify promising miRNA target sites within gene promoters. We identify many seed sequence matches within promoters and demonstrate that they are almost as common as those within 3′UTRs. We also identify many miRNA/promoter pairs that have unusually strong complementarity. These results can be used to rank order miRNA/promoter pairs for the demanding studies necessary to validate whether the potential for these interactions is biologically significant.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIGMS 77253 to DRC and R01CA 129632 to AP), The Robert A. Welch Foundation (I-1244), and an NIH Pharmacological Sciences Training Grant (GM07062 to STY). We thank A. Guillory for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Science. 2004;305:1289. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 2.Ting AH, Schuebel KE, Herman JG, Baylin SB. Nat Genet. 2005;37:906. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy DB, Shames DS, Minna JD, Corey DR. Nat Chem Biol. 2005;1:216. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 4.Li LC, Okino ST, Zhao H, Pookot D, Urakami S, Enokida H, Dahiya R. Proc Natl Acad Sci USA. 2006;103:17337. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Nat Chem Biol. 2007;3:166. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 6.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Nat Struct Mol Biol. 2006;13:787. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Nat Struct Mol Biol. 2006;13:793. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 8.Han J, Kim D, Morris KV. Proc Natl Acad Sci U S A. 2007;104:12422. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz JC, Younger ST, Nguyen N_B, Hardy DB, Corey DR. Nat Struct Mol Biol. 2008;15:842. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:85. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. Proc Natl Acad Sci USA. 2008;105:1608. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DH, Saetrom P, Snøve O, Jr, Rossi JJ. Proc Natl Aad Sci USA. 2008;105:16230. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. Nature. 2001;409:860. [Google Scholar]
- 17.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. Genome Res. 2002;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. NAR. 2008;36:D154. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. NAR. 2006;34:D140. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. NAR. 2004;32:D109. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark A, Brennecke J, Russell RB, Cohen SM. PLoS Biol. 2003;1:1. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai EC. Nat Genet. 2002;30:363. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 23.Needleman SB, Wunsch CD. J Mol Biol. 1970;48:443. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]