Abstract
A sparse population of thymocytes undergoes TCRα gene rearrangement early in development, before the double positive stage. The potential of these cells to contribute to the peripheral T cell pool is unknown. To examine the peripheral T cell compartment expressing a repertoire biased to early TCR gene rearrangements, we developed a mouse model in which TCRα rearrangements are restricted to the double negative stage of thymocyte development. These mice carry floxed RAG2 alleles and a Cre transgene driven by the CD4 promoter. As expected, conventional T cell development is compromised in such Cre(+) RAG2fl/fl mice, and the TCRαβ+ T cells that develop are limited in their TCRα repertoire, preferentially utilizing early-rearranging Vα genes. In the gut, the Thy-1+TCRαβ+ intraepithelial lymphocyte (IEL) compartment is surprisingly intact, while the Thy-1−TCRαβ+ subset is almost completely absent. Thus, T cells expressing a TCRα repertoire that is the product of early gene rearrangements can preferentially populate distinct IEL compartments. Despite this capacity, Cre(+) RAG2fl/fl T cell progenitors cannot compete with wild-type (WT) T cell progenitors in mixed bone marrow chimeras, suggesting that in normal mice, there is only a small contribution to the peripheral T cell pool by cells that have undergone early TCRα rearrangements. In the absence of WT competitors, aggressive homeostatic proliferation in the IEL compartment can promote a relatively normal Thy-1+ TCRαβ+ T cell pool from the limited population derived from Cre(+) RAG2fl/fl progenitors.
Keywords: T cells, T Cell Receptors, Repertoire Development, Gene Rearrangement, Mucosa
Introduction
The development of mature αβ T lymphocytes in the thymus depends on successful rearrangement of variable (V), diversity (D) and joining (J) gene segments to encode a functional αβ TCR. V(D)J gene recombination requires RAG1 and RAG2 gene products and is a tightly regulated, orderly process. Tcrb rearrangements occur first at the CD8−CD4− double negative (DN)3 stage of thymocyte development. Successful rearrangement and expression of a TCRβ chain that can pair with the pre-Tα chain drives a burst of proliferation and progression to the CD8+CD4+ double positive (DP) stage, through a process termed β-selection. Within the DP compartment, Tcra genes are rearranged, often on both alleles. This flexibility allows for multiple attempts at creating a functional αβ TCR, one that can promote positive selection, lineage commitment, loss of RAG expression and cessation of further gene rearrangements.
The complex organization of the murine TCRα/δ , with Dδ and Jδ genes situated between V and Jα gene segments, ensures that Tcrd genes are excised upon Vα-Jα gene rearrangement. Regulation of TCRα gene rearrangement is orchestrated by various promoters and enhancers that determine when rearrangement occurs and which V and J gene segments are used (1, 2). Primary, or early, Vα gene usage is biased to the 3’ end of the V gene array. Vα6 and Vα20 are at the 3’ end of the locus and are preferentially utilized in early rearrangements, while Vα12 and Vα19 are at the 5’ end and tend to be used in late rearrangements. In addition, Vα-Jα gene rearrangement is biased, with 3’ Vα genes preferentially rearranging to proximal 5’ Jα genes (3, 4).
While the majority of Tcra gene rearrangements occur in the DP stage, Tcra genes can be rearranged in early thymocytes, and transcripts have been found in the DN2, DN3 and DN4 compartments (5, 6). Although pTα−/− mice have thymic hypoplasia, they do have peripheral TCRαβ+ T cells, showing that TCRα chains expressed at the DN stage can pair with TCRβ chains to drive β-selection in the absence of pre-Tα expression (7). The timing of TCRα rearrangement may be a factor in lineage commitment. Some evidence suggests that premature expression of TCRαβ may divert cells into the TCRγδ lineage (8). In addition, aberrantly early TCRα expression in HY TCR transgenic mice promotes the expansion of the TCRαβ CD8αα+ subset of IELs in the gut (9). Delaying TCRα expression until the DP stage prevents the dramatic expansion of this compartment (10), suggesting that IELs may be the progeny of thymocytes that have undergone early TCRα rearrangements. In support of this notion, the IEL compartment is seeded early in development, a time when early-rearranging TCRs are overrepresented (4). TCRγδ+ and TCRαβ+ IELs are found in neonatal mice and in human fetal intestine (11). The rapid development of IELs is also supported by the fact that, in bone marrow radiation chimeras, repopulation of the IEL compartment occurs earlier than in the spleen or thymus (12). Based on these observations, we hypothesized that T cells bearing the products of early TCR rearrangements may constitute a specific T cell subset with the ability to populate certain compartments (such as IELs) preferentially.
To test this hypothesis and analyze the impact of a repertoire biased to early TCR gene rearrangements, we developed a mouse model in which TCR rearrangements are restricted to DN thymocytes. We crossed mice carrying floxed RAG2 alleles (13) with mice that are transgenic for Cre driven by the mouse CD4 promoter/enhancer/silencer (14). In Cre(+) RAG2fl/fl mice, Cre expression is initiated in the late DN3 stage (15), leading to deletion of RAG2 by the early DP stage. Therefore, T cells are able to progress efficiently through Tcrb gene rearrangement and β-selection but have a narrow window of opportunity in which to make productive TCRα rearrangements.
Analysis of Cre(+)RAG2fl/fl mice reveals that the TCRαβ+ T cell population in the spleen and peripheral lymph nodes (LNs) is significantly diminished, with an inversion of the CD4:CD8 ratio. In addition, the TCRα repertoire is oligoclonal, with preferential representation of TCRs utilizing early-rearranging Vα genes. In the small intestinal IEL compartment, however, the Thy-1+TCRαβ+ population is intact, while the Thy-1−TCRαβ+ population is nearly absent. In mixed chimeras, cells derived from WT bone marrow outcompete those derived from Cre(+) RAG2 fl/fl bone marrow, even in the gut. Thus, in the absence of WT competitors, aggressive homeostatic proliferation allows preferential repopulation of certain T cell compartments in the small intestine by cells expressing TCRs that are the products of early TCRα rearrangements.
Materials and Methods
Mice
RAG2fl/fl mice on the C57BL/6J (B6) background were a gift from K. Rajewsky (The CBR Institute for Biomedical Research) and were crossed to CD4p-Cre transgenic (C57BL/6NTac-TgCd4-cre) B6 mice (hereafter “Cre(+) RAG2+/+”), a gift from C. Wilson (University of Washington). Mice were bred to homozygosity. WT Ly5.2+ B6 and Ly 5.1+ B6.SJL (B6.SJL-PtprcaPepcb/BoyJ) mice were purchased from The Jackson Laboratory or bred in-house, and TCRα-deficient (Tcratm1Mom) mice were a gift from A. Rudensky (University of Washington). Controls were either Cre(−) RAG2fl/fl, Cre(+) RAG2+/+, or WT B6 mice, as a comparative analysis revealed no differences in cell numbers or phenotype between these strains (data not shown). All mice were bred and housed under specific pathogen-free conditions in accordance with institutional guidelines. Mice were used at 6–10 weeks of age, except bone marrow chimeras, which were used at 8–20 weeks of age. All experiments were performed in compliance with the University of Washington Institutional Animal Care and Use Committee.
Mixed bone marrow chimeras
Bone marrow was obtained from Cre(+) RAG2fl/fl (Ly5.2) and B6.SJL (Ly5.1) donors and was depleted of mature T cells using the EasySep biotinylated separation kit (Stem Cell Technologies) and the following biotin-conjugated antibodies: anti-CD4 (Rm4.4), anti-CD8 (53–6.7) and anti-Thy1.2 (30-H12), all from BD Biosciences. B6×B6.SJL (Ly5.1×5.2) hosts were lethally irradiated with 1000 rads at least 7 hours prior to reconstitution. Bone marrow was mixed 1:1 and 4×106 cells were injected in the lateral tail vein in 200µL sterile PBS. Hosts were maintained on antibiotic water (polymyxin B sulfate and neomycin sulfate) for 2 days prior to and 14 days post irradiation. Chimeras were analyzed >8 weeks post-injection.
Flow cytometry antibodies
Antibodies used to stain cell surface molecules were: PE-conjugated anti-CD19 (1D3), anti-CD25 (PC61) and anti-CD103 (M290), Allophycocyanin-(APC) conjugated anti-Thy1.2 (CD90.2; 53-2.1) and anti-CD62L (MEL-14), PerCP-, PE-Cy7- and APC AlexaFluor 750-(APCAF750) conjugated anti-CD4 (RM4–5), FITC- and PerCP-conjugated anti-CD8α (53-6.7), FITC-conjugated anti-pan TCRβ (H57-597.13) and anti-TCRγδ (GL3), biotin-conjugated anti-CD8.2β (53-5.8), PE- and PE-Cy7-conjugated anti-CD69 (H1.2F3), PE-Cy7- and APC-conjugated anti-CD44 (Pgp-1), PE-Cy7-conjugated anti-Ly5.1 (A20), and FITC- and APCAF750-conjugated anti-Ly5.2 (104), all from BD Biosciences or eBioscience. APCAF750-conjugated anti-CD8α (5H10) and PE-conjugated Streptavidin were from Caltag. TL-tetramer, used to detect CD8αα, was a gift from H. Cheroutre (La Jolla Institute for Allergy and Immunology) and was conjugated to PE-labeled Streptavidin (Molecular Probes).
Cell preparation
IELs were prepared as previously described with a few modifications (16). Briefly, small intestines were removed and all mesenteric fat cleaned. Peyer’s patches (PPs) were removed and processed. Intestines were opened longitudinally and washed of their contents. Intestines were cut into 0.5cm pieces and stirred at 450 rpm at 37°C for 20 min in CMF medium [HBSS (Ca++ and Mg++ free), 100mM HEPES, 250mM sodium bicarbonate, 2% FCS and 0.154mg/mL dithioerythriol]. Cell suspensions were filtered, washed and resuspended in 44% Percoll, underlaid with 67% Percoll, and centrifuged at 2800rpm for 20 min. Enriched IELs were extracted from the interface and washed. IEL preps were free of contaminating PP lymphocytes, as they consistently contained <1.3% CD19+ B cells. IELs, thymus, LNs (axial, brachial, inguinal and mesenteric), PPs and water-lysed spleen cells were processed using 70µM nitex filters and resuspended in HBSS/1%BSA. Single-cell suspensions were incubated with anti-CD16/CD32 blocking Ab (2.4G2; BD Biosciences) and were stained according to standard procedures. Cells were analyzed on a FACSCanto flow cytometer (Becton-Dickinson, San Jose, CA) and data were analyzed using FlowJo software (TreeStar Inc, Ashland, OR). For some experiments, spleen and LN T cells were enriched by negative selection using the EasySep Mouse T cell Enrichment kit (StemCell Technologies) and stained with antibodies to CD4, CD8α and TCRβ. Cells were sorted on a FACSAria cell sorter (BD Biosciences) to >97% purity. Where indicated, purified T cells were incubated with 5mM CFSE (Molecular Probes) for 10 min at 37°C and then washed twice in ice cold complete RPMI 1640 medium with 10% FCS before being placed in culture.
In vitro stimulation and intracellular cytokine staining
CFSE-labeled, purified CD4+ and CD8+ T cells from spleens and LNs of WT and Cre(+)RAG2fl/fl mice were cultured with anti-CD3/anti-CD28 Dynabeads (Invitrogen) at a 1:1 bead:cell ratio. Cells were harvested on days 2 or 3 and restimulated for 5 h with 50ng/ml PMA (Sigma) and 0.7µM ionomycin (Sigma) in the presence of the protein transport inhibitor Brefeldin A (BD Golgiplug, BD Biosciences). Washed cells were surface stained with anti-CD4 or anti-CD8α for 20 min at 4°C and then washed. Intracellular cytokine staining was performed using the BD Cytofix/Cytoperm buffers according to the manufacturer’s protocol.
Quantitative RT-PCR
Total mRNA was extracted from purified cells using the RNeasy mini kit (Qiagen) and first-strand cDNA synthesis was performed with oligo(dT) primers, using SuperScript III (Invitrogen) according to the manufacturer’s directions. RNase-treated cDNA was subjected to quantitative PCR performed on an ABI 7300 Real Time PCR System (Applied Biosystems, Foster City, CA) using Power SYBR Green PCR Master Mix (Applied Biosystems). PCRs consisted of a 10 min 95°C denaturation step followed by 45 cycles of 15 s at 95°C and 30 s at 60°C. The following primers were used: Vα6 [TRAV21, IMGT nomenclature] sense primer, 5’-ATGGCTTTCCTGGCTATTGC-3’, Vα12 [TRAV2, IMGT nomenclature] sense primer, 5’-CTGTTTATCTCTGCTGACCGG-3’, Vα19 [TRAV1, IMGT nomenclature] sense primer, 5’-TGGATGGTTTGAAGGACAGTG-3’, Vα20 [TRAV19, IMGT nomenclature] sense primer, 5’-GGAAGACGGAAGATTCACAGTT-3’, Cα anti-sense primer, 5’-TGGCGTTGGTCTCTTTGAAG-3’, hypoxanthine guanine phosphoribosyltransferase (HPRT) sense primer 5’-GTTGGATACAGGCCAGACTTTGTTG-3’, and HPRT anti-sense primer 5’-GAGGGTAGGCTGGCCTATAGGCT-3’. Reactions were run in triplicate and samples were normalized to the internal HPRT control.
TCR CDR3 length spectratyping
CDR3 length spectratyping was performed as previously described by with a few modifications (17). Primary PCR reactions were performed using the following previously described primers (18, 19): Vα6 sense primer, 5’-ATGGCTTTCCTGGCTATTGCC-3’, Vα12 sense primer, 5’-TCTGTTTATCTCTGCTGACC-3’, Vα19 sense primer 5’-TGCCTCATACCTCTGTGCTG-3’, Vα20 sense primer, 5’-TTCTCACTGCACATCACAGC-3’, and the Ca anti-sense primer as above. Cycling conditions were: 94°C for 45 s, 55–60°C for 45 s and 72°C for 1 min for 35 cycles. Secondary run-off PCR reactions using 25% of the primary product were performed using a Fam-labeled Ca anti-sense primer: 5’-ACACAGCAGGTTCTGGGTTC-3’. Cycling conditions were: 94°C for 30 s, 58°C for 30 s and 72°C for 1 min for 10 cycles. Run-off products were separated on the basis of length by the Genomics Facility at the Fred Hutchinson Cancer Research Center (Seattle, WA). Using the Genescan 3.7 software (Applied Biosystems), each PCR product is resolved as peaks, with each peak representing TCRs of different CDR3 length (in base pairs). Data were quantitated by determining the total peak height for each Vα-Cα pairing and expressing each fragment as a percentage of total height.
Results
Post-selection thymocyte development is severely impaired in Cre(+) RAG2fl/fl mice
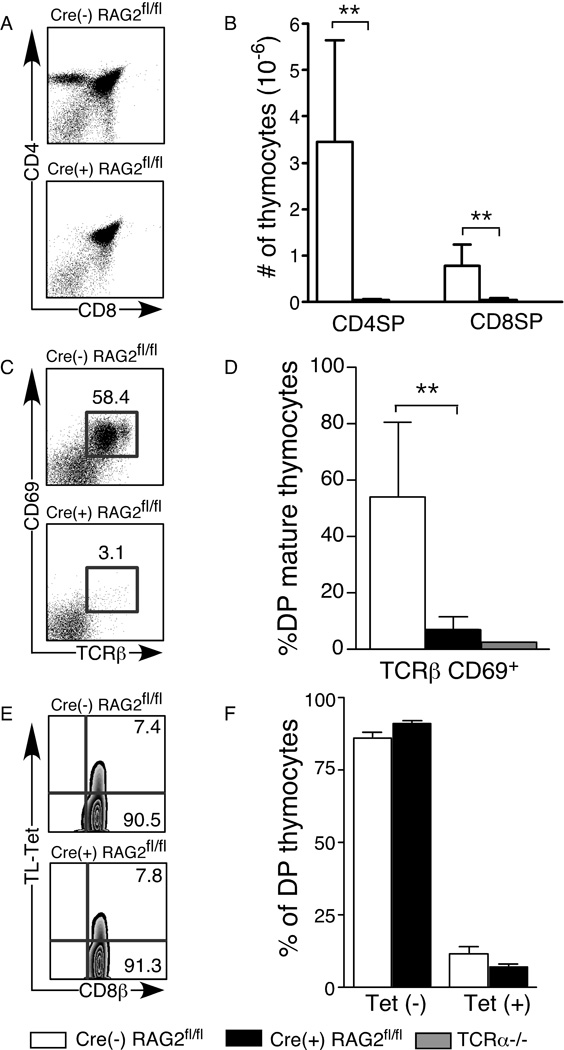
Flow cytometric analysis of thymocytes from Cre(+) RAG2fl/fl, Cre(−) RAG2fl/fl control, and TCRα-deficient (TCRα−/−) mice reveals that, while DN and DP compartments and total thymic cellularity are similar, there is a drastic reduction in both the percent and number of CD4 and CD8 single positive (SP) thymocytes in Cre(+) RAG2fl/fl mice, similar to that seen in mice lacking TCRα (Figure 1A and B and data not shown). In WT mice, the CD8loCD4lo population (“mature” DP) contains a high percentage of TCRαβ+CD69+ thymocytes that have undergone positive selection (20). In Cre(+) RAG2fl/fl mice, there is a greatly reduced percentage of mature DP thymocytes in comparison to controls, suggesting that the majority of DP thymocytes in these mice do not complete positive selection (Fig. 1C and D).
Figure 1. Thymocyte development is impaired in Cre(+) RAG2fl/fl mice.
Flow cytometric analysis of thymocytes from Cre(+) RAG2fl/fl, control Cre(−) RAG2fl/fl and TCRα−/− mice. Thymocytes were surface stained for CD4, CD8α, CD8β, CD69, pan TCRβ and with the TL-tetramer (to detect CD8αα). A) CD4/CD8 profile of total thymocytes. B) Numbers of mature SP (CD4+TCRβ+ and CD8+TCRβ+) thymocytes. C, D) Percent TCRβ+CD69+ of mature DP (CD4loCD8lo) thymocytes. E, F) Thymocytes were gated on CD4+CD8α+ DP cells and the percentage of CD8αβ+CD4+ (“Tet −”) and CD8αβ+CD8αα+CD4+ (“Tet +”) cells was calculated. Data are presented as mean±SD of 4–7 mice in each category and represent 7 independent experiments. Significant differences were calculated using an unpaired, two-tailed Student’s t test and denoted with stars (** p < 0.005).
Some CD4+CD8αβ+ DP thymocytes also express the CD8αα+ homodimer and give rise to the TCRαβ+CD8αα+IEL population (21). Using tetramers of the thymic leukemia (TL) antigen, that bind specifically to CD8ααand not CD8αβ(22), we observed that the percentage of CD4+CD8αβ+CD8αα+triple positive (TP) thymocytes in Cre(+) RAG2fl/fl mice is not significantly different from that in Cre(−) RAG2fl/fl control mice (Fig. 1E and F). Thus, while the thymic precursor pools of TCRαβ+ peripheral and gut T cells remain intact, the post-positive selection thymocyte compartment is greatly diminished in Cre(+) RAG2fl/fl mice.
The TCRαβ+ T cell compartment in secondary lymphoid organs is significantly reduced in Cre(+) RAG2fl/fl mice
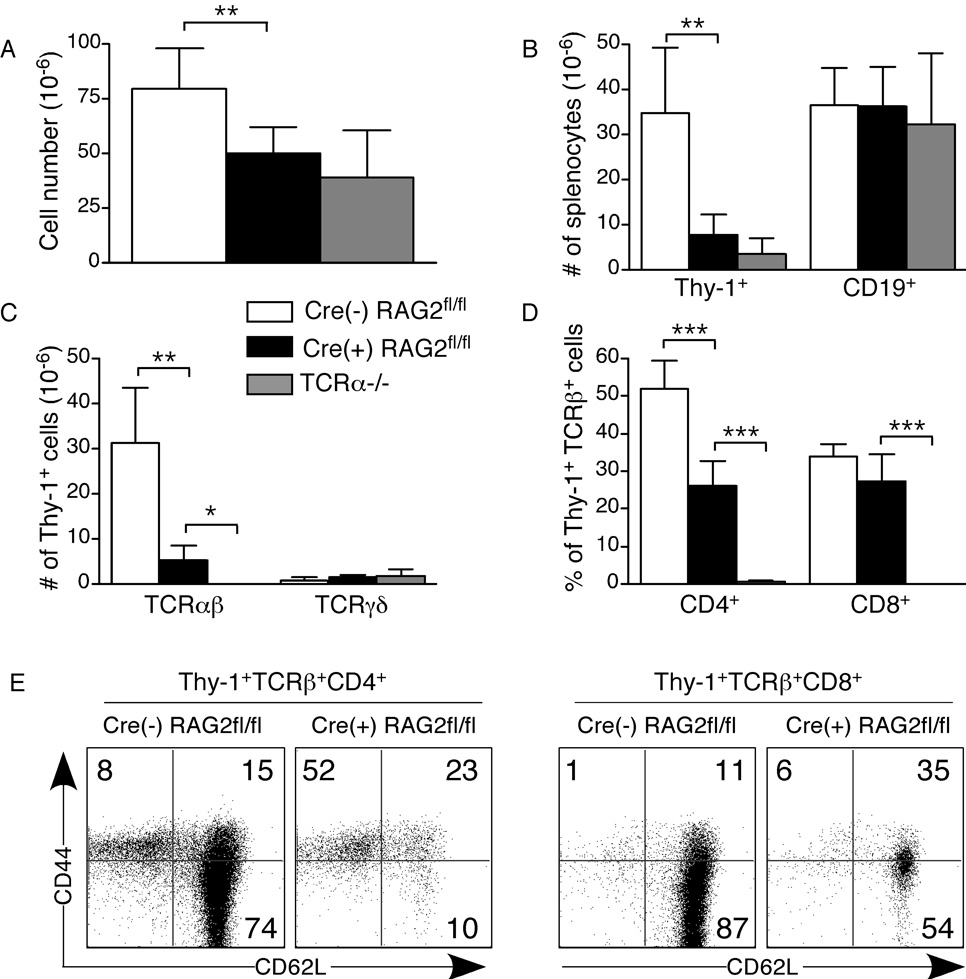
Although thymic output is greatly reduced in Cre(+) RAG2fl/fl mice, there is a small peripheral TCRαβ+ T cell compartment, comprising ~5–7% of total splenocytes (Fig. 2). A similarly reduced compartment is found in peripheral LNs and mesenteric LNs (MLNs) (data not shown). The cellularity of secondary lymphoid organs is reduced in Cre(+) RAG2fl/fl mice, a result of the small T cell compartment, as CD19+ B cell numbers in both spleen and LNs (including MLNs) are similar in Cre(+) RAG2fl/fl and Cre(−) RAG2fl/fl control mice (Fig. 2A and B and data not shown). Within the Thy-1+ compartment, the number of TCRγδ+ T cells is similar in Cre(+) RAG2fl/fl and Cre(−) RAG2fl/fl control mice (Fig. 2C). Interestingly, within the Thy-1+TCRαβ+ T cell compartment, the percentage of CD8+ T cells is similar to that of Cre(−) RAG2fl/fl control mice. In contrast, there is a ~50% reduction in the CD4+ subset, leading to a reduced CD4:CD8 ratio (Fig. 2D and data not shown). Flow cytometric analysis reveals that Thy-1+TCRαβ+ CD4+ and CD8+ splenocytes from Cre(+) RAG2fl/fl mice are CD44hi (Fig. 2E). The CD4+ T cell subset also contains a population that is CD62Llo, while the CD8+ T cell subset does not. Thus, the TCRαβ+ (but not the B cell or TCRγδ+) peripheral compartment is diminished in Cre(+) RAG2fl/fl mice and the remaining cells bear an activated/memory phenotype.
Figure 2. The peripheral TCRαβ+ T cell compartment is compromised in Cre(+) RAG2fl/fl mice.
Flow cytometric analysis of splenocytes from Cre(+) RAG2fl/fl, control Cre(−) RAG2fl/fl, and TCRα−/− mice. Splenocytes were surface stained for CD19, Thy-1.2, pan TCRβ, TCRγδ, CD4 and CD8. A) Total number of splenocytes. B) Numbers of Thy-1+ and CD19+ splenocytes. C) Numbers of Thy-1+TCRβ+ and Thy-1+TCRγδ+ splenocytes. D) Percentage of Thy-1+TCRβ+ splenocytes that are CD4+ or CD8+. E) Thy-1+TCRβ+ splenocytes were gated on CD4 and CD8 and analyzed for CD44 and CD62L expression. Data are presented as mean±SD for 4–10 mice in each category and represent 9 independent experiments. Significant differences were calculated using an unpaired, two-tailed Student’s t test and denoted with stars (* p < 0.05, **p<0.005, ***p<0.0005).
T cells from Cre(+) RAG2fl/fl mice are hyperproliferative and produce high levels of IFN-γ upon stimulation
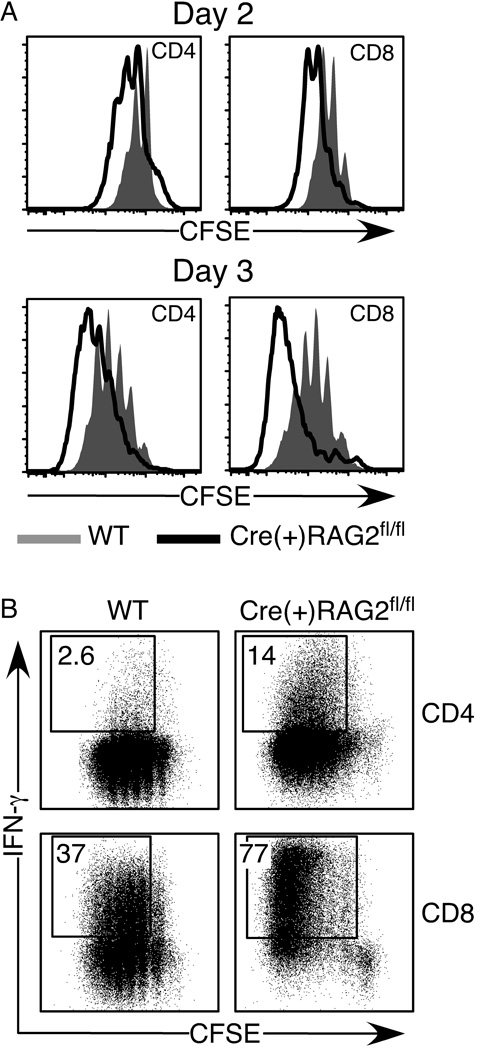
We next assessed whether splenic and LN T cells from Cre(+) RAG2fl/fl mice are functional. Within 48 hours of anti-CD3/anti-CD28 stimulation, both CD4+ and CD8+ T cells from Cre(+) RAG2fl/fl mice have proliferated more than cells from WT mice (Fig. 3A). By day 3, most Cre(+) RAG2fl/fl T cells have undergone more than 4 divisions. In addition to being hyperproliferative, the proportion of IFN-γ producers among Cre(+) RAG2fl/fl T cells is 2 to 5-fold higher than that of the WT cultures (Fig. 3B).
Figure 3. Cre(+) RAG2fl/fl T cells are hyperproliferative and display rapid effector function upon stimulation.
CFSE-labeled CD4+ and CD8+ splenic and LN T cells from WT and Cre(+) RAG2fl/fl mice were stimulated with anti-CD3/anti-CD28 beads for 2 and 3 days, and then stained for intracellular IFN-γ. Cells were gated as CD4+ or CD8+. A) Shaded histograms represent WT, and open histograms represent Cre(+) RAG2fl/fl cells. B) Numbers represent the percentage of IFN-γ producing cells among those cells that have undergone at least one division.
TCRαβ+ T cells in Cre(+) RAG2fl/fl mice display a biased TCRα repertoire
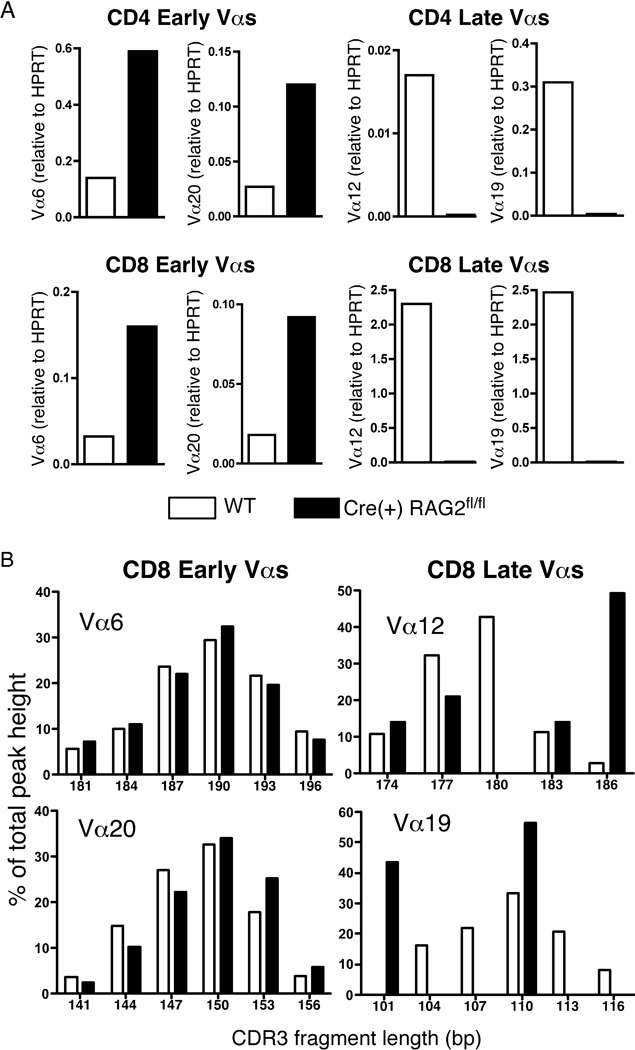
Quantitative RT-PCR on RNA from purified CD8+ and CD4+ T cells reveals that, while Vα6 and Vα20 expression levels are higher in Cre(+) RAG2fl/fl T cells compared to WT, Vα12 and Vα19 expression levels are significantly lower (Fig. 4A). All four genes are expressed at similar levels in WT T cells (data not shown) indicating that, in a mouse with a polyclonal repertoire, late rearranging Vα genes are well-represented. Spectratype analysis was performed to assess the diversity of CDR3 lengths in expressed TCRα chains in CD8+ splenocytes. This PCR-based assay allows the determination of CDR3 lengths within a specific Vα-Cα pairing and gives a clear indication of the clonality of the population (23). In line with our hypothesis, early TCRα rearrangements generate a Gaussian distribution of CDR3 lengths in both WT and Cre(+) RAG2fl/fl mice, while the CDR3 lengths for late Vα rearrangements are less diverse and less evenly distributed in Cre(+) RAG2fl/fl mice relative to WT (Fig. 4B).
Figure 4. TCR repertoire is skewed to early Vα usage in Cre(+) RAG2fl/fl mice.
A) Quantitative RT-PCR to determine “early” (Vα6, Vα20) and “late” (Vα12, Vα19) Va gene usage by CD4+ and CD8+ T cells from Cre(+) RAG2fl/fl and WT control mice. Data are presented as Vα expression relative to HPRT expression and are representative of 2 independent experiments for a total of 8 mice in each category. B) CDR3 length spectratype analysis of TCRα chains expressed by CD8+ splenocytes from Cre(+) RAG2fl/fl and WT mice. Data are expressed as a percentage of total peak height for each CDR3 fragment, whose lengths are denoted in base pairs (bp). Data are representative of 2 independent experiments for a total of 8 mice in each category.
The phenotype in the GALT is distinct from that in the periphery of Cre(+) RAG2fl/fl mice
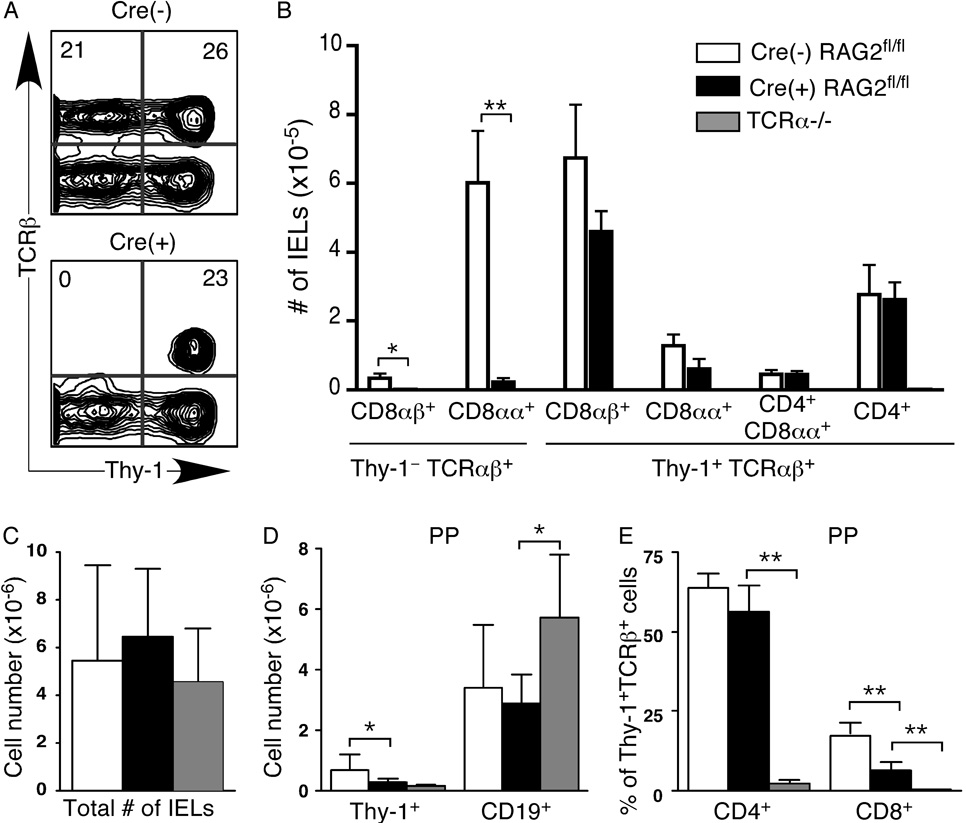
In clear contrast to the TCRαβ+ compartment in spleen and LNs, the Thy-1+TCRαβ+ IEL compartment is unimpaired in Cre(+) RAG2fl/fl mice, with no significant difference in the numbers of CD8αβ+, CD8αα+, CD4+CD8αα+ and CD4+ T cells (Fig. 5A and B). Surprisingly, the Thy-1−TCRαβ+ IEL compartment is almost completely absent in Cre(+) RAG2fl/fl mice (Fig. 5A and B). In contrast to the secondary lymphoid organs and PPs, the total numbers of IELs are not significantly different between Cre(+) RAG2fl/fl and Cre(−) RAG2fl/fl control mice (Fig. 5C). In the PPs of Cre(+) RAG2fl/fl mice, as in the spleen and LNs, the Thy-1+ compartment is significantly reduced, while the CD19+ B cell compartment appears normal in size (Fig. 5D).Within the Thy-1+TCRαβ+ compartment, the CD4+ population in Cre(+) RAG2fl/fl mice is similar in size to that of Cre(−) RAG2fl/fl controls, while the CD8+ compartment is reduced (Fig. 5E). Thus, in contrast to spleen and LNs, the PP CD4 compartment is less compromised in Cre(+) RAG2fl/fl mice. However, we find similar defects in total cellularity and a reduced TCRαβ+ T cell population in the PPs. Interestingly, the IEL compartment has a distinct phenotype in Cre(+) RAG2fl/fl mice. These data support the notion that Thy-1+TCRαβ+ IELs are the progeny of thymocytes that have undergone early TCR rearrangements, and that Thy-1−TCRαβ+ IELs represent a distinct T cell lineage.
Figure 5. The Thy-1−TCRαβ+ IEL population is absent in Cre(+) RAG2fl/fl mice.
Flow cytometric analysis of A–C) small intestinal IELs and D, E) PP cells from Cre(+) RAG2fl/fl, control Cre(−) RAG2fl/fl and TCRα−/− mice. Cells were surface stained for Thy-1.2, pan TCRβ, TCRγδ, CD4, CD8α, CD8.2β and CD19. A) Representative flow cytometric plots showing Thy-1 and TCRβ expression on small intestinal IELs. Numbers represent percent of cells in that quadrant. B) Number of IELs in each TCRβ+ IEL subset. C) Total number of IELs in the small intestine. D) Numbers of Thy-1+ and CD19+ cells in PPs. E) Percentage of Thy-1+TCRβ+ T cells in the PPs that are CD4+ or CD8+. Data are presented as mean±SD for 4–8 mice in each category and represent 7 independent experiments. Significant differences were calculated using an unpaired, two-tailed Student’s t test and denoted with stars (* p < 0.05, **p<0.005).
In the presence of competition, bone marrow from Cre(+) RAG2fl/fl mice does not contribute to the peripheral TCRαβ+ T cell population
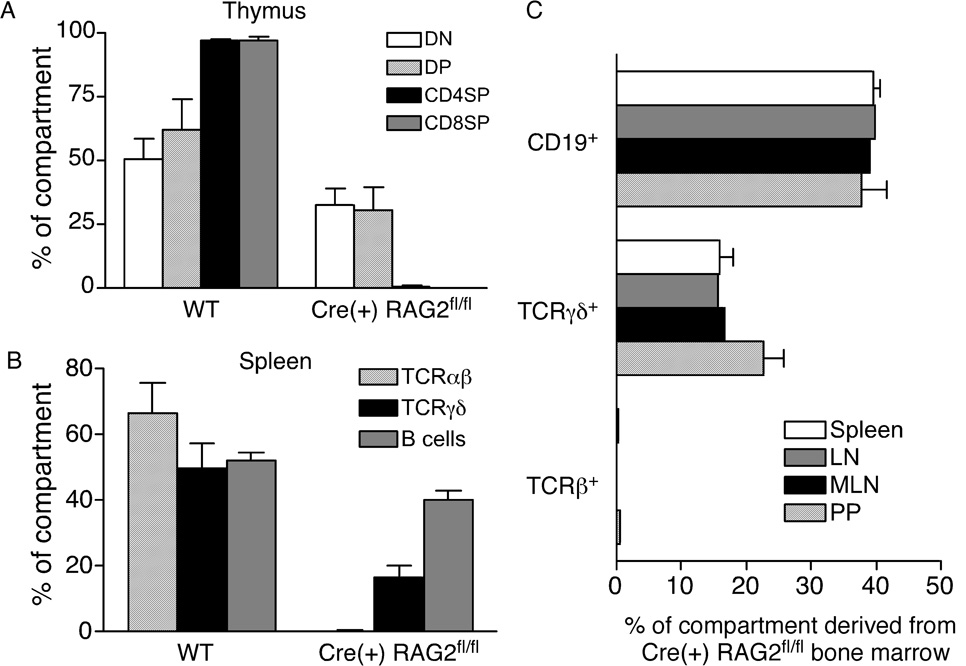
To determine whether T cells possessing early TCR rearrangements preferentially seed certain peripheral compartments, we made radiation chimeras using a 50:50 mixture of bone marrow from WT (Ly5.1+) and Cre(+) RAG2fl/fl (Ly5.2+) mice to reconstitute lethally irradiated WT hosts (Ly5.1+Ly5.2+). After 8 weeks, the contribution of each donor cell type to the thymus, spleen, LNs and PPs was determined. Cre(+) RAG2fl/fl and WT bone marrow repopulate the DN and DP thymocyte compartments equally well (Fig. 6A). In contrast, the CD4SP and CD8SP populations are comprised almost entirely of cells derived from WT bone marrow. In the spleen, LNs, MLNs and PPs, Cre(+) RAG2fl/fl bone marrow contributes only to the B cell pool and the TCRγδ+, but not TCRαβ+, T cell population (Fig. 6B and C).
Figure 6. Cre(+) RAG2fl/fl bone marrow does not contribute to the peripheral TCRαβ+ T cell population in mixed chimeras.
Flow cytometric analysis of reconstitution by WT and Cre(+) RAG2fl/fl donor bone marrow in mixed radiation chimeras. Cells were surface stained for Ly5.1 and Ly5.2 to distinguish between donor and host populations. A) Thymocytes from 6 individual chimeras were surface stained for CD4 and CD8 and gated on CD4−CD8− DN, CD4+CD8+ DP and CD4SP and CD8SP populations. B) Splenocytes from 6 individual chimeras were surface stained for CD19, pan TCRβ and TCRγδ. For panels A and B, data are expressed as the percentage of cells of WT or Cre(+) RAG2fl/fl origin. C) Cells from spleen (n=6), LN (n=2), MLN (n=2) and PP (n=4) were surface stained for CD19, pan TCRβ and TCRγδ. Data are expressed as the percentage of cells of Cre(+) RAG2fl/fl origin found in those populations.
Thymocytes derived from Cre(+) RAG2fl/fl bone marrow are found in the TP population, but do not develop into IELs
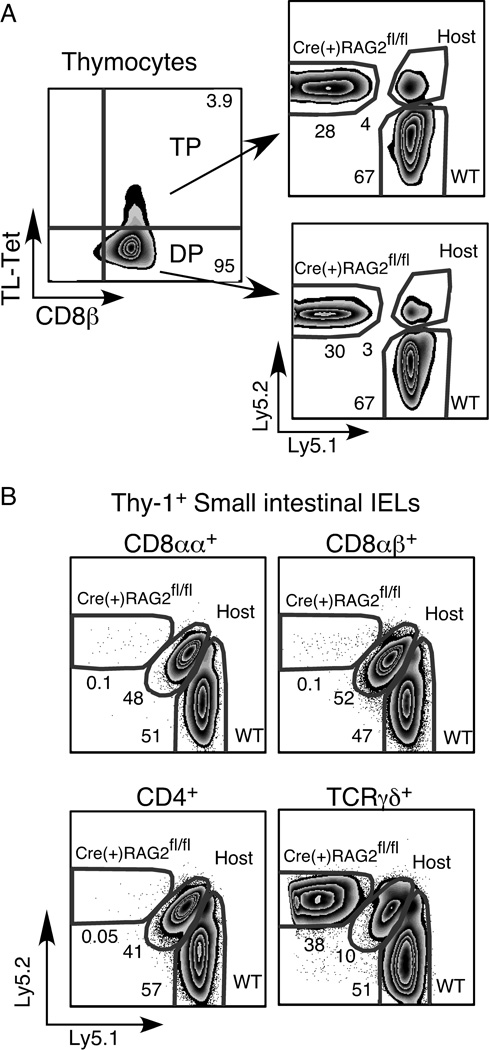
Although approximately 30% of the CD8αβ+CD4+CD8αα+ TP thymocyte population is derived from Cre(+) RAG2fl/fl bone marrow (Fig. 7A), TP thymocytes of Cre(+) RAG2fl/fl origin are unable to seed either the Thy-1+ or Thy-1− TCRαβ+ IEL compartments (Fig. 7B and data not shown). In contrast, Cre(+) RAG2fl/fl bone marrow readily contributes to the TCRγδ+ IEL population. Interestingly, while only 3–4% of thymocytes are of host origin, in the IEL population, 40–50% of the TCRαβ+ IELs are derived from the host.
Figure 7. In mixed chimeras, Cre(+) RAG2fl/fl bone marrow contributes to the TP thymocyte population but not to IEL.
Flow cytometric analysis of A) thymocytes, and B) IELs from mixed radiation chimeras. Cells were surface stained for Ly5.1 and Ly5.2 to distinguish between donor and host populations. A) Thymocytes were surface stained for CD4, CD8α, CD8.2β and with TL-tetramer and were gated on the CD4+CD8α+ DP population before subdividing the cells into DP and TP subsets as shown. Data are representative of 4 individual chimeras. B) IELs were surface stained for Thy-1.2, pan TCRβ, TCRγδ, CD4, CD8α and CD8β IELs were gated on TCRγδ+ or Thy-1+TCRβ+ and the CD8αα+, CD8αβ+ and CD4+ subsets were analyzed for reconstitution by donor bone marrow. Data are representative of 6 individual chimeras.
Discussion
We describe a mouse model in which peripheral T cells are the progeny of thymocytes that have undergone early TCR rearrangements. In these mice, the timing of RAG2 deletion is consistent with previous data showing that in CD4-Cre transgenic mice, Cre expression is initiated at the DN3 stage (15). Phenotypic analysis of the thymus in Cre(+) RAG2fl/fl mice and bone marrow chimeras shows that defects in thymocyte development first become noticeable at the mature DP stage, consistent with a loss of RAG2 in the late DN stage. Consistent with this phenotype, there is a drastic decrease in the CD4SP and CD8SP thymocyte populations. Despite this reduction in thymic output, there are measurable T cell populations in secondary lymphoid organs. Quantitative RT-PCR and spectratype analysis of select Vα genes shows that in Cre(+) RAG2fl/fl mice, deletion of RAG2 effectively limits TCRα rearrangements, maintaining a polyclonal, “early” repertoire in the spleen. Furthermore, the few peripheral T cells in Cre(+) RAG2fl/fl mice have not failed to excise RAG2, as the floxed alleles were excised in 91% and 94% of CD8+ and CD4+ T cells, respectively, as determined by PCR (data not shown). Overall, these data indicate that the majority of T cells in Cre(+) RAG2fl/fl mice bear early-rearranged TCRα chains.
Our data also show that Cre-mediated deletion of RAG2 is specific to T cells and that expression of Cre is not toxic to lymphocytes. B cell numbers are normal in intact Cre(+) RAG2fl/fl mice, and in bone marrow chimeras, Cre(+) RAG2fl/fl and WT bone marrow contribute equally to the B cell population. Despite reduced representation of CD4+ T cells in the spleen and LNs, the phenotype in the GALT suggests that continual Cre expression does not adversely affect cell development or homeostatic proliferation. Additionally, when in competition with WT stem cells, Cre(+) RAG+/+ transgenic bone marrow contributes equally to all T cell populations in mixed bone marrow chimeras (data not shown).
We reasoned that, due to decreased thymic output, the T cell compartment in the secondary lymphoid organs of Cre(+) RAG2fl/fl mice may be expanded by homeostatic proliferation. CD8+ T cells from Cre(+) RAG2fl/fl mice are CD44hiCD62Lhi, while the majority of CD4+ T cells are CD44hiCD62Llo, likely the result of lymphopenia-induced homeostatic proliferation. Indeed, Locksley and colleagues have shown a similar phenotype in their mouse model of lymphopenia, the CD4Cre/R-DTA mouse (24). Furthermore, stimulated CD4+ and CD8+ T cells from Cre(+) RAG2fl/fl mice are functional memory cells, undergoing rapid proliferation and higher levels of IFN-γ secretion compared to WT T cells.
In WT mice, the population of thymocytes undergoing TCRα gene rearrangements before the DP stage is presumably sparse and untrackable. We developed the Cre(+) RAG2fl/fl mouse model to determine whether this early lineage of T cells has the potential to seed certain compartments preferentially. This appears to be the case, as the Thy-1+TCRαβ+ IEL compartment of Cre(+) RAG2fl/fl contains relatively normal numbers, while the Thy-1-TCRαβ+ compartment does not develop. Although the biological function of Thy-1 remains controversial, studies in germ-free mice suggest that Thy-1 expression on at least some subsets of IELs is the result of TCR-mediated activation by external stimuli, most likely intestinal antigens (25). As many of our control and experimental mice are co-housed littermates maintained under specific pathogen free conditions, the difference in IEL Thy-1 expression between Cre(+) RAG2fl/fl and control mice is likely not solely due to antigenic stimuli. However it is possible that the skewed repertoire in Cre(+) RAG2fl/fl mice results in a population of IELs that are more responsive to activation by intestinal antigens. There is also evidence that Thy-1 may be involved in T cell costimulation and homeostasis (26). Thus the lack of a Thy-1− IEL compartment in Cre(+) RAG2fl/fl mice may be due to the need for these cells to undergo lymphopenia-induced homeostasis, leading to the maintenance of Thy-1 cell surface expression. This seems unlikely, given that in athymic nude mice, TCRαβ+CD8αα+ IELs are virtually absent, but nearly all are Thy-1−(27). Furthermore, in bone marrow chimeras rendered transiently lymphopenic by irradiation, a substantial Thy-1− IEL population is present from days 1 to 56 post reconstitution (12). In Cre(+) RAG2fl/fl mice, the TCRγδ+ IEL population has a substantial Thy-1− population, suggesting that Thy-1 expression is not an obligate product of the lymphopenic environment in these mice. Instead, it is more likely that the Thy-1− subset represents a separate T cell lineage whose development occurs later or whose function relies on a distinct TCR repertoire. This conclusion is further supported by evidence from LeFrancois and colleagues, who showed that CD8ααTCRαβ Thy-1− and Thy-1+ IELs exhibit differential requirements for IL-15 signaling, including the mechanism of IL-15 delivery (28).
Our data also point to the remarkable homeostatic properties of the IEL population. Despite lymphopenia in secondary lymphoid organs, Cre(+) RAG2fl/fl mice maintain a full IEL compartment. In addition, cells of host origin account for nearly 50% of the IEL pool in bone marrow chimeras, while less than 5% of thymocytes are of host origin. Together, these data suggest that the small populations of IELs and thymic emigrants can fill the available niche in adult lymphopenic mice. It is unclear how these populations would compete for seeding the IEL compartment as it is first established neonatally. The gut environment, where commensal flora provide ample foreign antigen, likely drives this dramatic homeostatic proliferation (29).
Lymphopenia, and the homeostatic proliferation driven by this condition, may lead to autoimmunity in both humans and mice (30). Despite being lymphopenic and possessing a limited TCR repertoire, aged Cre(+) RAG2fl/fl mice do not appear to suffer from systemic autoimmunity. We found no increase in the frequency of anti-nuclear antibodies in the serum of aged (>1 year) Cre(+) RAG2fl/fl mice relative to WT (data not shown). In addition, we found that the percentage of CD4+Foxp3+ T cells in young adult Cre(+) RAG2fl/fl mice was similar to that found in WT mice, except in the spleen, which contained a higher percentage of these regulatory cells (data not shown).
In summary, although comprising a sparse population, T cells bearing the products of early TCR rearrangements can populate the Thy-1+TCRαβ+ IEL compartment and undergo homeostasis-driven proliferation to fill this niche. This property may contribute to the oligoclonal nature of the IEL population and may explain why this oligoclonal repertoire varies, even between genetically identical mice exposed to the same environment (33). Our data suggest that a small number of IEL precursors may efficiently populate the gut, and this property may have also contributed to the historical confusion surrounding the extrathymic versus intrathymic derivation of the IEL compartment. The CD8αα+ IEL subset has several unique properties that suggest a non-conventional pathway of development. These cells appear early in ontogeny, and they have a self-reactive and oligoclonal repertoire (31–35). The presence of IELs in neonatally thymectomized and nude mice led to the hypothesis that the thymus may be dispensable for the development of this population. However, recent fate mapping (36) and adoptive transfer (21) experiments have shown that the majority of CD8αα+ IELs are the progeny of DP thymocytes. Development of IELs may continue post-thymically, given that IELs may develop from CD4−CD8−TCRβ− triple negative thymic emigrants in cryptopatches within the gut (37). Our data suggest that the capacity for homeostasis in the IEL compartment can mask defects by driving colonization of the gut by a small number of T cells, whether thymically- or extrathymically-derived.
Acknowledgments
We thank Drs. Klaus Rajewsky, Christopher Wilson, Alexander Rudensky and Hilde Cheroutre for reagents and mice, and Kristina Bavik and Dora Gyarmati for excellent technical assistance and helpful discussion.
Footnotes
This work was supported by National Institutes of Health Grants AG13078 and AI064318 (P.J.F.) and National Cancer Institute Basic and Cancer Immunology Grant (D.W.H.)
Abbreviations used in this paper: DN, double negative; DP, double positive; HPRT, hypoxanthine guanine phosphoribosyltransferase; IEL, intraepithelial lymphocyte; LN, lymph node; MLN, mesenteric lymph node; PP, Peyer’s patch; TL, thymic leukemia; WT, wild-type.
References
- 1.Hawwari A, Krangel MS. Regulation of TCR δand α repertoires by local and long-distance control of variable gene segment chromatin structure. J. Exp. Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jα promoters. Nat. Immunol. 2005;6:481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C-Y, Kanagawa O. Ordered and coordinated rearrangement of the TCRα locus: role of secondary rearrangement in thymic selection. J. Immunol. 2001;166:2597–2601. doi: 10.4049/jimmunol.166.4.2597. [DOI] [PubMed] [Google Scholar]
- 4.Pasqual N, Gallagher M, Aude-Garcia C, Loiodice M, Thuderoz F, Demongeot J, Ceredig R, Marche PN, Jouvin-Marche E. Quantitative and qualitative changes in V-J α rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J. Exp. Med. 2002;196:1163–1173. doi: 10.1084/jem.20021074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez-Munain C, Sleckman BP, Krangel MS. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity. 1999;10:723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- 6.Mancini SJ, Candeias SM, Di Santo JP, Ferrier P, Marche PN, Jouvin-Marche E. TCRα gene rearrangement in immature thymocytes in absence of CD3, pre-TCR, and TCR signaling. J. Immunol. 2001;167:4485–4493. doi: 10.4049/jimmunol.167.8.4485. [DOI] [PubMed] [Google Scholar]
- 7.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 8.Egawa T, Kreslavsky T, Littman DR, von Boehmer H. Lineage diversion of T cell receptor transgenic thymocytes revealed by lineage fate mapping. PLoS ONE. 2008;3:e1512. doi: 10.1371/journal.pone.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha B, von Boehmer H, Guy-Grand D. Selection of intraepithelial lymphocytes with CD8 α/α co-receptors by self-antigen in the murine gut. Proc. Natl. Acad. Sci. USA. 1992;89:5336–5340. doi: 10.1073/pnas.89.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin TA, Sandau MM, Jameson SC, Hogquist KA. The timing of TCRα expression critically influences T cell development and selection. J. Exp. Med. 2005;202:111–121. doi: 10.1084/jem.20050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheroutre H. IELs: enforcing law and order in the court of the intestinal epithelium. Immunol. Rev. 2005;206:114–131. doi: 10.1111/j.0105-2896.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 12.Mosley RL, Klein JR. Repopulation kinetics of intestinal intraepithelial lymphocytes in murine bone marrow radiation chimeras. Transplantation. 1992;53:868–874. doi: 10.1097/00007890-199204000-00030. [DOI] [PubMed] [Google Scholar]
- 13.Hao Z, Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 2001;194:1151–1163. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker qSM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 15.Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 16.Lefrancois L. Isolation of mouse small intestinal intraepithelial lymphocytes, Peyer's patch, and lamina propria cells. Curr. Protoc. Immunol. 1996:3.19.11–13.19.16. doi: 10.1002/0471142735.im0319s17. [DOI] [PubMed] [Google Scholar]
- 17.Pannetier C, Even J, Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 18.DiRienzo CG, Murphy GF, Jones SC, Korngold R, Friedman TM. T-cell receptor Vα spectratype analysis of a CD4-mediated T-cell response against minor histocompatibility antigens involved in severe graft-versus-host disease. Biol. Blood Marrow Transplant. 2006;12:818–827. doi: 10.1016/j.bbmt.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson G, Hare KJ, Jenkinson EJ. Positive selection of thymocytes: The long and winding road. Immunol. Today. 1999;20:463–468. doi: 10.1016/s0167-5699(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 21.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 23.Pannetier C, Cochet M, Carche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T cell receptor β chains vary as a function of the recombined germ-line segments. Proc. Natl. Acad. Sci. USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced "memory-like" T cells in constitutively T cell-depleted mice. J. Immunol. 2008;180:4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefrancois V, Goodman T. In vivo modulation of cytolytic activity and Thy-1 expression in TCR-γδ+ intraepithelial lymphocytes. Science. 1989;243:1716–1718. doi: 10.1126/science.2564701. [DOI] [PubMed] [Google Scholar]
- 26.Haeryfar SM, Hoskin DW. Thy-1: more than a mouse pan-T cell marker. J. Immunol. 2004;173:3581–3588. doi: 10.4049/jimmunol.173.6.3581. [DOI] [PubMed] [Google Scholar]
- 27.Rocha B. The extrathymic T-cell differentiation in the murine gut. Immunol. Rev. 2007;215:166–177. doi: 10.1111/j.1600-065X.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 28.Schluns KS, Nowak EC, Cabrera-Hernandez A, Puddington L, Lefrancois L, Aguila HL. Distinct cell tyes control lymphoid subset development by means of IL-15 and IL-15 receptor α expression. Proc. Natl. Acad. Sci. USA. 2004;101:5616–5621. doi: 10.1073/pnas.0307442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieper WC, Troy A, Burghardt JT, Ramsey C, Lee JY, Jiang H-Q, Dummer W, Shen H, Cebra JJ, Surh CD. Cutting edge: Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J. Immunol. 2005;174:3158–3163. doi: 10.4049/jimmunol.174.6.3158. [DOI] [PubMed] [Google Scholar]
- 30.King C, Ilic A, Koelsch K, Sarvetnick N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell. 2004;117:265–277. doi: 10.1016/s0092-8674(04)00335-6. [DOI] [PubMed] [Google Scholar]
- 31.Latthe M, Terry L, MacDonald TT. High frequency of CD8 αα homodimer-bearing T cells in human fetal intestine. Eur. J. Immunol. 1994;24:1703–1705. doi: 10.1002/eji.1830240737. [DOI] [PubMed] [Google Scholar]
- 32.Lin T, Matsuzaki G, Yoshida H, Kenai H, Omoto K, Umesue M, Singaram C, Nomoto K. Thymus ontogeny and the development of TCRαβ intestinal intraepithelial lymphocytes. Cell. Immunol. 1996;171:132–139. doi: 10.1006/cimm.1996.0183. [DOI] [PubMed] [Google Scholar]
- 33.Rocha B, Vassalli P, Guy-Grand D. The Vβ repertoire of mouse gut homodimeric α CD8+ intraepithelial T cell receptor α/β+ lymphocytes reveals a major extrathymic pathway of T cell differentiation. J. Exp. Med. 1991;173:483–486. doi: 10.1084/jem.173.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regnault A, Cumano A, Vassalli P, Guy-Grand D, Kourilsky P. Oligoclonal repertoire of the CD8 alpha alpha and the CD8 alpha beta TCR-alpha/beta murine intestinal intraepithelial T lymphocytes: evidence for the random emergence of T cells. J. Exp. Med. 1994;180:1345–1358. doi: 10.1084/jem.180.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arstila V, Arstila TP, Calbo S, Selz F, Malassis-Seris M, Vassalli P, Kourilsky P, Guy-Grand D. Identical T cell clones are located within the mouse gut epithelium and lamina propria and circulate in the thoracic duct lymph. J. Exp. Med. 2000;191:823–834. doi: 10.1084/jem.191.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eberl G, Littman DR. Thymic origin of intestinal αβ T cells revealed by fate mapping of ROR γt+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 37.Lambolez F, Arcangeli ML, Joret AM, Pasqualetto V, Cordier C, Di Santo JP, Rocha B, Ezine S. The thymus exports long-lived fully committed T cell precursors that can colonize primary lymphoid organs. Nat. Immunol. 2006;7:76–82. doi: 10.1038/ni1293. [DOI] [PubMed] [Google Scholar]