Abstract
DNA double-strand breaks repaired through nonhomologous end joining require no extended sequence homology as a template for the repair. A subset of end-joining events, termed microhomology-mediated end joining, occur between a few base pairs of homology, and such pathways have been implicated in different human cancers and genetic diseases. Here we investigated the effect of exposure of yeast and mammalian cells to ionizing radiation on the frequency and mechanism of rejoining of transfected unirradiated linear plasmid DNA. Cells were exposed to γ radiation prior to plasmid transfection; subsequently the rejoined plasmids were recovered and the junction sequences were analyzed. In irradiated yeast cells, 68% of recovered plasmids contained microhomologies, compared to only 30% from unirradiated cells. Among them 57% of events used ≥4 bp of microhomology compared to only 11% from unirradiated cells. In irradiated mammalian cells, 54% of plasmids used ≥4 bp of microhomology compared to none from unirradiated cells. We conclude that exposure of yeast and mammalian cells to radiation prior to plasmid transfection enhances the frequency of microhomology-mediated end-joining events in trans. If such events occur within genomic locations, they may be involved in the generation of large deletions and other chromosomal aberrations that occur in cancer cells.
INTRODUCTION
DNA double-strand breaks (DSBs) induced by ionizing radiation are the most severe form of DNA damage, and if not repaired they result in cell death (1). In yeast and mammalian cells, two major pathways direct the repair of DSBs, homologous recombination (HR) (2) and nonhomologous end joining (NHEJ) (3).
HR is a conventional DSB repair pathway that uses extensive sequence homology as the template for repair. In Saccharomyces cerevisiae, HR is the dominant mechanism for DNA DSB repair (4-6), whereas HR occurs infrequently in mammalian cells (7). NHEJ joins two broken DNA ends together in the absence of extended sequence homology (8). NHEJ is the predominant mechanism of repair in mammalian cells, but it is a minor mechanism of DSB repair in yeast and is evident only in the rad52 background (9) or in the absence of any sequence homology (10).
The NHEJ pathway has been shown to maintain genomic stability and to suppress tumorigenesis (11). In yeast, the main genetic factors of the NHEJ pathway include the endbinding Hdf1/Hdf2 (yKu70/yKu80) heterodimer, the Mre11/Rad50/Xrs2 complex, and Dnl4/Lif1 ligase complex (12). In mammalian cells, a central component of NHEJ repair is the presence of the DNA-PK complex, comprised of the Ku70 and Ku86 heterodimer (13) and DNA-PKcs repair complex (14). The Ku protein recruits DNA-PKcs and becomes phosphorylated, leading to the recruitment of other enzymes involved in repair (14). Broken DNA strands re-ligate through XRCC4-ligase IV complex action (15).
Microhomology-mediated end joining (MMEJ) repairs DSBs based on a few base pairs of homology between broken DNA ends and is distinct from the typical NHEJ pathway. In yeast, MMEJ is independent of Ku or Rad52 but is dependent on Rad1, Mre11 and Rad50 and partly dependent on Dnl4 (16-18). In mammalian cells, MMEJ is independent of Ku86, ligase IV and XRCC4 (19-21) and dependent on Nbs1 and Fen-1 (22). In addition, mismatch repair proteins such as Mlh1 and Msh2 modulate the occurrence of MMEJ (23, 24).
MMEJ is usually associated with the formation of deletions that may lead to chromosomal aberrations (25, 26). Genomic rearrangements generated by MMEJ are found in some human cancers and genetic diseases. In bladder tumor cells and head and neck cancer cells, DSBs rejoined by MMEJ contribute to high levels of genomic instability and promote tumor formation (27, 28). About 50% of large deletions associated with genetic diseases are products of MMEJ (29-31). Microhomology is often present at the junctions of radiation-induced genomic rearrangements (25, 26), suggesting a possible link between radiation-induced MMEJ and radiation-induced carcinogenesis.
In yeast, repair of HO-endonuclease-induced chromosomal DSBs showed microhomologies at the breakpoints (9, 32). Previous studies have shown that ionizing radiation enhances the frequency of nonhomologous integration in yeast and mammalian cells (33, 34). In addition, other damaging agents, including camptothecin, VP16, UV radiation and hydrogen peroxide, increase nonhomologous integration of transfected plasmids in mammalian cells (35-37). However, extensive sequence analysis of DNA damage-induced illegitimate recombination events has yet to be carried out.
We reported recently that radiation and restriction enzymes induce plasmid integration into the yeast genome using microhomology (38). However, in this paper, we investigate the effect of radiation on extrachromosomal MMEJ using a plasmid-based end-joining assay in yeast (39) and in mammalian cells (19). These experiments were designed to physically separate radiation damage from recombination substrate transfection to determine whether radiation-induced damages induce MMEJ events in trans.
MATERIALS AND METHODS
Yeast
Strains
Experiments were performed in the haploid Saccharomyces cerevisiae strain RSY12 (MATa leu2-3, 112 his3-11, 15 ura3Δ::HIS3), in which the entire URA3 open reading frame and promoter sequence was replaced by the HIS3 gene (40). E. coli strain DH5α was used for the maintenance and amplification of plasmid DNA.
Plasmids
YEplac195 contains the URA3 marker for selection, the 2-μm origin of replication, multi-cloning site (MCS), and the Ampr resistance gene was used to control for transformation efficiency (39).
Transformation
For yeast transformation the Lithium Acetate/Single Stranded DNA/PEG method was used (41).
Plasmid end-joining assay in yeast
To prepare the recombinational substrate for end joining, plasmid YEplac195, carrying a URA3 marker, a 2-μm origin of replication, and an AmpR resistance gene (39), was double-digested with HindIII and KpnI restriction endonucleases (within the MCS) at 37°C overnight. To test for complete digestion, an aliquot of the digest was re-ligated and transformed into E. coli, and transformants originated from partially digested DNA were counted. Complete double-digested DNA was purified by phenol:chloroform extraction and collected after ethanol precipitation. About 100-200 ng of HindIII/KpnI-linearized plasmid DNA was transformed into the wild-type RSY12 yeast strain lacking URA3 (40) using the transformation protocol described previously (41). In parallel, about 100 ng of uncut YEplac195 plasmid was transformed into the same strain to control for the transformation efficiency. After 4 days of growth, Ura+ transformants were selected. The efficiency of end joining was measured by normalizing the number of Ura+ transformants that had arisen after transformation with the linearized plasmid in comparison to that with the uncut plasmid.
Recovery of the recombined end-joining plasmids and sequencing of the junctions
Ura+ colonies were selected from the transformation with linearized plasmids and grown in synthetic complete medium lacking uracil. Yeast plasmid DNAs from each of the Ura+ transformants were purified using glass beads (42), transformed into E. coli DH5α, and plated onto LB+Amp plates to select colonies harboring the rescued YEplac195 plasmid. Plasmid DNA from single colonies was purified using a miniprep purification kit (Qiagen, Santa Clarita, CA). Isolated plasmids were first digested with BamHI, located between the HindIII and KpnI sites that should already be cut, to exclude any plasmid DNA that resulted from recircularization of a single-digested plasmid caused by partial HindIII or KpnI digestion. Selected plasmids were digested with HindIII, which generates two fragments, one containing the 1.1-kb URA3 fragment and a second fragment whose size was used to estimate for the size of the rejoined plasmid. The junction site of each rejoined plasmid was sequenced using primers located upstream and downstream of the MCS; forward primers 195-7 5′-ATACGCAAACCGCCTCTCC-3′ and 195-5153 5′-TTGGCATAATGGCGGAAA CTC-3′ and reverse primers 195-1103 5′-GCATTATAGAGCGCACAAAGG-3′ and 195-2106 5′-CAGCTTATCATCGGATCGATCC-3′.
Cell cycle arrest
Yeast cells were grown overnight, diluted in YPD, and exposed to methylbenzimadazole-2-yl-carbamate (MBC) (Fisher Scientific, Houston, TX). MBC was prepared as a 10-mg/ml solution in DMSO and added to cells at a final concentration of 100 μg/ml. MBC-treated cells were incubated at 30°C for 4 h and monitored for G2-arrested cells by microscopic observation (43).
γ irradiation in yeast
Yeast were irradiated with γ rays from a Mark I 137Cs irradiator (J. L. Shepherd & Associates, Glendale, CA) at a dose rate of 5.3 Gy/min. Spun-down yeast cells were exposed to 50 Gy, which resulted in about 60% survival (34), 3 h before the heat-shock step of the transformation procedure (41).
Mammalian Cells
Cells
Cells of the Chinese hamster ovary (CHO) cell line CHO-K1 were purchased from ATCC (Manassas, VA) (44). Cells were grown in F12 medium (Invitrogen, Palo Alto, CA) containing 10% v/v fetal bovine serum (Invitrogen), 1% v/v penicillin/streptomycin, and 1% v/v glutamine (Invitrogen).
Plasmid end-joining assay in mammalian cells
Plasmid pCMS-end (Fig. 1), an end-joining recombination substrate used in mammalian cells, has been described recently (19). A DSB was introduced in pCMS-end DNA at the MCS by double digestions using XhoI-BamHI, EcoRI-XmaI, and XhoI-XmaI restriction enzymes (Fig. 1), and digestions were incubated overnight at 37°C (19, 22). The next day, digestions were heat-inactivated for 20 min at 65°C, purified through Micropure-EZ (Millipore, Bedford, MA) columns, and tested for complete digestions (19). For transfection experiments, CHO-K1 cells were seeded 24 h before the transfection at 2 × 105 cells per T-25 flask (25-cm2 tissue culture flasks) (BD Falcon). About 2 μg of digested and purified DNA per flask was transfected into the cells using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocol. Then, 24 h later, transfected CHO-K1 cells were trypsinized, spun down, resuspended in 0.5 ml of medium, and analyzed for recombination efficiency by FACS analysis.
FIG. 1.
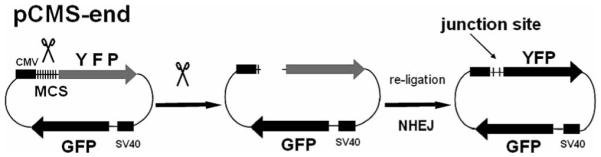
Schematic diagram of pCMS-end plasmid construct and NHEJ assay in mammalian cells. The plasmid pCMS-end was designed to monitor nonhomologous end joining (NHEJ) by FACS analysis. The multicloning site (MCS) is positioned between CMV enhancer/promoter region and coding sequence for EYFP; the EGFP sequence is under control of SV40 promoter (left). When the plasmid is digested with restriction enzyme(s) that only cut in MCS, linear DNA expresses EGFP but not EYFP (middle). Both colors can be detected by FACS analysis if repair of the DSB has taken place (right).
Recovering of the recombined end-joining plasmids and sequencing of the junctions
Low-molecular-weight DNA was purified from the excess of transfected cells left over after FACS analysis. Cells were spun down at 3,000 rpm for 10 min, the supernatant was aspirated, and DNA was harvested from the cell pellet using a mini-prep purification kit (Qiagen). Low-molecular-weight DNA was purified in a total volume of 35 μl of elution buffer. Next, 1 to 2 μl of this plasmid DNA was transformed into E. coli 10B electro-competent cells (Invitrogen) to obtain single clones. Plasmid DNA was purified from individual colonies and digested with SalI restriction enzyme, which cuts in the between two enzymes used originally to eliminate undigested plasmids that passed through. Combinations of XmnI, XmnI/BglII, XmnI/PvuI, XmnI/HindIII enzymes were used to analyze the sizes of recovered clones and to determine which DNA primers were closest to the junction sites and to use for sequencing through the repair junctions (19). The UCLA Core Sequencing Lab Facility performed sequencing reactions. The length of microhomology was quantified for each successfully sequenced clone.
Radiation sensitivity of CHO-K1 cells and clonogenic survival assay
CHO-K1 cells in exponential growth were harvested with 0.05% trypsin-EDTA (Invitrogen), counted and plated as 5 × 102 cells in 25-cm2 tissue culture flasks (BD Falcon). After 18 h, cells were irradiated in the Mark I irradiator as described above. To test cells for clonogenic survival, different doses of radiation were applied; after 7 days of postirradiation growth, cells were washed with PBS and stained with 2% methyl blue, and colonies of >40 cells were scored. Separate experiments at each dose were performed at least four times in duplicate. A dose of 3 Gy was found to kill 28% of cells, and this dose was used in later experiments in which mammalian cells were irradiated with 3 Gy 12 min prior to plasmid transfection.
Yeast and Mammalian Cells
Molecular techniques
Standard methods were followed. All enzymes were purchased from New England Biolabs (Ipswich, MA).
Statistical analysis
The frequencies of end joining before and after irradiation were compared by Student’s t test. The frequency of microhomology use in MMEJ events in relation to the random occurrence of these events among the analyzed clones was calculated by the formula described by Roth et al. (45). Significance for these events was calculated by the χ2 test with Yates’s correction and Fisher’s exact test if needed.
RESULTS
Yeast
1. γ rays enhance the frequency of end-joining events in yeast
We investigated the effect of γ radiation on end joining using a plasmid end-joining assay in yeast, similar to other plasmid recircularization assays (46). Plasmid YEplac195 contains a 2-μm origin of replication, a URA3 gene marker, and an AmpR resistance gene. First YEplac195 was linearized with HindIII and KpnI to generate incompatible DNA ends. When 98.5% to 99% of DNA was completely digested, linearized plasmid DNA was purified and transformed into the wild-type RSY12 strain lacking the URA3 gene with and without prior γ irradiation. We exposed yeast cells to 50 Gy of γ rays 3 h before the heat-shock step of transformation. In parallel, uncut YEplac195 plasmid was transformed into yeast cells to control for transformation efficiency. In both cases, after 4 days of growth, we selected Ura+ transformants and calculated the frequency of end joining. The frequency of end joining in unirradiated yeast cells was about 1.22 ± 0.08% and increased to 3.42 ± 0.20% in irradiated yeast cells, indicating that radiation caused a 2.8-fold increase in the end-joining frequency (P < 0.005, by Student’s t test).
2. γ rays induce MMEJ in trans in yeast
To investigate the mechanism of radiation-induced end-joining events, we isolated yeast plasmid DNA from individual Ura+ transformants and transformed them into E. coli. We randomly selected and analyzed 37 clones of unirradiated and 28 clones of irradiated yeast cells. Junctions were sequenced using primers upstream and downstream of the MCS. In plasmids from unirradiated yeast cells, 11 out of 37 (30%) rejoined plasmids contained 2 to 5 bp of microhomology at the junctions (Table 1); however, in plasmids from irradiated yeast cells, 19 out of 28 (68%) rejoined plasmids displayed 2 to 6 bp of microhomology at the junctions (P < 0.005) (Table 1, Fig. 2. In particular, 16 out of 28 (57%) rejoined plasmids from irradiated yeast cells used ≥4 bp of microhomology, whereas only 4 out of 37 (11%) of the rejoined plasmids from unirradiated cells used ≥4 bp (P < 0.0001). In three independent experiments, one junction 237-673 occurred 12 times in irradiated cells; because each of these colonies was selected from a different plate, such events represent independent endjoining events and could be at a hotspot of MMEJ with 4 bp. These results indicate that radiation induces the MMEJ pathway of end joining in yeast. The expected random occurrences of 2, 3, 4, 5 and 6 bp of microhomology in an unbiased sequence are 10.5, 3.5, 1.1, 0.3 and 0.1% (45). We found that the distribution of MMEJ events using 2, 3, 4 and 5 bp of microhomology in unirradiated cells is 19, 0, 8.1 and 2.7% (29.8% ≥2 bp), which is not significantly different from random occurrence. In contrast, the observed distribution of MMEJ using microhomologies of 2, 3, 4, 5 and 6 bp in irradiated cells is 3.6, 7.1, 50, 0 and 7.1%, respectively (67.8% ≥2 bp), which is significantly different from random occurrence (P < 0.0001). These results indicate that radiation induces a MMEJ pathway in trans to the radiation-induced damage in yeast cell DNA.
TABLE 1. End-Joining Events in Unirradiated and Irradiated Yeast Cells.
| Junctiona | Length of microhomology (bp)b | Overlapped basesc | Deletiond from |
||
|---|---|---|---|---|---|
| 5′ PSS end | 3′ PSS end | ||||
| Unirradiated clones | |||||
| C13 | 237-1226 | 5 | AGCTT | 0 | 953 |
| C2, C26 | 237-279 | 4 | AGCT | 0 | 6 |
| C18 | 237-1629 | 4 | AGCT | 0 | 1356 |
| C1, C27, C44 | 234-673 | 2 | TT | 3 | 400 |
| C8, C11, C17 | 234-676 | 2 | AG | 0 | 403 |
| C35 | 141-822 | 2 | GC | 96 | 549 |
| C4, C7, C14, C16, C22, C23, C36, C37 | 236-277 | 1 | G | 1 | 4 |
| C42 | 238-282 | 1 | A | 1 | 9 |
| C6, C32 | 134-277 | 1 | G | 103 | 4 |
| C20 | 142-274 | 1 | A | 95 | 1 |
| C25 | 237-279 | 0 | — | 0 | 6 |
| C37 | 236-277 | 0 | — | 1 | 4 |
| C9, C12, C29, C30, C38 | 237-276 | 0 | — | 0 ( 1) | 3 |
| C10 | 237-276 | 0 | — | 0 | 3 |
| C21, C41 | 237-281 | 0 | — | 0 | 8 |
| C40 | 237-282 | 0 | — | 0 | 9 |
| C3 | 142-277 | 0 | — | 95 | 4 |
| C24, C33 | 142-278 | 0 | — | 98 | 5 |
| Irradiated clones | |||||
| R24, R50 | 232-276 | 6 | AGCT*GG | 0 | 3 |
| R5, R11, R16, R17, R23, R25, R28, R30, R33, R34, R39, R41 | 237-673 | 4 | AG*TT | 0 | 400 |
| R14 | 236-747 | 4 | GCTT | 1 | 474 |
| R19 | 237-375 | 4 | AGCT | 0 | 102 |
| R9 | 142-280 | 3 | AGC | 95 | 7 |
| R26 | 143-676 | 3 | AAG | 94 | 403 |
| R29 | 219-441 | 2 | TC | 18 | 165 |
| R31 | 236-277 | 1 | G | 1 | 4 |
| R48 | 142-675 | 1 | A | 95 | 402 |
| R7 | 141-1023 | 0 | — | 96 | 750 |
| R8 | 142-276 | 0 | — | 95 | 3 |
| R32 | 141-276 | 0 | — | 96 | 3 |
| R15 | 229-273 | 0 | — | 8 | 2 |
| R6 | 237-277 | 0 | — | 0 | 4 |
| R35 | 236-276 | 0 | — | 1 | 3 |
| R36 | 139-279 | 0 | — | 98 | 6 |
Asterisks (*) represent mismatched nucleotides within the microhomology region. Junction sequences of MMEJ events containing mismatches are shown in Fig. 4.
The location of the junctions with respect to YEplac195 plasmid sequences.
The length of microhomology used during end joining.
The sequence of the overlapping bases. — represents events with no microhomology.
The number of nucleotides deleted at the 5′ PSS and 3′ PSS ends. The number of nucleotides inserted is shown in parentheses.
FIG. 2.
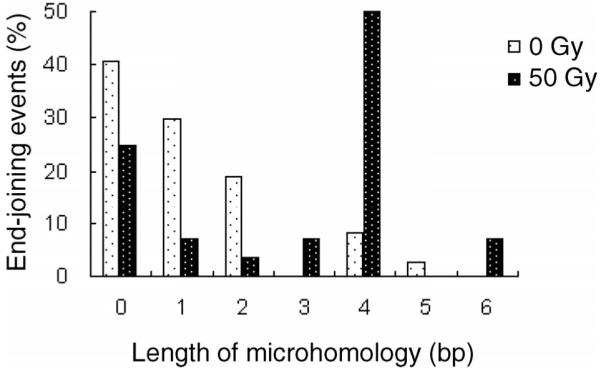
The distribution of microhomology used during end joining in yeast in the absence and presence of 50 Gy of γ radiation.
3. Radiation-induced MMEJ contains mismatches within the microhomologies
None of the rejoined plasmids from unirradiated yeast cells exhibited mismatched nucleotides within the region of microhomology, whereas 14 out of 28 end-rejoined plasmids from irradiated yeast cells contained one mismatched nucleotide within the microhomology sequences (Table 1 and Fig. 4) (P < 0.00001). Among MMEJ events that contain mismatches, R24 and R50 had the recipient strand sequences 5′-CCGAGCT-3′and R16, R17, R28, R30, R39 and R41 contained a different recipient strand 5′-AATCT-3′ in the rejoined plasmids while R5, R11, R23, R25, R33 and R34 contained the sequence of the invading strand 5′-AAGCT-3′. Thus there is no preference for the invading or recipient strand in error-prone MMEJ.
FIG. 4.
MMEJ events in yeast containing mismatches. The top line is the junction sequence recovered from the rejoined plasmids; the location of the sequence is indicated. The second line shows the sequence up-stream of the 5′ HindIII-linearized end. The third line shows the sequence at the site of MMEJ either close to the KpnI-linearized end or distant from the end. Overlapping sequences between lines 2 and 3 are shown as dashed lines, and the microhomologies used are shown as boldface letters. The mismatched nucleotides within the microhomologies are identified with a star on top of the nucleotides.
4. Radiation-induced end-joining events exhibit larger deletions at the 3′ protruding single-stranded (PSS) ends
Nine out of 37 rejoined plasmids from unirradiated yeast cells had large deletions between 400 and 1356 nucleotides (nt) at the 3′ PSS ends (KpnI-linearized), whereas 18 out of 28 plasmids from irradiated yeast cells had large deletions between 102 and 750 nt at the 3′ PSS ends (P < 0.005) (Table 1). In contrast, seven out of 37 plasmids from unirradiated yeast cells had large deletions of greater than 90 nt at the 5′ PSS end (HindIII-linearized) compared to seven out of 28 plasmids from irradiated yeast cells, which is not significantly different.
All nine plasmids from unirradiated yeast cells with large deletions at the 3′ PSS ends contained microhomologies at the junctions, whereas only one out of seven events with large deletions at the 5′ PSS end was mediated by MMEJ (P < 0.005). In irradiated yeast cells, 16 out of 18 rejoined plasmids with large deletions at the 3′ PSS ends contained microhomologies, whereas three out of seven plasmids with large deletions at the 5′ PSS ends used microhomologies (P < 0.05). These results indicate a correlation between microhomology use and large deletions at the 3′ PSS ends and suggest that the 5′ PSS ends remain intact and may be used for microhomology search during spontaneous and radiation-induced MMEJ.
5. Induction of MMEJ events does not depend on the G2 phase of the cell cycle in yeast
Since ionizing radiation has been shown to arrest yeast cells at the G2 phase of the cell cycle (47), it was possible that the induced MMEJ events observed in irradiated cells are dependent on G2 phase rather than being a consequence of ionizing radiation. To test this possibility, yeast cells were exposed to 50 Gy of γ rays, resuspended in YPD or lithium acetate (the transformation buffer), and incubated at 30°C. The cell cycle distribution of yeast cells were analyzed 3 h after irradiation by microscopic observation, the time when cells were heat-shocked during the transformation process. We found that yeast cells that were incubated in YPD or buffer were not arrested in G2 at this time (data not shown).
We further tested the effect of G2-arrested cells on end joining. Yeast cells were first treated with the anti-microtubule drug MBC for 4 h to arrest cells at G2. Then the HindIII, KpnI-linearized YEplac195 plasmid was transformed into MBC-treated and untreated cells and Ura+ transformants were selected. The efficiency of end joining in MBC-treated cells was 25.6 ± 2.1%, whereas the efficiency of end joining in cells without treatment was 27.2 ± 3.5%, which is not significantly different by Student’s t test. These results exclude the possibility that the induced end-joining events observed in irradiated cells are dependent on G2-phase arrest triggered by radiation and support the interpretation that the end-joining pathway, in particular MMEJ, is induced as a consequence of ionizing radiation.
Mammalian Cells
1. Effect of γ rays on end-joining events in CHO-K1 cells
For experiments with radiation, both nonirradiated and irradiated CHO-K1 cells (previously exposed to 3 Gy) were simultaneously transfected with XhoI-BamHI, EcoRI-XmaI, and XhoI-XmaI-linearized pCMS-end plasmids (19). Cells were analyzed 24 h later by FACS for the intensity of fluorescence representing frequencies of end joining. Unlike yeast, irradiated CHO-K1 cells did not show a significant difference from nonirradiated cells in the frequency of end-joining events (data not shown).
2. γ rays induce MMEJ in trans in CHO-K1 cells
The junction sequences of the end-joining events recovered from mammalian cells were analyzed when plasmid DNA was recovered from the same nonirradiated and irradiated cells after FACS analysis. Excess cells were spun down and low-molecular-weight DNA was purified from the cells. DNA from each digest was transformed into E. coli 10B electro-competent cells, and individual clones were collected. From randomly selected clones, plasmid DNA was analyzed to reveal sequences at the junctions. A total of 23 clones from nonirradiated cells were successfully sequenced (Table 2, nonirradiated cells). Among these clones, 10 (43%) were rejoined without any use of microhomologies, eight clones (35%) with one nucleotide, three clones (13%) with two nucleotides, and two clones (9%) with three nucleotides (Fig. 3, gray bars). There were no clones found with more then three nucleotides of overlap (Fig. 3). From irradiated CHO-K1 cells, 13 clones were analyzed and sequenced (Table 2, irradiated cells). Among them, 31% were repaired without any microhomology, 15% with three, 15% with four, 8% with five, 8% with six, 15% with eight, and 8% of clones with ten nucleotides of overlap (Fig. 3, black bars). Fifty-four percent of irradiated CHO-K1 cells used ≥4 bp of overlap, while no such clones were recovered from nonirradiated CHO-K1 cells (Fig. 3), which is highly significantly different (P < 0.00001).
TABLE 2. End-Joining Events in Unirradiated and Irradiated CHO-K1 Cells.
| Junctiona | Length of microhomologyb (bp) | Overlapped basesc | Deletiond from |
||
|---|---|---|---|---|---|
| 5′ PSS end | 3′ PSS end | ||||
| Unirradiated clones | |||||
| CHO-K1-22 (ApaI) | 885-2284 | 3 | TGG | 265 | 1133 |
| CHO-K1-10 (X/B) | 962-4359 | 3 | TCT | 145 | 3205 |
| CHO-K1-14 (X/X) | 972-1144 | 2 | TG | 135 | 24 |
| CHO-K1-12 (X/X) | 692-3318 | 2 | GG | 415 | 2168 |
| CHO-K1-17 (X/X) | 1047-1167 | 2 | CG | 60 | 17 |
| CHO-K1-03 (E/X) | 820-3869 | 1 | G | 303 | 2719 |
| CHO-K1-07 (X/B) | 954-4865 | 1 | G | 153 | 3711 |
| CHO-K1-09 (X/B) | 391-4381 | 1 | G | 716 | 3227 |
| CHO-K1-18 (ApaI) | 1056-3183 | 1 | G | 94 | 2032 |
| CHO-K1-23 (ApaI) | 413-1941 | 1 | G | 737 | 790 |
| CHO-K1-05 (E/X) | 989-2947 | 1 | C | 134 | 1797 |
| CHO-K1-13 (X/X) | 513-3599 | 1 | C | 594 | 2449 |
| CHO-K1-16 (X/X) | 850-1350 | 1 | A | 257 | 200 |
| CHO-K1-01 (E/X) | 291-4356 | 0 | — | 832 | 3386 |
| CHO-K1-02 (E/X) | 452-4197 | 0 | — | 671 | 3047 |
| CHO-K1-04 (E/X) | 4-3263 | 0 | — | 1119 | 2113 |
| CHO-K1-06 (X/B) | 1094-3919 | 0 | — | 13 | 2765 |
| CHO-K1-08 (X/B) | 1141-4222 | 0 | — | 325 | 334 |
| CHO-K1-11 (X/X) | 1053-1545 | 0 | — | 54 | 395 |
| CHO-K1-15 (X/X) | 782-1484 | 0 | — | 325 | 334 |
| CHO-K1-19 (ApaI) | 594-1644 | 0 | — | 556 | 493 |
| CHO-K1-20 (ApaI) | 807-1503 | 0 | — | 343 | 352 |
| CHO-K1-21 (ApaI) | 982-1395 | 0 | — | 168 | 244 |
| Irradiated clones | |||||
| CHO-K1-IR-06 (X/B) | 402-3459 | 10 | CAAGTCCGCC | 705 | 2305 |
| CHO-K1-IR-11 (X/B) | 404-3062 | 8 | TCCGCCCC | 703 | 1098 |
| CHO-K1-IR-13 (X/B) | 675-3530 | 8 | CCCGC*CCG | 432 | 2376 |
| CHO-K1-IR-10 (X/B) | 405-3198 | 6 | CGCC*CC | 702 | 2044 |
| CHO-K1-IR-07 (X/B) | 692-4188 | 5 | GGCGG | 415 | 3034 |
| CHO-K1-IR-04 (X/B) | 242-4548 | 4 | CCGC | 865 | 3394 |
| CHO-K1-IR-05 (X/B) | 235-2044 | 4 | AAAT | 872 | 850 |
| CHO-K1-IR-08 (X/B) | 128-3511 | 3 | GAC | 979 | 2357 |
| CHO-K1-IR-02 (X/B) | 256-3530 | 3 | CCC | 851 | 2376 |
| CHO-K1-IR-01 (X/B) | 987-4306 | 0 | — | 120 | 3152 |
| CHO-K1-IR-03 (X/B) | 670-4423 | 0 | — | 437 | 3269 |
| CHO-K1-IR-09 (X/B) | 963-3732 | 0 | — | 144 | 2578 |
| CHO-K1-IR-12 (X/B) | 262-2103 | 0 | — | 845 | 949 |
Asterisk (*) represents a mismatched nucleotide within the microhomology region.
Clone name and original restriction enzymes used for linearization of pCMS-end.
The length of microhomology used during end joining.
The sequence of the overlapping bases. — represents no microhomology.
The number of nucleotides deleted at the 5′ PSS and 3′ PSS ends.
FIG. 3.
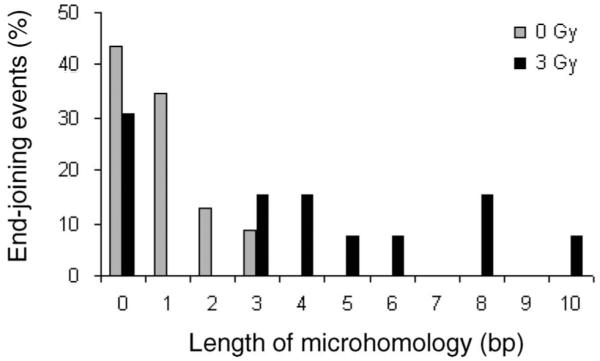
The distribution of microhomology use during end joining in CHO-K1 mammalian cells in the absence and presence of 3 Gy of γ radiation; cells were irradiated prior to transfection of the end-joining substrates.
The expected probabilities of random occurrence of 2, 3 and 4 bp of microhomology in an unbiased sequence are 10.5, 3.5, 1.1, 0.3 and 0.1% (45). In our experiment, the frequency of distribution of MMEJ events using 2 and 3 nt of microhomology in unirradiated cells is 13, and 9 (32% ≥ 2 bp), which is not significantly different from random occurrence. In contrast, in our experiment, the frequency of distribution of MMEJ using microhomologies of 2, 3, 4, 5, 6, 7, 8, 9 and 10 bp in irradiated cells is 0, 8, 15, 15, 15 and 8, 8, 0, 15, 0 and 8%, respectively (69% ≥2 bp), which is significantly different from random occurrence (P < 0.0001). These results indicate that radiation induces an MMEJ pathway in trans to the radiation-induced damage in mammalian cells.
Two out of 13 clones collected from irradiated CHO-K1 cells contained one mismatched nucleotide each. This difference is not significant but is in agreement with the data in yeast, where the difference was highly significant. Our results indicate that radiation induces MMEJ events in yeast as well as mammalian cells to repair DSBs in trans to radiation damage. Deletion size among clones in mammalian cells has a large range (from one to 4200 nt) regardless of whether plasmids were recovered from nonirradiated or irradiated cells (Table 2), indicating that unlike yeast, there is no correlation between irradiation and the size of the deletions in mammalian cells.
To address the possibility that a change in cell cycle distribution causes the increase in microhomology use, we measured the cell cycle distribution of CHO-K1 cells at different times in nonirradiated and 3 Gy-irradiated CHO-K1 cells. We found (Table 3) that there was a slight increase in the fraction of cells in G2/M phase 4-8 h after irradiation cells then return to distributions similar to those observed in nonirradiated cells. We suggest that this small increase in the percentage of cells in G2/M phase after irradiation could not cause the observed increase (more than fourfold) in the fraction of clones with more than 2 bp of microhomology. Since only an additional 10% of the cells are arrested by the radiation, this arrest should account for only a 10% increase in clones using microhomology assuming that all events in arrested cells are due to MMEJ. This could not explain a 40% increase in such events. Rather, a new mechanism triggered by radiation should be responsible for such events.
TABLE 3. Cell Cycle Distribution of CHO-K1 Cells at Different Times after Irradiaton with 3 Gy.
| Time after irradiation | Fraction of cells in G2/M phase |
|---|---|
| Control (unirradiated cells) | 0.248 ± 0.040 |
| 2 h | 0.245 ± 0.059 |
| 4 h | 0.355 ± 0.062 |
| 8 h | 0.352 ± 0.009 |
| 16 h | 0.295 ± 0.029 |
| 24 h | 0.204 ± 0.0014 |
DISCUSSION
We analyzed the effect of radiation on end joining of linearized plasmid substrates in yeast and mammalian cells. Based on sequence analysis of the rejoined junctions, we found that radiation increased the frequency of MMEJ in both organisms. Since cells were irradiated before introduction of the linearized plasmids, the radiation-induced MMEJ events are in trans to the radiation-induced primary damage.
DSBs Induce MMEJ
DSB repair in yeast was studied by overexpression of HO-endonuclease (9, 16, 32) or other plasmid end-joining assays (16). Many of these studies reported microhomologies at the junctions of the recombined products, suggesting that DSBs are repaired by an MMEJ pathway. In previous studies, human cells carrying a pHAZE shuttle vector were exposed to α particles, and 60% of the rejoined junctions revealed microhomologies (48). However, these studies were unable to distinguish between MMEJ events at the targeted DSB sites and MMEJ events in trans at nontargeted sites. Other studies have analyzed X-ray- and α-particle-induced deletions at the hprt locus in human lymphoblasts (49, 50). Based on mapping of deletions by Southern analysis and PCR amplification, these investigators found that radiation-induced events exhibited a larger extent of deletions compared to spontaneous events. Since these studies examined only the induced mutations at the site-specific locus, radiation-induced deletions at nontargeted sites have not been investigated. It has been shown previously that radiation and restriction enzymes induce plasmid integrations into the genome using microhomology in yeast (38). These data, however, did not prove that the induced recombination events were in trans to the genomic DNA damage. Therefore, current experiments were carried out so that the radiation exposure was physically separated from the nonirradiated recombination substrate, proving that radiation induces MMEJ in trans.
Potential Mechanisms of MMEJ in Yeast and Mammalian Cells
Our yeast data showed a larger extent of deletions at the 3′ PSS ends of the rejoined plasmids in radiation-induced end-joining events compared to spontaneous events. In contrast, the extent of deletions at the 5′ PSS ends was not induced by radiation. Among the events mediated by MMEJ, 10 out of 11 unirradiated clones and 16 out of 19 irradiated clones had the 5′ PSS end remained intact or contained just 1 to 5 nt of deletions. However, the majority of these MMEJ events had large deletions at the 3′ PSS ends, showing a significant correlation between microhomology use and the size of deletions at the 3′ PSS ends. Based on these results, it is likely that the 5′ PSS end is used for microhomology search and strand invasion of the microhomology regions distant from the 3′ PSS end results in homologous pairing; presumably an endonuclease is involved in the removal of the displaced single-stranded DNA, causing a large deletion at the 3′ PSS end. Alternatively, strand resection of the 3′ PSS end of the break facilitates exposure of microhomologies for annealing (17). Because 3′ PSS ends are used for strand invasion in homologous recombination in yeast, it is likely that resection of breaks occurs in the other direction to generate 5′ PSS ends for strand invasion in MMEJ. Further studies on end joining using different linearized ends are essential to investigate the mechanism of MMEJ in yeast.
In contrast to the data for yeast, the deletion size among clones recovered from mammalian cells has a large range regardless of radiation exposure of cells. These results could be explained by the presence of high levels of nuclease activities in some mammalian cell lines (53). Presumably, both the 5′ and 3′ DNA ends are degraded by exonucleases, and the two ends are joined based on microhomology. A similar model of MMEJ in mammalian cellshas been proposed (51). In this regard, Mre11 has been shown to degrade DNA ends, and its exonuclease activity is inhibited when a microhomology is encountered (52).
The Radiation-Induced MMEJ Pathway May Be Associated with an Inducible Process
Radiation-induced recombination occurs during the repair process of DSBs while all the radiation-induced damage is reported to be fixed within 2 h postirradiation (53). However, radiation-induced integration is active days after irradiation (54). In our experiments, transfection of mammalian cells (addition of linearized DNA in the presence of Lipofectamine 2000) was done very soon (12 min) after radiation, but the process of linearized DNA entering into the mammalian cells could take up to 2 h after transfection. Moreover, the yeast transformation procedure takes much longer, yeast cells were heat-shocked about 3 h after irradiation (41). Thus, in the mammalian and yeast cell systems, it is likely that most or all DSBs had already been repaired when the plasmids entered the cells. Our data show that the rejoined plasmids, which were transfected into cells after exposure to radiation, contain radiation-induced recombination events that are in trans to radiation-induced primary lesions. Radiation-induced effects that are not due to the initial damage, including delayed reproductive genetic instability and nontargeted effects have been observed previously in vitro (55) and in vivo (56).
The MMEJ Pathway may Contribute to Radiation-Induced Genomic Instability
Our results showed that radiation-induced end-joining events in yeast exhibit a larger extent of deletions at the 3′ PSS ends than spontaneous events. Of the 18 radiation-induced events containing large deletions at the 3′ PSS end, 16 events were mediated by MMEJ. These data strongly suggest that MMEJ could be the mechanism responsible for generating radiation-induced large deletions (25, 26, 49). The human and hamster hprt gene and the hamster aprt gene analyzed for deletions showed that radiation-induced deletions are generated randomly throughout the genome (57). Together with our previous results showing that radiation and restriction enzymes induce plasmid integrations into the genome using microhomology (38), our current findings of an in trans effect of radiation on MMEJ could mean that radiation induces genomic rearrangements at random sites, which supports the notion that one chromosomal DSB can invade other genomic locations through MMEJ and lead to large deletions and translocations.
MMEJ is Associated with Human Disorders and Cancer
MMEJ causes large deletions and misrejoining of DSBs, leading to human disorders such as muscular dystrophy and Fabry’s disease (29-31). In addition, a preference using MMEJ to repair DSBs in bladder tumor cells and head and neck cancer cells is linked to its high level of genomic instability and tumor progression (27, 28). MMEJ has been observed in mammalian cells that are defective in ATM, Ku70, Ku86, ligase IV and XRCC4 (19-21, 51), and some of these mutants were characterized by high levels chromosomal aberrations, including translocations, aneuploidy and early onset of cancer (11, 58). These observations suggest that MMEJ is a potential mechanism that leads to genomic instability and carcinogenesis.
ACKNOWLEDGMENTS
This research was supported in part by project 1 to RHS of NIH grant 1 U19 AI 67769-01 to William McBride and a fellowship of the UCLA Chemistry-Biology Interface Training Program with USPHS National Research Service Award GM08496 and a research fellowship of the UC Toxic Substances Research and Teaching Program, both to CYC. KH was partially funded by a National Institute of Biomedical Imaging and Bioengineering training grant and a NASA graduate student fellowship.
REFERENCES
- 1.Haber JE. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 2.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ. and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukamoto Y, Kato J, Ikeda H. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiestl RH, Zhu J, Petes TD. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson LH. Evidence that mammalian cells possess homologous recombinational repair pathways. Mutat. Res. 1996;363:77–88. doi: 10.1016/0921-8777(96)00008-0. [DOI] [PubMed] [Google Scholar]
- 8.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst.) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol. Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Secretan MB, Scuric Z, Oshima J, Bishop AJ, Howlett NG, Yau D, Schiestl RH. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat. Res. 2004;554:351–364. doi: 10.1016/j.mrfmmm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Smith J, Riballo E, Kysela B, Baldeyron C, Manolis K, Masson C, Lieber MR, Papadopoulo D, Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howlett NG, Scuric Z, D’Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst.) 2006;5:251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Smith JA, Waldman BC, Waldman AS. A role for DNA mismatch repair protein Msh2 in error-prone double-strand-break repair in mammalian chromosomes. Genetics. 2005;170:355–363. doi: 10.1534/genetics.104.039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bannister LA, Waldman BC, Waldman AS. Modulation of error-prone double-strand break repair in mammalian chromosomes by DNA mismatch repair protein Mlh1. DNA Repair (Amst.) 2004;3:465–474. doi: 10.1016/j.dnarep.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Nohmi T, Suzuki M, Masumura K, Yamada M, Matsui K, Ueda O, Suzuki H, Katoh M, Ikeda H, Sofuni T. Spi(-) selection: An efficient method to detect gamma-ray-induced deletions in transgenic mice. Environ. Mol. Mutagen. 1999;34:9–15. doi: 10.1002/(sici)1098-2280(1999)34:1<9::aid-em2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Morris T, Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl. Acad. Sci. USA. 1993;90:1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin KH, Kang MK, Kim RH, Kameta A, Baluda MA, Park NH. Abnormal DNA end-joining activity in human head and neck cancer. Int. J. Mol. Med. 2006;17:917–924. [PubMed] [Google Scholar]
- 29.Love DR, England SB, Speer A, Marsden RF, Bloomfield JF, Roche AL, Cross GS, Mountford RC, Smith TJ, Davies KE. Sequences of junction fragments in the deletion-prone region of the dystrophin gene. Genomics. 1991;10:57–67. doi: 10.1016/0888-7543(91)90484-v. [DOI] [PubMed] [Google Scholar]
- 30.Krawczak M, Cooper DN. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 31.Kornreich R, Bishop DF, Desnick RJ. Alpha-galactosidase A gene rearrangements causing Fabry disease. Identification of short direct repeats at breakpoints in an Alu-rich gene. J. Biol. Chem. 1990;265:9319–9326. [PubMed] [Google Scholar]
- 32.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens CW, Zeng M, Cerniglia GJ. Ionizing radiation greatly improves gene transfer efficiency in mammalian cells. Hum. Gene Ther. 1996;7:1727–1734. doi: 10.1089/hum.1996.7.14-1727. [DOI] [PubMed] [Google Scholar]
- 34.Kiechle M, Manivasakam P, Eckardt-Schupp F, Schiestl RH, Friedl AA. Promoter-trapping in Saccharomyces cerevisiae by radiation-assisted fragment insertion. Nucleic Acids Res. 2002;30:e136. doi: 10.1093/nar/gnf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shcherbakova OG, Filatov MV. Camptothecin enhances random integration of transfected DNA into the genome of mammalian cells. Biochim. Biophys. Acta. 2000;1495:1–3. doi: 10.1016/s0167-4889(99)00151-2. [DOI] [PubMed] [Google Scholar]
- 36.Fujimaki K, Aratani Y, Fujisawa S, Motomura S, Okubo T, Koyama H. DNA topoisomerase II inhibitors enhance random integration of transfected vectors into human chromosomes. Somat. Cell Mol. Genet. 1996;22:279–290. doi: 10.1007/BF02369567. [DOI] [PubMed] [Google Scholar]
- 37.Stevens CW, Cerniglia GJ, Giandomenico AR, Koch CJ. DNA damaging agents improve stable gene transfer efficiency in mammalian cells. Radiat. Oncol. Investig. 1998;6:1–9. doi: 10.1002/(sici)1520-6823(1998)6:1<1::aid-roi1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 38.Chan CY, Kiechle M, Manivasakam P, Schiestl RH. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–5059. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 40.Manivasakam P, Schiestl RH. Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1736–1745. doi: 10.1128/mcb.18.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams GDE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- 43.Galli A, Schiestl RH. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics. 1998;149:1235–1250. doi: 10.1093/genetics/149.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins AR. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat. Res. 1993;293:99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- 45.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartek J, Lukas L, Strauss M. Control mechanisms of cell transition from the G1 phase to the S phase-the R point. Cas. Lek. Cesk. 1996;135:634–635. [PubMed] [Google Scholar]
- 48.Lutze LH, Cleaver JE, Morgan WF, Winegar RA. Mechanisms involved in rejoining DNA double-strand breaks induced by ionizing radiation and restriction enzymes. Mutat. Res. 1993;299:225–232. doi: 10.1016/0165-1218(93)90099-y. [DOI] [PubMed] [Google Scholar]
- 49.Bao CY, Ma AH, Evans HH, Horng MF, Mencl J, Hui TE, Sedwick WD. Molecular analysis of hypoxanthine phosphoribosyltransferase gene deletions induced by alpha- and X-radiation in human lymphoblastoid cells. Mutat. Res. 1995;326:1–15. doi: 10.1016/0027-5107(94)00152-u. [DOI] [PubMed] [Google Scholar]
- 50.Nelson SL, Giver CR, Grosovsky AJ. Spectrum of X-ray-induced mutations in the human hprt gene. Carcinogenesis. 1994;15:495–502. doi: 10.1093/carcin/15.3.495. [DOI] [PubMed] [Google Scholar]
- 51.Ganesh A, North P, Thacker J. Repair and misrepair of site-specific DNA double-strand breaks by human cell extracts. Mutat. Res. 1993;299:251–259. doi: 10.1016/0165-1218(93)90101-i. [DOI] [PubMed] [Google Scholar]
- 52.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother. Oncol. 1989;14:307–320. doi: 10.1016/0167-8140(89)90143-6. [DOI] [PubMed] [Google Scholar]
- 54.Stevens CW, Puppi M, Cerniglia GJ. Time-dose relationships in radiation-enhanced integration. Int. J. Radiat. Biol. 2001;77:841–846. doi: 10.1080/09553000110053882. [DOI] [PubMed] [Google Scholar]
- 55.Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: I. Radiation-induced genomic instability and bystander effects in vitro. Radiat. Res. 2003;159:567–580. doi: 10.1667/0033-7587(2003)159[0567:nadeoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 56.Morgan WF, Day JP, Kaplan MI, McGhee EM, Limoli CL. Genomic instability induced by ionizing radiation. Radiat. Res. 1996;146:247–258. [PubMed] [Google Scholar]
- 57.Hutchinson F. Analysis of deletions induced in the genome of mammalian cells by ionizing radiation. J. Mol. Biol. 1995;254:372–380. doi: 10.1006/jmbi.1995.0624. [DOI] [PubMed] [Google Scholar]
- 58.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]



