Abstract
Initial studies of the mammalian hSAGA transcriptional coactivator complex identified the acetyltransferase hGCN5/PCAF as the only known enzymatic subunit. Recently we and others demonstrated that the ubiquitin hydrolase USP22 comprises a second enzymatic subunit of hSAGA, which is required for activator-driven transcription. USP22 is expressed with polycomb ubiquitin ligases in an 11 gene signature that defines therapy-resistant tumors. At the biochemical level, these Polycomb proteins function as global transcriptional repressors by catalyzing the ubiquitylation of histone H2A. In yeast, the USP22 homolog functions as a transcriptional coactivator by removing ubiquitin from a distinct core histones, H2B. Given that USP22 is expressed in cancer as part of an 11 gene signature that includes transcriptional repressors which ubiquitylate H2A, it seemed possible that USP22 might activate transcription in part via the deubiquitylation of this same substrate. As reported here, biochemical analysis of the substrate specificity of USP22 reveals that it deubiquitylates histone H2A in addition to H2B. This finding supports a model in which the H2A ubiquitin hydrolase USP22 is coordinately expressed with Polycomb H2A ubiquitin ligases in order that the transcription of certain critical transforming genes be maintained in the face of the global repression mediated by Polycomb.
Keywords: ubiquitin, USP22, hSAGA, histone, H2A, H2B, polycomb
Introduction
Glinsky et al., recently identified an 11 gene mRNA expression profile that they termed a “death from cancer signature”.1 The pattern of expression of the genes within this signature was able to identify highly aggressive tumors in both humans and animal models. Tumors with this expression signature were consistently resistant to therapy and had a high incidence of recurrence after surgical resection. The behavior of these tumors, coupled with the finding that most of the genes within the signature are regulated by the Polycomb group protein BMI-1, led the authors to suggest that the signature might identify tumors with cancer stem cell properties. 2,3 This suggestion was supported by a number of recent studies implicating Polycomb proteins in stem cell function in both normal tissues and tumors.4 In addition to being regulated by Polycomb proteins, the death-from-cancer signature also includes the Polycomb gene Ring1B/RNF2 among its 11 members.1 Ring1B/RNF2 resides within a multi-protein complex termed PRC1,5 that participates in global transcriptional repression by ubiquitylating histone H2A.6
We and others recently reported that another protein encoded by the death-from-cancer signature, USP22, is a subunit of the hSAGA coactivator complex.3,7 We also showed that USP22 that can remove ubiquitin from another of the core histones, H2B.7 Our analysis of H2B as a potential substrate for USP22 was prompted by reports from S. cerevisiae showing that a USP22 ortholog termed Ubp8p functions to activate transcription largely via the deubiquitylation of H2B.8,9 Yeast have nearly insignificant levels of histone H2A ubiquitylation. 10 In contrast, approximately 10–15% of H2A molecules are ubiquitylated in human cells.11 Given that USP22 can deubiquitylate another core histone (i.e., H2B), that ubiquitylated H2A levels are relatively high in mammals, and that other members of the USP22/polycomb signature function by regulating the levels of H2A ubiquitylation, we asked whether USP22 is capable of deubiquitylating histone H2A. The data presented below show that human USP22 can hydrolyze ubiquitin that has been conjugated to either histone H2A or H2B.
Results
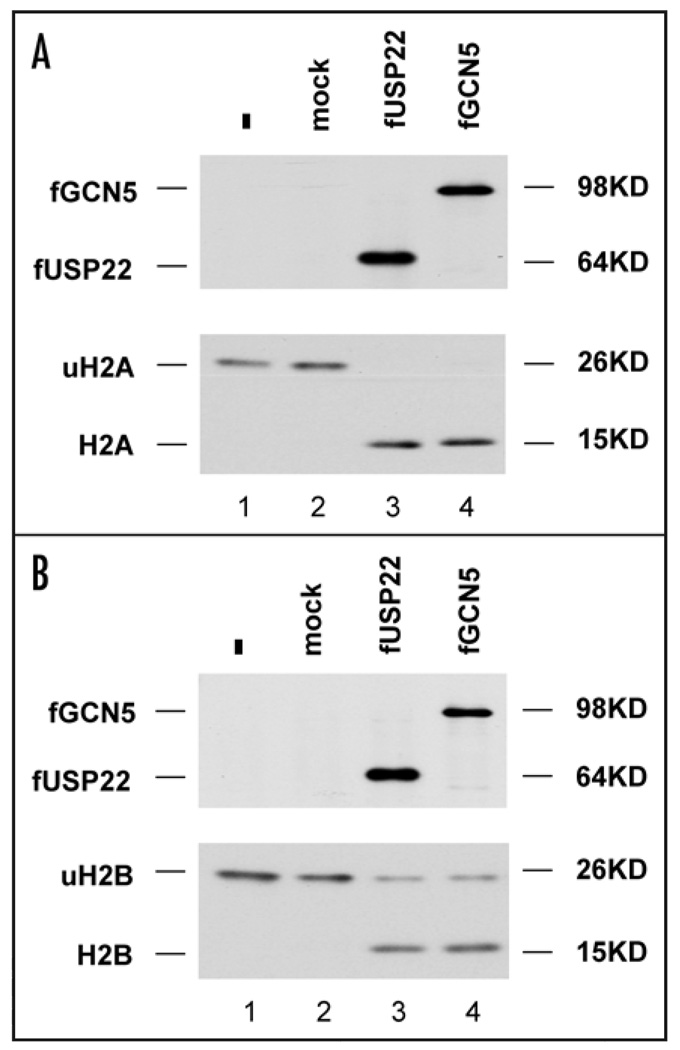
In order to assess whether USP22 can deubiquitylate histone H2A, epitope-tagged USP22 was purified after transfection of the H1299 human lung cancer cell line. In parallel, H1299 cells were transfected with an expression vector for an epitope-tagged version of the other known enzymatic hSAGA subunit, the acetyltransferase GCN5. Mock-transfected cells served as a negative control. After purification of USP22 or the holo-hSAGA complex (via GCN5), proteins were incubated in vitro with ubiquitylated histone H2A or ubiquitylated H2B that had been purified from mammalian cells.12 These in vitro reactions were incubated and the products resolved and detected by western blotting (Fig. 1). Blotting for the FLAG epitope shared by USP22 and GCN5 revealed that both proteins were specifically expressed and purified. Probing of reaction products for histones H2A and H2B demonstrated that both purified USP22 and the holo-hSAGA complex were capable of deubiquitylating histone H2B, as we and others recently reported.7,17 When these protein complexes were exposed to ubiqutitylated H2A as a potential substrate, both USP22 and the holo-hSAGA complex exhibited deubiquitylating activity. No deubiquitylating activity on either H2A or H2B was evident in reactions containing eluates from mock transfected cells. In addition, the accumulation of unit length non-ubiquitylated histones in the reactions suggests that the enzymatic activity detected is not simply due to the presence of nonspecific proteases. These data suggest that USP22, whether purified directly or as part of the holo hSAGA complex, possesses histone H2A deubiquitylating activity. The ubiquitylation of histone H2A is catalyzed at least in part by proteins within the Polycomb group of transcriptional repressors. The implications of shared substrate specificity between USP22 and Polycomb PRC1 components are discussed below.
Figure 1.
USP22 catalyzes the deubiquitylation of histones H2A and H2B in vitro. (A) FLAG-tagged USP22 or GCN5 were expressed in 293T cells and affinity purified under nondenaturing conditions. The purification of USP22 and hGCN5 was confirmed by western blotting for the FLAG epitope (upper). Purified USP22 or hSAGA complex was incubated in vitro with ubiquitylated H2A (uH2A) isolated from calf in deubiquitylation assay buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 1 mM DTT and 5% glycerol). After incubation at 37°C for 2 hr, reactions were stopped by the addition of SDS-PAGE sample buffer. Samples were then run on 4%–20% SDS-PAGE gels (Invitrogen) and analyzed by western blot with an anti-H2A antibody from Upstate (lower). In addition to deubiquitylation reactions that included the USP22 complex (lane 3), or hSAGA (lane 4), a mock reaction was analyzed in parallel (lane 2). Ubiquitylated H2A was run as a control (lane 1). (B) The same experiment was performed as described in (A), except that the ubiquitylated H2B (uH2B) isolated from porcine thymus was used as the substrate in the deubiquitylation assay. Samples were analyzed by western blot with an anti-H2B antibody from Upstate.
Discussion
We and others recently demonstrated that USP22 is a ubiquitin hydrolase that is present within the human SAGA transcriptional co-activator complex.7,17 Previously Glinsky et al., demonstrated that USP22 is one of 11 genes linked to the polycomb group that are misexpressed in therapy-resistant tumors.1 Among the genes in this 11 gene signature is Ring1B/RNF2, which resides in the PRC1 complex that represses transcription via the ubiquitylation of histone H2A. The data presented here demonstrate that USP22 can remove this same modification from H2A. These observations suggest a model in which the simultaneous induction of polycomb ubiquitin ligases, such as Ring1B/RNF2 and the ubiquitin hydrolase USP22, is critical during cancer progression because USP22 recruitment allows certain essential cell cycle genes to be transcriptionally activated in the face of the global transcriptional repression catalyzed by the PRC1 complex. Several lines of indirect evidence support this model. For example, both the PRC1 subunit BMI-1 and the hSAGA subunit USP22 are required for appropriate transit through the cell cycle,13 despite their antagonistic enzymatic activities. In addition, USP22 is required for MYC function and BMI-1 was originally discovered as an oncogene capable of cooperating with MYC during transformation.14,15 Thus the biological function of the H2A ubiquitin hydrolase USP22 and the H2A ubiquitin ligase PRC1 complex are largely overlapping, despite their direct antagonism on H2A ubiquitylation.
Among the issues that remain to be explored is whether H2A, H2B or some other substrate is the relevant target of USP22-mediated deubiquitylation during transcription, cell cycle progression and cancer. While the precise answer to this question must await the development of reagents capable of identifying USP22 substrates and selectively blocking their deubiquitylation, the presence of USP22 within the “death from cancer signature” suggests that developing inhibitors of its enzymatic activity might have a therapeutic benefit. This is particularly true given that targeted therapies directed at cancer stem cells are now recognized as a goal worth pursuing.16
Acknowledgements
We thank Drs. Alex Mazo, Ramin Shiekhattar and Amanda Norvell for helpful discussions. This work was supported by grants from the NIH: CA090465 and CA098172 (S.B.M.). In addition, this work was partially supported by funds from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
References
- 1.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4:1171–1175. doi: 10.4161/cc.4.9.2001. [DOI] [PubMed] [Google Scholar]
- 3.Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- 4.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The Putative Cancer Stem Cell Marker USP22 Is a Subunit of the Human SAGA Complex Required for Activated Transcription and Cell Cycle Progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow PS, Schuster T, Finley D. A conserved sequence in histone H2A which is a ubiquitination site in higher eucaryotes is not required for growth in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4905–4911. doi: 10.1128/mcb.10.9.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: a tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- 12.Thorne AW, Sautiere P, Briand G, Crane-Robinson C. The structure of ubiquitinated his-tone H2B. Embo J. 1987;6:1005–1010. doi: 10.1002/j.1460-2075.1987.tb04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goebl MG. The bmi-1 and mel-18 gene products define a new family of DNA-binding proteins involved in cell proliferation and tumorigenesis. Cell. 1991;66:623. doi: 10.1016/0092-8674(91)90106-9. [DOI] [PubMed] [Google Scholar]
- 14.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 15.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 16.Kakarala M, Wicha MS. Cancer stem cells: implications for cancer treatment and prevention. Cancer Journal. 2007;13:271–275. doi: 10.1097/PPO.0b013e318156da4e. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]