Summary
The recognition of specific DNA binding sites by transcription factors is a critical yet poorly understood step in the control of gene expression. Members of the Hox family of transcription factors bind DNA by making nearly identical major groove contacts via the recognition helices of their homeodomains. In vivo specificity, however, often depends on extended and unstructured regions that link Hox homeodomains to a DNA-bound cofactor, Extradenticle (Exd). Using a combination of structure determination, computational analysis, and in vitro and in vivo assays, we show that Hox proteins recognize specific Hox-Exd binding sites via residues located in these extended regions that insert into the minor groove, but only when presented with the correct DNA sequence. Our results suggest that these residues, which are conserved in a paralog-specific manner, confer specificity by recognizing a sequence-dependent DNA structure instead of directly reading a specific DNA sequence.
Introduction
For biological systems to function, defined combinations of macromolecules must interact selectively at the correct time and place, and assemble into productive higher order complexes. One context in which the specificity of macromolecular interactions is critical is in the recognition of DNA sequences by transcription factors, which must select a small subset of relevant binding sites from the large excess of potential binding sites that are typically present in eukaryotic genomes. Atomic-resolution structural studies have provided high resolution views of how the various classes of DNA binding domains recognize their cognate DNA binding sites (Garvie and Wolberger, 2001; Harrison and Aggarwal, 1990; Pabo and Sauer, 1992). However, despite these insights, how transcription factors discriminate between closely related, yet biologically distinct, DNA sequences is not well understood.
Here, we address this problem for the Hox family of homeodomain-containing transcription factors, which assign morphological identities along the antero-posterior (AP) axis of both vertebrates and invertebrates. To execute their functions, Hox proteins regulate many types of target genes, some of which are specific for a particular Hox paralog (Pearson et al., 2005). In Drosophila, for example, the Hox protein Sex combs reduced (Scr) is the only paralog that is able to initiate salivary gland development (Andrew et al., 1994; Panzer et al., 1992). In contrast, other Hox functions and targets are shared by multiple Hox paralogs. For example, many Hox proteins share the ability to repress the antennal-specifying gene homothorax (hth) and at least two Hox paralogs have the capacity to repress the appendage-specifying gene Distalless (Dll) (Casares and Mann, 1998; Vachon et al., 1992; Yao et al., 1999). This distinction between paralog-specific and shared Hox target genes implies that some Hox binding sites must be specific for a particular Hox paralog, while other binding sites may not require the same degree of specificity. Confounding the Hox specificity problem is that this family of proteins, which are encoded by eight Hox paralogs in Drosophila that have been duplicated to a total of 39 Hox genes in humans, all bind to very similar DNA sequences (Gehring et al., 1994; Mann, 1995).
A growing list of directly regulated Hox target genes supports the view that Hox proteins use a variety of mechanisms to recognize their in vivo binding sites (Pearson et al., 2005). At one extreme, the regulation of some target genes requires that Hox proteins bind DNA cooperatively with cofactors, notably, Extradenticle (Exd; Pbx in vertebrates) and Homothorax (Hth; Meis in vertebrates) (Mann and Affolter, 1998; Mann and Chan, 1996; Moens and Selleri, 2006). For example, to initiate salivary gland development, Scr binds with Exd to a paralog-specific Scr-Exd binding site to activate its target fork head (fkh) (Ryoo and Mann, 1999). At the other extreme, Hox proteins are capable of regulating some target genes via clusters of what appear to be Hox monomer binding sites, without the help of Exd or Hth (Galant et al., 2002; Hersh and Carroll, 2005; Lohmann et al., 2002). One possibility is that paralog-specific Hox functions are more dependent on cofactor interactions than functions that are not paralog-specific.
Complementing our knowledge of in vivo Hox binding sites are studies on chimeric Hox proteins, which identified the protein domains required for their specific in vivo functions (Chauvet et al., 2000; Zhao and Potter, 2002); reviewed by (Mann, 1995; Mann and Morata, 2000). These studies reveal that Hox homeodomains, in particular their N-terminal arms, are critical for specificity. However, homeodomain N-terminal arms, especially the first four residues, are typically disordered in the existing NMR and crystal structures (Billeter et al., 1993; Fraenkel and Pabo, 1998; Fraenkel et al., 1998; Hirsch and Aggarwal, 1995; Hovde et al., 2001; Li et al., 1995; Passner et al., 1999; Piper et al., 1999; Tucker-Kellogg et al., 1997; Wolberger et al., 1991). Hox N-terminal arms are also mostly disordered in two X-ray structures of Hox-Exd/Pbx-DNA ternary complexes (Passner et al., 1999; Piper et al., 1999). Adding to this paradox, all of these structures show the third α-helix (the so-called recognition helix) of these homeodomains making nearly identical base-specific contacts in the DNA major groove. Thus, while the existing structures explain how Hox proteins recognize AT-rich sequences, they do not account for their in vivo specificities. Further, although the Hox-Exd/Pbx ternary structures reveal how these proteins interact with each other while bound to DNA, they fail to explain the observed sequence specificities of Hox-Exd/Pbx dimers (LaRonde-LeBlanc and Wolberger, 2003; Passner et al., 1999; Piper et al., 1999). As suggested by our results, a likely reason for this shortcoming is that the binding sites present in these structures are not derived from a specific in vivo target, but instead are high affinity consensus sites derived from in vitro selection experiments.
In the present study, we use X-ray crystallography to solve the structures of two Hox-Exd-DNA ternary complexes. In one complex, an Scr-Exd dimer is bound to a DNA sequence (fkh250), derived from the fkh gene, that shows a preference for Scr-Exd over other Hox-Exd dimers, both in vitro and in vivo (Ryoo and Mann, 1999). In the second complex, an Scr-Exd dimer is bound to a variant of fkh250 (fkh250con*) in which the Scr-Exd binding site was replaced by a consensus Hox-Exd site. These two structures thus provide a direct comparison between Scr-Exd bound to paralog-specific and non-specific Hox-Exd binding sites. Although the overall arrangement of the proteins is similar to each other and to previous Hox-Exd/Pbx structures, additional Scr residues, in the N-terminal arm and adjacent linker region, are ordered in the fkh250 complex. Among these, an arginine and a histidine, which are conserved among Scr orthologs, insert into a narrow region of fkh250's minor groove. We show that these residues are important for Scr to bind fkh250 and carry out Scr-specific functions in vivo. Analysis of the fkh250 sequence suggests that its minor groove geometry is an intrinsic property of its sequence. As a consequence of its shape, the fkh250 minor groove creates an enhanced negative electrostatic environment ideally suited for binding basic residues. Thus, by correctly positioning Scr's N-terminal arm and linker residues, Exd facilitates an interaction with the specific conformation of the fkh250 binding site. Together with previous results, we suggest that Hox proteins recognize ‘generic’ binding sites through major groove–recognition helix interactions, but that N-terminal arm and linker residues select among these sites by reading the structure and electrostatic potential of the minor groove.
Results
Scr binds cooperatively with Exd to the Hox-Exd binding site in fkh250, AGATTAATCG, while other Hox-Exd dimers fail to bind this sequence as well (Ryoo and Mann, 1999). To understand the basis for this specificity, we generated cocrystals and solved the structures of Scr and Exd bound to 20 bp overhanging oligonucleotides containing either the fkh250 and fkh250con* binding sites. fkh250con* is identical to fkh250 except in three base pairs which were changed to match the Hox-Exd/Pbx consensus sequence present in the Ubx-Exd ternary structure (Passner et al., 1999) (see Experimental Procedures) (Figure 1B). For the crystallizations, we expressed Scr (residues 298 to 385), which includes the Exd-interacting peptide YPWM, the linker region between YPWM and the homeodomain, and the full-length homeodomain (Figure 1A). For Exd, we expressed the full-length homeodomain (residues 238 to 300).
Figure 1. Overview of structures and sequences.
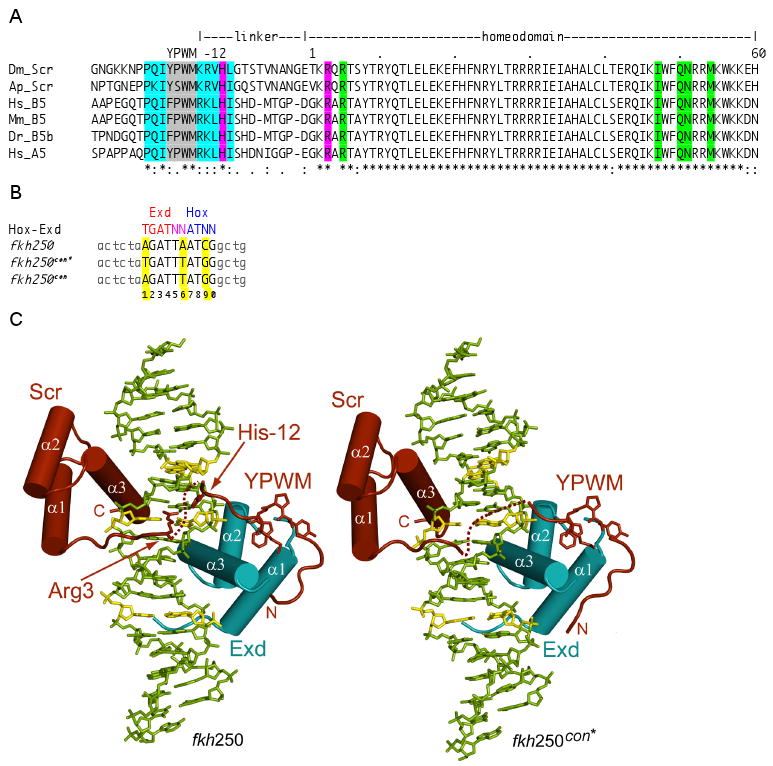
A. Comparison of the YPWM, linker, and homeodomain sequences of a subset of Scr orthologs. Residue numbering is relative to the first homeodomain residue, which is +1. Residues highlighted in cyan distinguish Scr and its orthologs from other Hox paralogs. His-12 and Arg3 are highlighted in magenta. DNA contacting residues that are shared by all Hox paralogs are highlighted in green. Dm, Drosophila melanogaster; Ap, Apis melifera; Hs, Homo sapiens; Mm, Mus musculus; Dr, Danio rerio.
B. Sequences of a generalized Hox-Exd site, and the fkh250con*, fkh250con, and fkh250 binding sites (in capital letters; identical flanking base pairs are shown in light grey). fkh250 and fkh250con*, which is a better match to the Hox-Exd consensus than fkh250con, were used for the crystallography. The base pairs in the Hox-Exd binding site are numbered 1 to 10. The Scr-Exd binding site in fkh250 is 100% conserved in all of the sequenced Drosophila genomes except for D. virilis, where only the last G has been changed to an A.
C. Overview of the fkh250 (left) and fkh250con* (right) complexes. The Scr (red) and Exd (cyan) homeodomains bind to opposite faces of the DNA. Arg3 and His-12 are seen in the fkh250 complex but not in the fkh250con* complex. Base pairs colored in yellow are those that differ between fkh250 and fkh250con*.
The structures of the Scr-Exd-fkh250 and Scr-Exd-fkh250con* complexes
The cocrystals of both complexes (hereafter referred to as the fkh250 and fkh250con* complexes, respectively) diffracted to 2.6 Å and each contained one complex per asymmetric unit (Supp. Table 1). Scr and Exd bind fkh250 and fkh250con* in a very similar manner using overlapping DNA sites in a head-to-tail orientation on opposite faces of the DNA (Figure 1C). For the segments of the proteins that are visible in both structures, the protein main chains superimpose with a Cα-RMSD of only 0.8 Å. Most of the contacts to the DNA backbone are also shared in the two structures, except that there are more water-mediated contacts to the DNA backbone in the fkh250 complex (Figure 2A, B).
Figure 2. Protein-DNA contacts.
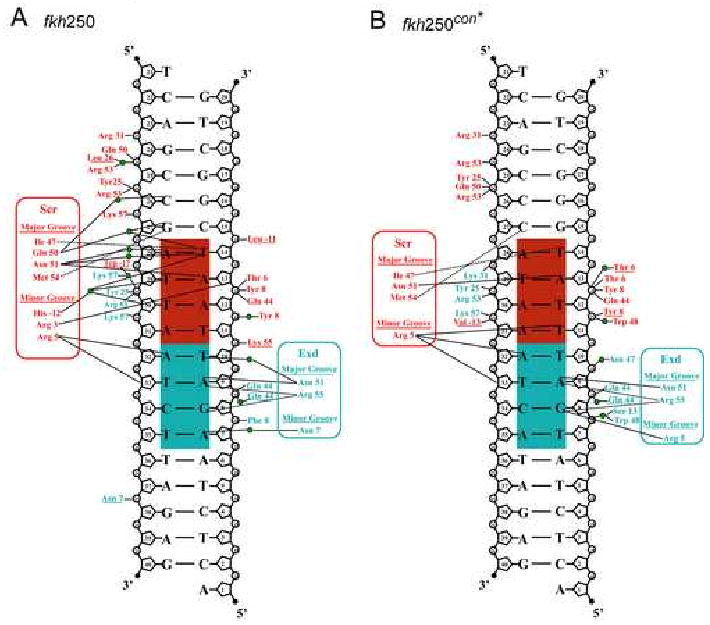
Protein-DNA contacts for the (A) fkh250 and (B) fkh250con* complexes. The Exd half site is shaded cyan, the Hox half site is shaded red. Hydrogen bonds are represented by solid lines and non-polar interactions by dotted lines. Interactions involving the protein main chain are underlined. Green circles are visualized water molecules. The fkh250 complex has more water mediated contacts from residues such as Gln50, that is oriented differently in the two complexes, and Trp-17, that is positioned identically in the two complexes.
Scr can be divided into its homeodomain (residues 1-60), composed of a tri-α-helical core (residues 10-60) and N-terminal arm (residues 1-9), and an extended linker region containing the YPWM motif (residues -25 to -1). Compared to most homeodomains, the Exd/Pbx homeodomain (residues 1-22, 23a-c, 24-60) has an additional three residues (23a, 23b and 23c) in the loop between helices α1 and α2. As seen previously (LaRonde-LeBlanc and Wolberger, 2003; Passner et al., 1999; Piper et al., 1999), these residues form part of the hydrophobic pocket that receives the Scr YPWM motif in both the fkh250 and fkh250con* complexes. Protein-DNA interactions arise primarily from the Scr and Exd homeodomains, whose recognition helices (α3s) project along the DNA major groove while their N-terminal arms (residues 5 to 9 for fkh250con* and 3 to 9 for fkh250) wind along the minor groove. The major groove interactions (Figures 2A,B) are similar to those described previously for the Ubx-Exd ternary complex (Passner et al., 1999) and are consistent with the patterns observed for other homeodomain protein-DNA complexes (Billeter et al., 1993; Fraenkel et al., 1998; Gehring et al., 1994).
In both the fkh250 and fkh250con* complexes, the N-terminal segment that includes the YPWM motif is ordered (residues -26 to -11 in fkh250 and residues -27 to -13 in fkh250con*). The YPWM motif folds into a classical type I reverse turn and, as in the Ubx-Exd and Hoxb1-Pbx complexes, a hydrogen bond is formed between the first (Tyr) and the fourth (Met) residues of the turn. As in other complexes, the Trp plays the dominant role in Hox-Exd/Pbx interactions, mediating many of the contacts within the Exd hydrophobic pocket including a hydrogen bond to the main carbonyl of Leu23a of Exd. The segments N- and C-terminal to the YPWM motif assume extended conformations and are positioned at right angles to each other. These segments contribute to the Scr-Exd interaction, in which Pro-22 and Ile-20 of Scr make hydrophobic contacts with Ile60 of Exd, and the backbone carbonyl of Lys-15 in Scr makes a hydrogen bond with Lys57 of Exd.
A unique feature of the fkh250 complex is the entry of Arg3 and His-12 into the DNA minor groove (Figure 3). The contacts made by these two residues are especially interesting in light of the fact that they are highly conserved among all Scr orthologs, both in vertebrates and invertebrates (Figure 1A). Arg3 lies approximately equidistant between the sugar-phosphate backbones of the two DNA strands that line the narrow minor groove. The guanidinium nitrogens are approximately 4 Å away from the bases (N3 atom of Ade13 and O2 atom of Thy30) and do not make direct or water mediated hydrogen bonds with the bases. His-12 lies deeper within the groove and its Nε2 atom is linked to a water molecule, which in turn makes hydrogen bonds to three thymines (Thy14, Thy29, and Thy30) within the Scr recognition sequence (Figure 3B). Hydrogen bonds to Thy14 and Thy29 are made with the base O2 atoms, while the bond to Thy30 is made with the sugar O4′ atom. The Arg3 guanidinium group and the His-12 imidazole ring lie approximately in the same plane, and a hydrogen bond between the groups (Nη1-H…Nδ1) suggests some synergy in the interactions of Arg3 and His-12 with DNA. In contrast to the fkh250 complex, neither Arg3 nor His-12 are seen in the fkh250con* complex.
Figure 3. Minor groove insertion of Scr residues His-12 and Arg3 in fkh250.
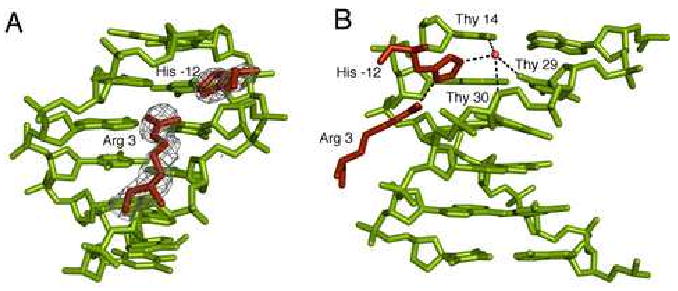
A. Electron densities for Arg3 and His-12 in the fkh250 complex, based on a simulated annealing Fo-Fc omit map (contoured at 3.0σ)
B. Details of the His-12–Arg3 interaction and water-mediated interactions with Thy14, Thy29, and Thy30 of fkh250. The red circle marks a water molecule and dotted lines represent putative hydrogen bonds.
Since the proteins are identical in both complexes, the differences between the two structures are likely a consequence of the different DNA sequences. As seen in the crystal structures, the minor groove in the fkh250 complex is generally narrower than in the fkh250con* complex, particularly where Arg3 and His-12 insert (Figure 4A-D). Arg5, which is ordered in both complexes, also inserts into a narrow region of the minor groove present in both fkh250 and fkh250con*. AT-rich regions often lead to narrow minor grooves due in large part to negative propeller twisting (Crothers and Shakked, 1999). Consistently, the average value for propeller twist in these regions is -14.6° for fkh250 and -12.2° for fkh250con*, and the pattern of inter-base pair contacts in the major groove is similar to that observed in other AT-rich sequences (Nelson et al., 1987) (see Discussion).
Figure 4. Differences in minor groove geometries and electrostatic potentials between fkh250 and fkh250con*.
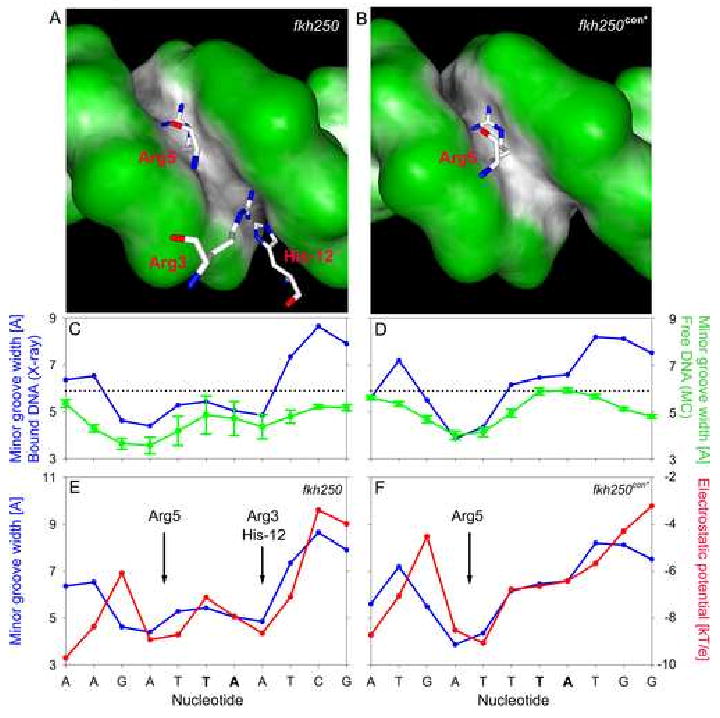
A,B: The surfaces of the (A) fkh250 and (B) fkh250con* DNAs, color-coded according to shape using GRASP (Nicholls et al., 1991). Black/gray and green surfaces represent concave and convex surfaces, respectively. Side chains for Arg5 (A and B) and Arg3/His-12 (A, only) entering the minor groove are shown in magenta.
C,D: Graphs comparing minor groove widths seen in the crystal structures (blue curves) with those predicted by the MC simulations (green curves) for fkh250 (E) and fkh250con* (F). The sequences of the two binding sites are below; TpA steps are shown in bold lettering.
E,F: Graphs comparing minor groove widths (Å) and electrostatic potential (kT/e) for fkh250 (E) and fkh250con* (F), based on the crystal structures. The positions of Arg5 and Arg3/His-12 minor groove insertion are indicated.
These observations suggest that the differences in minor groove shape seen in the two crystal structures arise as a consequence of the known conformational preferences of the two DNA sequences. The differences in minor groove shape were also observed from all-atom Monte Carlo (MC) simulations of the free DNAs (see Experimental Procedures) (Rohs et al., 2005b). In agreement with the crystal structures, these simulations predict a single minor groove width minimum in fkh250con* and two minima in fkh250 (Figure 4C,D). Similar agreement exists between the MC simulations and X-ray structures for a second DNA helical parameter, roll, in the two DNAs (Supp. Figure 1). In addition, we carried out MC simulations of DNAs with all possible combinations of the three base pair differences between the fkh250 and fkh250con* binding sites (Supp. Figs. 2 and 3). These analyses suggest that the differences at positions 6 and 9 play a more important role than the difference at position 1 in distinguishing between the structures of these two binding sites.
Electrostatic potentials correlate with minor groove width
Electrostatic potentials are affected by the shape and charge distribution of macromolecules (Honig and Nicholls, 1995). To determine if differences in electrostatic potential provide the physical basis for the interaction between basic amino acids and the minor groove seen in the fkh250 complex, we used the DelPhi program to calculate this parameter for both DNA sequences. Strikingly, for both sequences, there is a near-perfect correlation between minor groove width and the magnitude of the negative electrostatic potential (Figure 4E,F). In both complexes, Arg5 inserts into the minor groove in a region where there is a minimum both in groove width and in electrostatic potential. In the fkh250 structure there is a second minimum for both parameters where the His-12/Arg3 pair inserts. In contrast, in the corresponding location in the fkh250con* complex, the minor groove is wide and the electrostatic potential is less negative. The difference in the electrostatic potential between the two sites, about 2.2 kT/e, corresponds to about an order of magnitude in affinity for a single positive charge. This difference can account for the observation that His-12/Arg3 inserts into the minor groove of fkh250 but not into the groove of fkh250con*. In both cases there is a net electrostatic attraction between the DNA and these basic amino acids, but this attraction must be large enough, as it appears to be in fkh250, to overcome the entropy loss associated with inserting an unstructured peptide into a specific location on the surface of DNA.
His-12 and Arg3 of Scr are important for binding fkh250
To test if His-12 and Arg3 contribute to Scr's affinity for fkh250, we mutated these residues either individually or in the same protein to alanines to generate three mutants, ScrHis-12A, ScrArg3A, and ScrHis-12A,Arg3A, and carried out DNA binding studies. For these experiments, we compared fkh250 with fkh250con, whose distinct properties have been characterized both in vitro and in vivo (Ryoo and Mann 1999) (Figure 1B). In particular, unlike fkh250, fkh250con binds to multiple Hox-Exd dimers equally well (Ryoo and Mann 1999). Moreover, although a lacZ reporter gene made with fkh250 (fkh250-lacZ) is specifically activated in vivo by Scr and exd, an analogous reporter gene made with fkh250con (fkh250con-lacZ) is activated by multiple Hox proteins in vivo (Ryoo and Mann 1999).
We carried out several experiments to test if Arg3 and His-12 were required for Scr's ability to bind fkh250. These DNA binding studies were all done in the presence of Exd and the HM domain of Hth (HthHM), to best mimic the in vivo requirement for these cofactors (Noro et al., 2006) (see Experimental Procedures). First, we compared the ability of wild type Scr (ScrWT) and the double mutant (ScrHis-12A,Arg3A) to bind fkh250 and fkh250con over multiple protein concentrations. Using concentrations where ScrWT and ScrHis-12A,Arg3A bound similarly to fkh250con, ScrHis-12A,Arg3A bound fkh250 ∼30% as strong as ScrWT (Figure 5A,B). Second, we measured the Kds for all four proteins to both binding sites (Figure 5C). These measurements indicate that ScrWT and ScrHis-12A have similar affinities to both fkh250 and fkh250con (all within ∼8 to 15 nM). ScrHis-12A,Arg3A also had a similar affinity for fkh250con (∼18 nM). Significantly, both ScrHis-12A,Arg3A and ScrArg3A had lower affinities to fkh250 (∼55 and 47 nM, respectively) (Figure 5C). Together, these measurements suggest that His-12 and Arg3 of Scr are more important for binding to fkh250 than to fkh250con. Further, although Arg3 is more critical than His-12, both of these residues appear to contribute to this interaction because the double mutant has a lower affinity for fkh250 than the single Arg3A mutant.
Figure 5. Binding of Scr and Scr mutants to fkh250 and fkh250con DNAs.
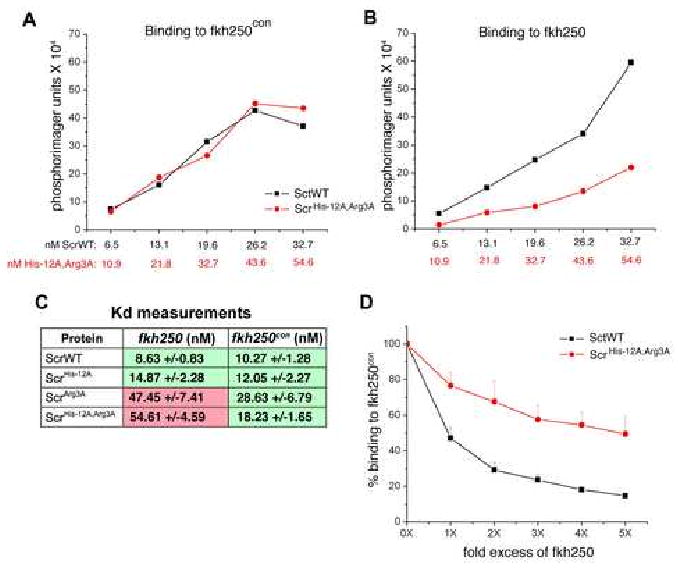
A, B: Binding of ScrWT and ScrHis-12A,Arg3A to fkh250con (A) and fkh250 (B). Protein concentrations were chosen to give equivalent binding of ScrWT and ScrHis-12A,Arg3A to fkh250con (A), and the same concentrations were used to measure binding to fkh250 (B). All DNA binding measurements were done in the presence of HthHM-Exd (see Experimental Procedures).
C: Binding affinities (Kds in nM) of the indicated proteins for fkh250 and fkh250con. Green and pink shaded boxes indicate interactions that can and cannot activate the respective reporter gene in vivo (data from Figure 6F-O).
D: Competition experiment. Unlabeled fkh250 oligo at the relative concentrations indicated on the X axis was used to compete for the formation of ScrWT (black curve) and ScrHis-12A,Arg3A (red curve) complexes formed on a labeled fkh250con probe.
As a third test to see if these residues are required for DNA binding specificity, we carried out a DNA binding competition assay with ScrWT and ScrHis-12A,Arg3A (Figure 5D). In this experiment, ScrWT or ScrHis-12A,Arg3A was bound to 32P-labeled fkh250con in the presence of varying concentrations of unlabeled fkh250. In agreement with our previous results, fkh250 was a much better competitor of ScrWT complex formation than of ScrHis-12A,Arg3A complex formation (Figure 5D). Thus, ScrWT is better able to discriminate between fkh250 and fkh250con than ScrHis-12A,Arg3A, suggesting that His-12 and Arg3 are critical for Scr's DNA binding specificity.
Although Arg3 of Scr is critical for Scr-Exd to bind fkh250, this residue is not unique to this Hox paralog. In particular, Ubx, Antp, and Abdominal-A (Abd-A) all have Arg3 (and Arg5), but do not bind fkh250 well with Exd (Ryoo and Mann, 1999). In Scr, Arg3 is part of an RQR motif, whereas it is part of an RGR motif in these other Hox proteins, suggesting that the context Arg3 may be important. Although we have little structural information about Hox RGR motifs, crystal structures are available for several other transcription factors that contain a DNA-bound RGR motif (Cheetham et al., 1999; Huth et al., 1997; Meinke and Sigler, 1999). In these cases the Gly inserts into the minor groove in a region where it is not narrow, while the arginines splay out in very different directions than they do in Scr. Based on these observations, we mutated Gln4 in Scr to Gly (thus changing RQR to RGR) and measured the affinity of this mutant to fkh250 and fkh250con. This point mutation reduced Scr-Exd's affinity for fkh250 by about six-fold, but only reduced affinity for fkh250con by ∼two-fold (Supp. Table 2). The resulting Kds are very similar to those of ScrArg3A, and are remarkable because the side chain of Gln4 makes no DNA contacts.
His-12 and Arg3 are important for Scr to execute its specific functions in vivo
The above results provide biochemical evidence that His-12 and Arg3 are important for Scr to bind its specific binding site, fkh250. To test if these residues are important for Scr's activity in vivo, we generated transgenic lines of Drosophila capable of mis-expressing ScrWT and the three Scr mutants described above. For these in vivo assays, a full length form Scr was used. Each of these proteins was N-terminally tagged with a hemagglutinin (HA) epitope, allowing us to compare transformants that express similar levels of nuclear-localized protein (data not shown). As described previously (Gibson et al., 1990), uniform expression of ScrWT during embryogenesis resulted in a characteristic and Scr-specific transformation of the second and third thoracic segments (T2 and T3) towards the identity of the first thoracic segment (T1), where Scr is normally expressed (Figure 6A,B). A weaker transformation of the anterior abdominal segments (A1 to A3) was also evident. This transformation is readily recognized by the acquisition of a normally T1-specific pattern of small hairs (the T1 beard) in each transformed segment. To quantify this transformation, we counted the number of hairs present in each affected segment after ubiquitous expression of ScrWT and the Scr mutants. ScrHis-12A,Arg3A was severely compromised in its ability to produce this transformation (Figure 6C). ScrHis-12A was able to produce this transformation as efficiently as ScrWT, while ScrArg3A was partially compromised in this assay (best seen in A1 to A3).
Figure 6. Compromised in vivo activities of Scr mutants.
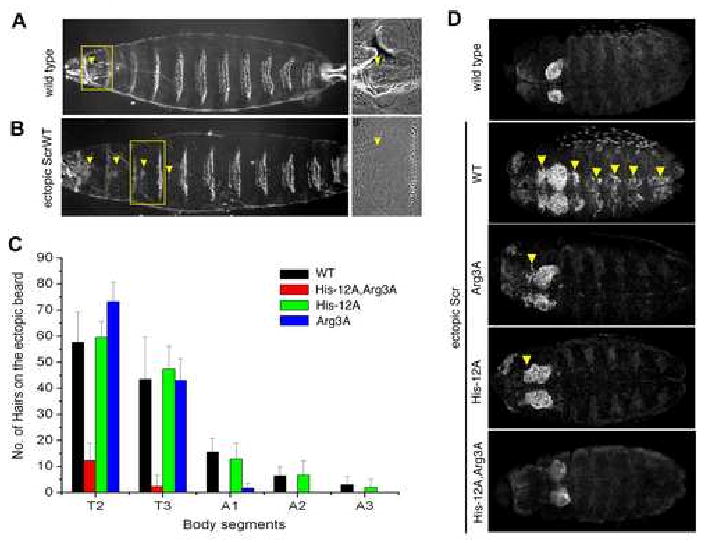
A: Ventral surface of wild type first instar larval cuticle. A′: higher magnification of wild type T1 segment showing a T1 beard (arrowhead).
B: Ventral surface of a first instar cuticle after ubiquitous expression of ScrWT. B′: higher magnification of the T3 segment with an ectopic T1 beard (arrowhead).
C: Quantification of ectopic T1 beard formation in thoracic (T2 and T3) and abdominal (A1 to A3) segments due to ubiquitous expression of ScrWT and Scr mutants. For comparison, the number of beard hairs in a wild type T1 segment is 128.2 +/-8.4.
D: Ventral views of wild type embryos, or embryos ubiquitously expressing ScrWT or the indicated Scr mutant, stained for dCrebA. Ectopic activation is indicated by arrowheads.
Another Scr-specific function is to initiate the formation of the salivary gland, and a good marker for this structure is dCrebA (Andrew et al., 1994; Panzer et al., 1992). ScrWT, but not ScrHis-12A,Arg3A, was able to induce ectopic CrebA expression, while ScrHis-12A and ScrArg3A were able to weakly activate this marker (Figure 6D). Taken together, these results demonstrate that His-12 and Arg3 are important for Scr to execute its specific functions in vivo. They also highlight that for some readouts, such as dCrebA, both residues are critical, whereas for other readouts, such as the T1-specific cuticle pattern, both residues must be mutated to eliminate activity (Supp. Table 2).
His-12 and Arg3 are required for Scr to activate fkh250-lacZ and fkh
The above in vivo data demonstrate that His-12 and Arg3 are required for Scr to generate an Scr-specific transformation of segmental identity. If this hypothesis is correct, we would expect ScrHis-12A,Arg3A to be unable to activate the fkh250-lacZ reporter gene. In wild type embryos, fkh250-lacZ is expressed in parasegment (PS) 2 (Figure 7F) where it is dependent on Scr and exd activities (Ryoo and Mann, 1999). When ScrWT was ubiquitously expressed during embryogenesis, ectopic activation of fkh250-lacZ was observed in segments anterior and posterior to PS2 (Figure 7G). ScrHis-12A also activated fkh250-lacZ (Figure 7I). In contrast, ubiquitous expression of ScrHis-12A,Arg3A or ScrArg3A failed to activate this reporter gene (Figure 7H,J). Similar results were obtained when the expression of the endogenous fkh gene was monitored, although in this case ScrHis-12A was a less potent activator than ScrWT (Figure 7A-E). Thus, His-12 and Arg3 are critical for Scr's ability to activate a paralog-specific target gene in vivo. As with the in vitro binding data, these results suggest that Arg3 is more critical than His-12 for the activation of these genes (Supp. Table 2).
Figure 7. ScrHis-12A,Arg3Ais unable to execute specific Scr functions in vivo.
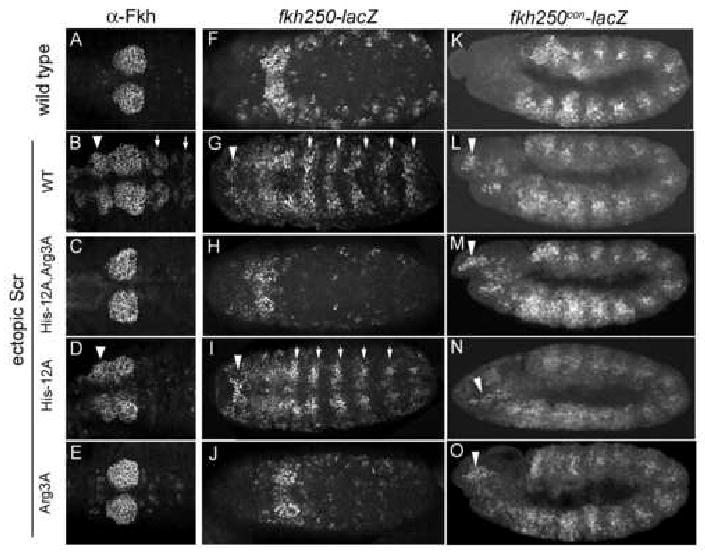
A-E: Wild type embryos or embryos ubiquitously expressing ScrWT or the indicated Scr mutants, stained for Fkh protein. Ectopic activation in anterior (arrowheads) and posterior (arrows) segments is indicated. Ventral views of PS1 to PS4 are shown.
F-J: Expression of fkh250-lacZ, revealed by anti-β-gal antibody staining, in wild type embryos or embryos ubiquitously expressing ScrWT or Scr mutants. Ectopic activation in anterior (arrowheads) and posterior (arrows) segments is indicated. Shown are ventral views.
K-O: Expression of fkh250con-lacZ, revealed by anti-β-gal antibody staining, in wild type embryos or embryos ubiquitously expressing ScrWT or Scr mutants. Ectopic activation in the head is indicated (arrowheads). Due to the already broad expression pattern in wild type embryos (K), lateral views are shown to visualize the ectopic expression.
His-12 and Arg3 are not required for Scr to activate fkh250con-lacZ
Based on the above results, we suggest that His-12 and Arg3 are important for Scr to execute its specific functions, such forming the T1 beard, inducing salivary gland development, or activating fkh250-lacZ. Our observation that 1) His-12 and Arg3 are not ordered in the fkh250con* complex and 2) mutating these two residues to alanines only weakly affects Scr's ability to bind to the non-paralog specific Hox-Exd binding site in fkh250con, suggests that these residues may be less important for Scr to execute shared Hox functions. A prediction based on this idea is that ScrHis-12A,Arg3A should still be able to activate the fkh250con-lacZ reporter gene, which can be activated by multiple Hox proteins in vivo (Ryoo and Mann, 1999). Consistently, ScrWT and all three Scr mutants were able to activate this reporter gene in vivo (Figure 7K-O).
Discussion
It is well established that homeodomain-DNA recognition utilizes hydrogen bonds formed between recognition helix side chains and base-specific moieties in the major groove (Gehring et al., 1994; Mann, 1995). However, the residues making these contacts are identical in all Hox proteins. While some N-terminal arm residues have been seen in the minor groove, these interactions have not been sufficient to account for specificity differences among Hox proteins. In particular, although Arg5 is often observed in the minor groove, it is common to all homeodomains. Conversely, residues 1 to 4 are important for Hox specificity, but are often not observed in homeodomain–DNA structures. The structure reported here, of a complex formed between a Scr-Exd dimer and an in vivo paralog-specific binding site, fkh250, reveals Hox–DNA contacts that provide new insights into the molecular basis of Hox specificity. We show that minor groove contacts from linker (His-12) and N-terminal arm (Arg3) residues are critical for Scr's specific in vitro and in vivo properties. Moreover, both residues insert into an unusually narrow region of the minor groove, which in turn creates a local dip in electrostatic potential through the phenomenon of electrostatic focusing (Honig and Nicholls, 1995; Klapper et al., 1986). In contrast, in the fkh250con* complex, the minor groove does not have these features, and, like many of the previous structures, there are no DNA contacts N-terminal to Arg5.
Based on these findings, we suggest that there are two conceptually separable components to Hox–DNA binding. First, contacts between the DNA major groove and the recognition helix are sufficient to target Hox homeodomains to ‘AT-rich’ DNA sequences. Second, contacts made between the DNA minor groove and N-terminal arm/linker residues help to discriminate among AT-rich binding sites. Unlike recognition-helix residues in the major groove, the residues that insert into the minor groove recognize a specific DNA structure instead of forming base-specific hydrogen bonds. Below we discuss the implications of these findings for binding site recognition by Hox proteins as well as other DNA binding proteins.
Sequence-dependent DNA structure
Consecutive ApA, TpT, or ApT base pair steps are known to result in a narrow minor groove due to negative propeller twisting that is stabilized by inter-base pair interactions in the major groove (Crothers and Shakked, 1999). In contrast, due to poor base stacking interactions, TpA steps tend to widen the minor groove (Burkhoff and Tullius, 1988; Stefl et al., 2004) and, for example, produce significant unwinding in the case of the TATA box (Kim et al., 1993). We suggest that these sequence-dependent effects on DNA structure can account for the conformations of the two DNAs observed here. The fkh250con* binding site is TGATTTATGG (TpA steps are underlined). ATTT is expected to have the observed narrow minor groove where Arg5 binds. The AT sequence 3′ to the TpA step is too short to produce the pattern of inter-base pair contacts required for minor grove narrowing. Moreover, the minor groove that is widened by this TpA step remains wide, in part due to the 3′ guanines which introduce amino groups into the minor groove. In contrast, the fkh250 binding site is AGATTAATCG. Here, the ATT and AAT sequences flanking the TpA step both have the pattern of inter-base pair contacts and propeller twisting required for minor groove narrowing. Consequently, two minor groove width minima are observed (Figure 4). The second minimum, where His-12/Arg3 insert, is reinforced by a positive roll introduced by a 3′-CpG step (Hizver et al., 2001; Rohs et al., 2005b).
The DNA conformations observed in the crystal structures were qualitatively reproduced by our MC simulations, and the importance of the TpA steps and 3′ flanking G-C base pairs in affecting DNA structure were supported by the simulations of DNAs containing individual base pair differences (Supp. Figs. 2 and 3). Interestingly, the standard deviations observed in these simulations are different for fkh250 and fkh250con (Figure 4C,D). This difference, which may reflect an inherent difference in flexibility, is also consistent with known sequence-dependent properties of DNA (Faiger et al., 2007). The fkh250con* sequence, which shows a smaller standard deviation, is expected to be rigid due to the presence of an “A-tract”, a sequence that consists of at least three consecutive ApA, ApT or TpT steps (Crothers and Shakked, 1999). In contrast, the larger deviations seen in the fkh250 simulations indicate greater conformational flexibility that can be attributed to the absence of an A-tract and the presence of a TpA step in the middle of the sequence.
Arg3 and the N-terminal arm
The N-terminal arm has been known for some time to play an important role in Hox specificity. Consistent with this idea, we find Arg3 and Arg5 in the minor groove of fkh250. However, Arg5 is conserved in all homeodomains and Arg3 is present in many Hox proteins, raising the question of what makes Scr's N-terminal arm unique. One answer is that other N-terminal arm differences are important for Scr's properties. In agreement with this notion, we found that changing RQR to RGR reduced the affinity for fkh250 by ∼six-fold, similar to the effect observed when Arg3 was mutated to Ala. These data suggest that, unlike RQR of Scr, it is energetically unfavorable for the RGR motifs of Antp, Ubx, and AbdA to assume the conformation of the RQR motif as seen in the fkh250 complex. This may be due in part to the increased entropic cost associated with fixing a Gly in any given conformation but also to the fact that its lack of a Cβ precludes the formation of the hydrophobic contact formed between Gln4 and Thr6 in the fkh250 complex (the distance between the Cδ of Gln4 and the Cγ of Thr6 is about 4.7 Å).
Taken together, our results suggest that the conformational preferences of Hox N-terminal arms are an important determinant of Hox specificity. However, there is clearly more to the story because, like Scr, Deformed (Dfd) also has an RQR motif in its N-terminal arm, but Dfd does not activate fkh250-lacZ in vivo. Thus, while the sequence of the N-terminal arm plays an important role, and allows Hox proteins to be categorized into RGR and RQR subgroups, other specificity-determining factors must also exist. Based on our results, and as discussed below, we suggest that other important contributors are the paralog-specific residues neighboring the YPWM motif.
Paralog-specific signature residues
His-12 is located in Scr's linker region, four residues away from its YPWM motif. Interestingly, not only is His-12 conserved in all Scr orthologs, residues on both sides of its YPWM motif are also well conserved (Figure 1A). This pattern is not unique to Scr and its orthologs: residues in the vicinity of Hox YPWM motifs are generally conserved in a paralog-specific manner (Supp. Figure 4) (Mann, 1995; Sharkey et al., 1997). In fact, the evolutionarily conserved sequences in the vicinity of YPWM are sufficient to distinguish between Hox paralogs, and can even discriminate between Scr and Deformed (Dfd), which, like Scr, also has a His in the same position relative to its YPWM motif (Supp. Figure 4). These observations suggest that paralog-specific residues near the YPWM motif, together with the N-terminal arm, may be considered as specificity-determining ‘signature’ residues. Analogous to our findings with Scr-fkh250, we suggest that these paralog-defining residues in other Hox proteins are critical for the recognition of specific binding sites in vivo. These residues may, as shown here for His-12 and Arg3 of Scr, contact DNA. Alternatively, as shown here for Scr's Gln4, they may be important for specifying the correct conformation of the DNA-contacting residues. A general role for linker and N-terminal arm residues in Hox specificity is supported by the in vivo specificities of Hox protein chimeras (Chauvet et al., 2000); reviewed by (Mann, 1995; Mann and Morata, 2000).
Although His-12 is conserved among all Scr orthologs, mutating it to an Ala had, for most readouts, only a partial effect on binding or in vivo activity. In contrast, the Arg3 to Ala mutation had a much larger effect, and the strongest effect was observed when both His-12 and Arg3 were mutated to Ala (Supp. Table 2). Some simple considerations can in principle account for the data. First, we suggest that the main contribution of His-12/Arg3 is to provide a positive charge and, consequently, a favorable electrostatic interaction between Scr and fkh250. Second, given the N-N distance of 2.9 Å in the His-Arg hydrogen bond, His-12 is likely neutral in the fkh250 complex, so that the net charge for both residues is +1. In the double mutant this charge is lost. The His-12 to Ala mutation leaves Arg3 intact and the net charge unchanged. The Arg3 to Ala mutation would likely result in the protonation of His-12 given the negative electrostatic environment in the minor groove, also leaving the net charge of the protein unchanged. While these considerations can explain why the effect of the double mutant is stronger than of either single mutant, they do not explain why ScrArg3A binds more weakly to fkh250 than ScrWT or ScrHis-12A. One possibility is that there is an unfavorable free energy cost of proton uptake to His-12 when it is bound to DNA since, as opposed to Arg3, the free His is only partially protonated.
The role of Hox-Exd dimer formation
Our results suggest that the interaction of Hox proteins with Exd/Pbx through the YPWM motif is important, not only because the presence of two homeodomains allows for a larger and more specific DNA sequence readout in the major groove, but also because it favors conformations of the linker and N-terminal arm residues such that they can recognize structural patterns in the minor groove. Indeed, it appears that these residues are unable to assume these conformations in the absence of Exd/Pbx. That these residues have not been observed in two other Hox-Exd/Pbx ternary complexes may suggest that their intrinsic flexibility is designed to inhibit binding to the wrong DNA site. That is, only when the protein sequence is compatible with the structure of the minor groove will the stabilizing interaction be strong enough to overcome the entropic loss associated with binding.
Studies on homeodomain–DNA binary complexes also suggest that the N-terminal arm has a tendency to be disordered, unless presented with a DNA structure that provides sufficient stabilizing interactions to compete with conformational entropy. For example, residues 1 to 4 are not observed in the Antp and Engrailed X-ray complexes (Fraenkel and Pabo, 1998; Fraenkel et al., 1998). In contrast, most of the N-terminal arm is structured in an Even-skipped–DNA complex where, notably, both Arg3 and Tyr4 insert into the minor groove (Hirsch and Aggarwal, 1995). In that complex the minor groove is quite narrow where Arg3 inserts, consistent with the idea that a narrow groove is required to structure a region of the protein which is intrinsically disordered. In the HoxA9–Pbx–DNA ternary complex, the N terminal arm is also ordered but in that case, a very short linker severely limits the conformational freedom of the N-terminal arm (LaRonde-LeBlanc and Wolberger, 2003).
The role of DNA structure in conferring specificity
As seen in the crystal structure, binding of Scr-Exd to fkh250con* involves residues that are present in all Hox proteins, thus providing an explanation for why this site is not specific for a particular paralog. As discussed above, the answer to the inverse question, of why fkh250 preferentially binds Scr-Exd, involves the insertion of His-12 and Arg3 into the minor groove, which is narrower than the equivalent region in fkh250con*. That a narrow groove is an inherent feature of the fkh250 site suggests the more general idea that Hox proteins recognize their specific binding sites by reading a sequence-dependent DNA structure which, in turn, enhances the negative electrostatic potential and attracts the positively charged Arg/His pair. Thus, local differences in electrostatic potential provide an explanation for why sequence-dependent DNA conformations can attract basic amino acids. This shape-dependent DNA recognition mechanism is distinct from “direct readout” mechanisms that involve specific hydrogen bond formation and hydrophobic contacts between amino acid side chains and bases. It is also distinct from “indirect readout” where protein binding is influenced by the global shape of a DNA molecule or by sequence-dependent DNA bending and deformability (Hizver et al., 2001; Lavery, 2005; Rohs et al., 2005b; Zhurkin et al., 2005).
Scr's ability to recognize the shape of the minor groove via basic residues may provide an example of a more general class of protein-DNA recognition mechanisms. For example, an Arg of phage 434 repressor inserts into the minor groove of its operator (Aggarwal et al., 1988) and a His in the DNA binding domains of interferon regulatory factors (IRFs) inserts into a compressed minor groove (Escalante et al., 1998; Fujii et al., 1999; Panne et al., 2004). Moreover, the sequence (either FGR, RGR or RGGR) in the minor groove binding region of monomeric human estrogen related receptors, hERR, is an important specificity determinant for that family of transcription factors (Gearhart et al., 2003; Meinke and Sigler, 1999). The analogy between Hox and hERR2, a nuclear receptor, is particularly striking as the Zn finger domain of nuclear receptors makes major groove contacts while a normally extended peptide expands the binding site by making minor groove contacts. It will be interesting to determine if, as suggested here for Hox proteins, other families of DNA binding proteins use a common set of major groove contacts to recognize large sets of degenerate binding sites with individual family members distinguishing among these sites via more specific minor groove contacts. For Hox proteins, we suggest that such a two-tiered recognition system gives them the flexibility to bind both shared and paralog-specific binding sites.
Experimental Procedures
Additional information is provided in the Supplementary material.
Structure determination
Both cocrystals were obtained from solutions containing 10-14% polyethylene glycol 4000, 20% MPD, 0.1 M Tris (pH 8.7-8.9), 0.2 M Sodium Acetate and 0.2 M KCl. The best cocrystals with fkh250 were obtained with the overhanging 20-mer (5′-TCAGCCGATTAATCTTAGAG-3′/5′-ACTCTAAGATTAATCGGCTG-3′) and Scr (residues 298-384) and Exd (238-300). These Scr-Exd-fkh250 cocrystals belonged to space group C2221 with unit cell dimensions of a=88.8 Å, b= 92.6 Å, c= 78.8 Å. The best cocrystals with fkh250con* were obtained with the overhanging 20-mer (5′-TCAGCCCATAAATCATAGAG-3′/5′-ACTCTATGATTTATGGGCTG-3′) with the same Scr and Exd proteins. These Scr-Exd-fkh250con* cocrystals belonged to space group P43212, with unit cell dimensions of a=b= 65.3 Å, c= 200.3 Å. The consensus Hox-Exd binding site in this sequence is identical to the site present in the Ubx-Exd structure (Passner et al., 1999). Data were measured at the Advanced Photon Source (Beamline 19-ID) and the Brookhaven National Laboratory (Beamline X25).
Computational
DNA geometry was analyzed with the Curves algorithm (Lavery and Sklenar, 1989). The structures of fkh250 and fkh250con* or variants were predicted using an all-atom, force-field based MC algorithm (Rohs et al., 2005a; Rohs et al., 2005b; Sklenar et al., 2006).
Electrostatic potentials were calculated using the DelPhi program (Rocchia et al., 2002). Figures 4E and 4F plot the potential in the minor groove at the midpoint of a line connecting the O4′ atoms of nucleotide i+1 on the 5′ strand and nucleotide i-1 on the 3′ strand (the reference is approximately located in the plane of base pair i).
Protein-DNA binding assays
The fkh250 (GATCTCAATGTCAAGATTAATCGCCAGCTGTGGGACGAGG) and fkh250con (GATCTCAATGTCAAGATTTATGGCCAGCTGTGGGACGAGG) probes and electrophoretic mobility shift assays (Ryoo and Mann, 1999) and Kd measurements (LaRonde-LeBlanc and Wolberger, 2003) were carried out as previously described. All DNA binding experiments were carried out with nearly full-length forms of Scr (residues 2 to 406) and full-length Exd, both 6XHis-tagged, expressed, and purified from E. coli. Because expression of fkh250-lacZ does not require the Hth homeodomain, but does require HthHM (Noro et al., 2006), we included HthHM, which binds to Exd, in the reactions. The HthHM-Exd dimer was co-purified from E. coli and added at 150 ng/reaction.
In vivo analyses
Drosophila embryos were stained using anti-CrebA, anti-β-gal, and anti-fkh antibodies as described (Andrew et al., 1994; Noro et al., 2006; Ryoo and Mann, 1999). Ectopic expression of ScrWT and Scr mutants in embryos was via the AG11 driver crossed to the expression-matched UAS-Scr line. Embryonic cuticles were analyzed by standard methods.
Supplementary Material
Acknowledgments
We thank D. Andrew and S. Beckendorf for reagents, D. Andrew for advice on the Creb and Fkh stains, and D. Andrew, L. Shapiro and T. Jessell for comments on the manuscript. We thank P. Wright for pointing out the importance of RGR and RGGR motifs in human orphan estrogen receptors. This work was supported by NIH grants AI41706 and GM62947 to A.K.A., GM074105 and GM54510 to R.S.M., and an NIH U54 CA121852 MAGNet grant.
Footnotes
Accession numbers: The coordinates have been deposited to the RCSB Protein Data Bank with accession codes 2R5Z (fkh250) and 2R5Y (fkh250con).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Horner MA, Petitt MG, Smolik SM, Scott MP. Setting limits on homeotic gene function: restraint of Sex combs reduced activity by teashirt and other homeotic genes. Embo J. 1994;13:1132–1144. doi: 10.1002/j.1460-2075.1994.tb06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M, Qian YQ, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- Burkhoff AM, Tullius TD. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988;331:455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Chauvet S, Merabet S, Bilder D, Scott MP, Pradel J, Graba Y. Distinct hox protein sequences determine specificity in different tissues. Proc Natl Acad Sci U S A. 2000;97:4064–4069. doi: 10.1073/pnas.070046997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- Crothers DM, Shakked Z. DNA bending by adenine-thymine tracts. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structures. London: Oxford University Press; 1999. pp. 455–470. [Google Scholar]
- Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- Faiger H, Ivanchenko M, Haran TE. Nearest-neighbor non-additivity versus long-range non-additivity in TATA-box structure and its implications for TBP-binding mechanism. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel E, Pabo CO. Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat Struct Biol. 1998;5:692–697. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- Fraenkel E, Rould MA, Chambers KA, Pabo CO. Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures. J Mol Biol. 1998;284:351–361. doi: 10.1006/jmbi.1998.2147. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. Embo J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129:3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. 2003;327:819–832. doi: 10.1016/s0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Gibson G, Schier A, LeMotte P, Gehring WJ. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- Harrison SC, Aggarwal AK. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hersh BM, Carroll SB. Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development. 2005;132:1567–1577. doi: 10.1242/dev.01737. [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Aggarwal AK. Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. Embo J. 1995;14:6280–6291. doi: 10.1002/j.1460-2075.1995.tb00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine--thymine tract and its role in gene regulation. Proc Natl Acad Sci U S A. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Hovde S, Abate-Shen C, Geiger JH. Crystal structure of the Msx-1 homeodomain/DNA complex. Biochemistry. 2001;40:12013–12021. doi: 10.1021/bi0108148. [DOI] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase: effects of ionic strength and amino-acid modification. Proteins. 1986;1:47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R. Recognizing DNA. Q Rev Biophys. 2005;38:339–344. doi: 10.1017/S0033583505004105. [DOI] [PubMed] [Google Scholar]
- Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- Lohmann I, McGinnis N, Bodmer M, McGinnis W. The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell. 2002;110:457–466. doi: 10.1016/s0092-8674(02)00871-1. [DOI] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat Struct Biol. 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. Embo J. 2004;23:4384–4393. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S, Weigel D, Beckendorf SK. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development. 1992;114:49–57. doi: 10.1242/dev.114.1.49. [DOI] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- Rohs R, Bloch I, Sklenar H, Shakked Z. Molecular flexibility in ab initio drug docking to DNA: binding-site and binding-mode transitions in all-atom Monte Carlo simulations. Nucleic Acids Res. 2005a;33:7048–7057. doi: 10.1093/nar/gki1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005b;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 1999;13:1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M, Graba Y, Scott MP. Hox genes in evolution: protein surfaces and paralog groups. Trends Genet. 1997;13:145–151. doi: 10.1016/s0168-9525(97)01096-2. [DOI] [PubMed] [Google Scholar]
- Sklenar H, Wustner D, Rohs R. Using internal and collective variables in Monte Carlo simulations of nucleic acid structures: chain breakage/closure algorithm and associated Jacobians. J Comput Chem. 2006;27:309–315. doi: 10.1002/jcc.20345. [DOI] [PubMed] [Google Scholar]
- Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci U S A. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Kellogg L, Rould MA, Chambers KA, Ades SE, Sauer RT, Pabo CO. Engrailed (Gln50-->Lys) homeodomain-DNA complex at 1.9 A resolution: structural basis for enhanced affinity and altered specificity. Structure. 1997;5:1047–1054. doi: 10.1016/s0969-2126(97)00256-6. [DOI] [PubMed] [Google Scholar]
- Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Yao LC, Liaw GJ, Pai CY, Sun YH. A common mechanism for antenna-to-Leg transformation in Drosophila: suppression of homothorax transcription by four HOM-C genes. Dev Biol. 1999;211:268–276. doi: 10.1006/dbio.1999.9309. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Potter SS. Functional comparison of the Hoxa 4, Hoxa 10, and Hoxa 11 homeoboxes. Dev Biol. 2002;244:21–36. doi: 10.1006/dbio.2002.0595. [DOI] [PubMed] [Google Scholar]
- Zhurkin VB, Tolstorukov MY, Xu F, Colasanti AV, Olson WK. Sequence-Dependent Variability of B-DNA: An Update on Bending and Curvature. In: Ohyama T, editor. DNA Conformation and Transcription. Springer; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


