Summary
The development of the Drosophila leg requires both Decapentaplegic (Dpp) and Wingless (Wg), two signals that establish the proximo-distal (PD) axis by activating target genes such as Distalless (Dll). Dll expression in the leg depends on a Dpp- and Wg-dependent phase and a maintenance phase that is independent of these signals. Here, we show that accurate Dll expression in the leg results from the synergistic interaction between two cis-regulatory elements. The Leg Trigger (LT) element directly integrates Wg and Dpp inputs, and is only active in cells receiving high levels of both signals. The Maintenance (M) element is able to maintain Wg- and Dpp-independent expression, but only when in cis to LT. M, which includes the native Dll promoter, functions as an autoregulatory element by directly binding Dll. The ‘trigger-maintenance’ model describes a novel mechanism by which secreted morphogens act combinatorially to induce the stable expression of target genes.
Introduction
Drosophila leg development requires the elaboration and coordination of three body plan axes, antero-posterior (AP), dorso-ventral (DV), and proximo-distal (PD). The process of leg development begins during embryogenesis, when a small number of cells in each thoracic hemisegment are specified to become the leg imaginal disc. Once formed, the leg disc is comprised mainly of a single sheet of epithelial cells, which continue to proliferate during larval development (reviewed by (Cohen, 1993)). Both DV and PD information in the leg disc is derived from two secreted morphogens, Wg and Dpp. Wg, expressed ventrally, and Dpp, expressed dorsally, function combinatorially to create the leg’s PD axis (Campbell et al., 1993; Diaz-Benjumea et al., 1994). Genetic experiments suggest that these signals are not only required to initiate PD axis formation, but that different levels of Wg and Dpp are responsible for creating different fates along the PD axis (Lecuit and Cohen, 1997). Moreover, for both the initiation and specification of PD fates, both signals are required; neither the Wg nor Dpp pathways are sufficient, even when maximally activated (Abu-Shaar and Mann, 1998; Diaz-Benjumea et al., 1994; Lecuit and Cohen, 1997). Genetic experiments also demonstrate that the requirement for Wg and Dpp activities is transient; by ~72h of development Wg and Dpp are no longer required to generate a complete PD axis (Diaz-Benjumea et al., 1994; Galindo et al., 2002). Although these results are well supported by in vivo genetic experiments, we currently have very little understanding of the underlying molecular mechanisms by which the leg’s PD axis is established by Wg and Dpp.
Two targets of Wg and Dpp in the leg, Distalless (Dll) and dachshund (dac), serve as markers for different PD fates (Diaz-Benjumea et al., 1994; Mardon et al., 1994). Dll is activated by high levels of Wg plus Dpp signaling and, consequently, is expressed in distal regions of the leg. In contrast, dac is activated by lower levels of these two signals and is expressed in medial positions along the PD axis (Lecuit and Cohen, 1997). As transcriptional regulatory elements controlling Dll or dac in the leg disc have not been described, it is not known if Wg and Dpp directly regulate these genes during leg development. In fact, somewhat paradoxically, Dll expression in the leg disc responds to Wg and Dpp differently than it does in the embryonic leg primordia, where Dll is activated by Wg but repressed by Dpp (Cohen et al., 1993; Cohen, 1990; Goto and Hayashi, 1997). One scenario that would account for this difference, and is supported by our results, is that Dll expression is governed by a different set of cis-regulatory elements in the leg disc and embryo. Consistent with this idea, the best characterized Dll regulatory element, Dll304, is active only early in embryogenesis, when Dll is first expressed in the leg primordia (Vachon et al., 1992), but is not active in the leg disc (our unpublished observations). Alternatively, it is plausible that Wg and Dpp indirectly control Dll expression in the imaginal disc. Further, once activated by these signals, Dll expression is maintained by an unknown mechanism.
To gain further insights into the control of PD target gene expression by Wg and Dpp, we have characterized Dll cis-regulatory elements that are active in the leg disc. One element, which we call the Leg Trigger (LT), is active only in response to high levels of Wg plus Dpp. Consequently, an LT-lacZ reporter gene is expressed in a small subset of Dll-expressing cells in the center of the leg disc, where the Wg and Dpp expression domains abut. We also describe a second element, called Maintenance (M), which includes the Dll promoter. Although M, on its own, is only weakly active in leg discs, it is capable of synergizing with LT to produce accurate and robust Dll-like expression. Consistent with genetic analyses, LT directly integrates positive inputs from Wg and Dpp by binding the signal-activated transcription factors Tcf and Mothers against Dpp (Mad), respectively. LT also directly integrates negative input from the Dpp pathway by binding Brinker (Brk), a transcription factor known to repress Dpp target genes in other contexts (Campbell and Tomlinson, 1999; Jazwinska et al., 1999; Minami et al., 1999). Further, we show that M requires direct binding by Dll for full activity, suggesting that maintenance depends in part on an autoregulatory mechanism. Thus, Dll expression in the leg disc is controlled in a two-step manner by separable ‘trigger’ and ‘maintenance’ cis-regulatory elements that cooperate with each other to integrate Wg and Dpp inputs during an early phase and Dll input during a maintenance phase.
Results
Identification of a Dll element that integrates Wg and Dpp signaling
We used a transgenic reporter gene assay to search for Dll cis-regulatory elements that were active in the leg disc. Altogether, we scanned ~14 kb 5′ to the Dll transcription initiation site (Fig. 1A). From these experiments, we identified a ~1 kb fragment located ~12 kb 5′ of the Dll transcription initiation site, which we named the “Leg Trigger” (LT) element (Fig. 1A). The LT element drove high levels of reporter gene (LT-lacZ) expression in a subset of the Dll domain in third instar ventral (leg, antennal, and genital) discs, but was not active in dorsal (wing and haltere) imaginal discs (Fig. 1 and Supp. Fig. 1). LT was the only element within this 14 kb that, when cloned into a standard reporter gene (with a heterologous, minimal promoter; see Experimental Procedures), drove strong expression in leg or antennal discs (Figure 1B and data not shown).
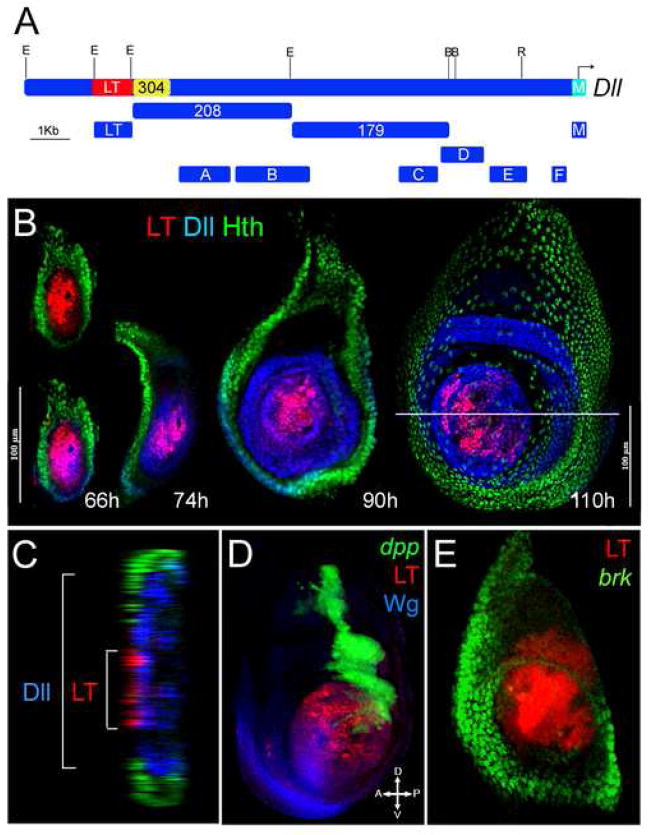
Figure 1. The LT enhancer.
(A) The Dll 5′ cis-regulatory region. DNA fragments were cloned based on sequence conservation to other Drosophilids and assayed in transgenic reporter genes for expression in imaginal discs. E, Eco-R1, B, Bam-H1 and R, RSR II. LT is in red, 304 in yellow and M in light blue. Of the fragments tested in a standard reporter gene (using the minimal promoter from the hsp43 gene), only LT drove expression in discs. Although fragments LT (previously 215), 304, 208, and 179 were originally cloned by Vachon et al., (1992), no imaginal disc expression was reported.
(B) Wild type leg discs at various stages of development stained for LT-lacZ (red), Dll (blue) and Homothorax (Hth) (green). The age of the larvae (+/− 6h) is indicated below each disc. Early in development (66h +/− 6h or before) LT was active in all the cells that express Dll. Later in development LT was active in a subset of Dll expressing cells.
(C) Cross-section image of the 110h leg disc from (B). LT was only active in a subset of Dll expressing cells.
(D) Wild type third instar leg disc stained for LT-lacZ (red), dpp-Gal4; UAS-GFP (green), and Wg (blue). LT was active in the center of the disc where the Wg and Dpp expression domains meet.
(E) Wild type early third instar leg disc (~96h AEL) stained for LT-Gal4; UAS-GFP (red) and brk-lacZ (green). LT was active in cells that have no or very low Brk levels.
Early in larval development (prior to ~ 72h after egg laying (AEL)), LT drove expression in all Dll-expressing cells of the leg disc (Fig. 1B). This time approximately coincides with the time when Dll is dependent on Wg and Dpp. As the leg disc continues to grow, Dll becomes independent of Wg and Dpp, and its expression expands beyond the cells in which LT was active (Fig. 1B and C). In a mature third instar leg disc (~110h AEL), Dll expression covered the future distal leg (tibia and tarsi), whereas LT was active only at the distal tip, close to where the Wg and Dpp expression domains meet (Fig. 1B, C, D and Supp. Fig. 2). Notably, these cells also had little or no expression of the Dpp pathway repressor, Brk, which is expressed in lateral and ventral regions of the leg disc (Fig. 1E). Taken together, these results suggest that LT is only active in cells that receive both Wg and Dpp inputs.
LT responds continuously to Wg and Dpp inputs
To test the idea that LT integrates Wg and Dpp inputs, we generated clones of cells expressing either an activated form of the β-catenin homolog Armadillo (Arm*) or an activated form of the Dpp receptor Thickveins (TkvQD), respectively. Activation of the Wg pathway using Arm* resulted in the cell-autonomous expression of LT-lacZ, but only in dorsal regions of the leg disc, where high levels of endogenous Dpp are present (Fig. 2A). Likewise, activation of the Dpp pathway by TkvQD resulted in the cell-autonomous expression of LT-lacZ, but only in ventral regions of the leg disc, where high levels of endogenous Wg are present (Fig. 2C). Thus, as for Dll, LT is activated only when both signaling pathways converge. Consistently, co-expression of Arm* and TkvQD resulted in LT-lacZ activation in both ventral and dorsal clones (Supp. Fig. 3).
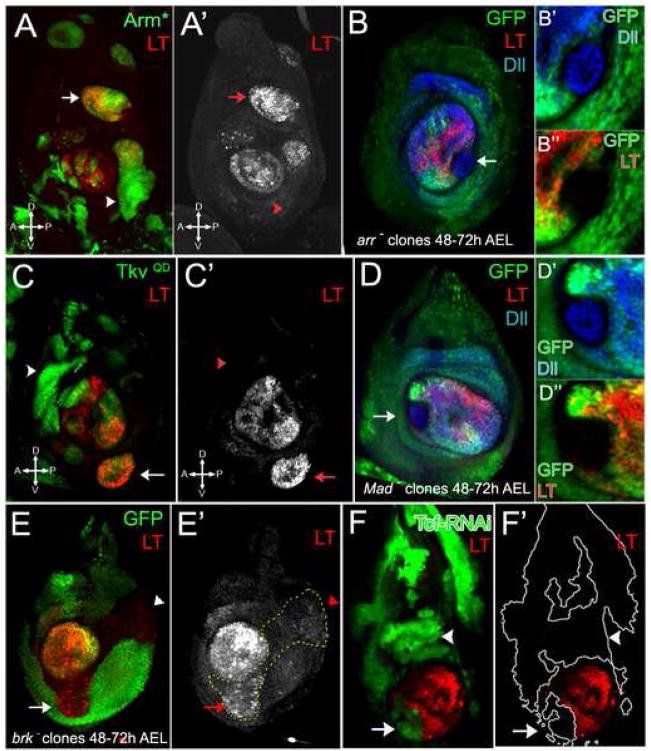
Figure 2. LT continuously requires the Wg and Dpp pathways.
(A – A′) Clones expressing Arm* marked by GFP (green) activated LT-lacZ (red) in dorsal (arrow), but not ventral (arrowhead) regions of the disc.
(B – B″) An arr− clone induced between 48–72 hours after egg laying (h AEL), marked by absence of GFP (green), had no LT-lacZ expression (red), but maintained Dll expression (blue).
(C – C′) Clones expressing TkvQD marked by GFP (green) activated LT-lacZ (red) in ventral (arrow), but not dorsal (arrowhead) regions of disc.
(D – D″) A Mad− clone induced between 48–72 hours AEL, marked by absence of GFP (green), had no LT-lacZ expression (red), but maintained Dll expression (blue).
(E – E′) A brk− clone induced 48–72h AEL, marked by absence of GFP (green), de-repressed LT-lacZ (red) in the ventral disc, close to the source of Wg (arrow), but not in dorsal regions (arrowhead).
(F – F′) Clones expressing Tcf-RNAi, marked by GFP (green), did not de-repress LT-lacZ expression (red) in dorsal (arrowhead) or lateral regions of the leg disc. LT-lacZ was not expressed in Tcf-RNAi expressing cells (arrow).
To test for the necessity of Wg and Dpp inputs for LT-lacZ expression, we generated by mitotic recombination clones of cells that were unable to transduce the Wg or Dpp signals. LT-lacZ expression was lost in clones mutant for the Wg co-receptor arrow (arr) or mutant for the Dpp pathway transcriptional effector Mad (Fig. 2B and D). No effect on Dll expression was observed, because Dll was independent of these signals at the time these clones were generated (48 to 72h AEL or later) (Fig. 2B and D). These results confirm that LT continuously requires the combined inputs of Wg and Dpp to be active, while Dll becomes independent of these signals by the third instar.
Due to its role in repressing Dpp target genes in Drosophila wing development, we next examined the role of brk in the control of LT-lacZ expression. brk− null clones located close to the source of Wg in the ventral region of the leg disc were able to derepress LT-lacZ (Fig. 2E), suggesting that Brk is normally a repressor of LT activity. However, the level of LT-lacZ derepression in brk− clones was significantly weaker than the amount of expression seen in TkvQD-expressing clones (compare Fig. 2C and E). As brk is repressed by Dpp signaling in the leg as in the wing (Supp. Fig. 4), these observations suggest that to activate LT-lacZ, Dpp signaling does more than repress brk. One possibility is that, in addition to repressing brk, Dpp signaling is working through Mad to activate LT-lacZ. Consistently, LT-lacZ is not expressed in Mad−; brk− double mutant clones (Supp. Fig. 5A and B). These results suggest that Dpp signaling is activating LT both via repressing brk and activating Mad.
Analogous to the role that Brk plays in Dpp signaling, Wg pathway components, in particular the effector transcription factor Tcf, have the potential to repress Wg target genes in the absence of pathway activation (Cavallo et al., 1998). Accordingly, Tcf could potentially be a Dll repressor in the dorsal leg disc, away from the source of Wg. To test this idea we generated clones of cells expressing a Tcf hairpin construct to induce RNAi and knock-down Tcf levels. In the center of the leg disc, Tcf RNAi clones eliminated LT-lacZ expression, demonstrating the efficacy of the Tcf RNAi and confirming the requirement for Wg input for LT activity (Fig. 2F). However, we failed to observe any LT-lacZ derepression in Tcf RNAi clones in the dorsal or lateral regions of the leg disc (Fig. 2F). This experiment suggests that Tcf is not a repressor of LT and, therefore, that Wg input into LT is not mediated by derepression. Instead, these results suggest that Wg may activate LT directly, a conclusion that is supported below.
The Dpp and Wg pathways directly regulate LT
To understand how Dpp and Wg control LT expression at the molecular level, we generated a series of ~100 bp deletions of LT and searched for putative binding sites for the transcription factors Mad, Brk, and Tcf (Fig. 3A). Candidate binding sites were tested for their ability to bind recombinant proteins in electrophoretic mobility shift assays (EMSAs), and sites that bound were mutated to destroy binding (Fig. 3B and D). To assess the contribution of identified binding sites to LT’s activity, each mutant or deleted LT element was tested for its ability to drive lacZ expression in vivo using a standard reporter gene assay. Most of the deletions that resulted in a loss or reduction of LT activity removed either a Mad or Tcf binding site (Fig. 3A). In all, we discovered four Tcf binding sites (Fig. 3A and B). Mutation of each site in isolation had weak or no impact on LT activity; however, simultaneous disruption of all four Tcf sites (LTTcf−-lacZ) resulted in the near elimination of LT activity (Fig. 3E). In addition, consistent with the results obtained by inducing Tcf RNAi, none of the reporter genes with mutant Tcf binding sites showed any derepression, confirming that Tcf is not repressing LT activity in the absence of Wg signaling. We also used chromatin immunoprecipitation (ChIP) to test if Tcf was bound to LT in vivo. Compared to control immunoprecipitations (IPs), an anti-Tcf antibody specifically IPd LT DNA from Drosophila leg and wing imaginal discs (Fig. 3C). Moreover, anti-Tcf IPd LT from leg discs, where LT is active, significantly better than it did from wing plus haltere discs, where LT is inactive (Fig. 3C). The enrichment of IPd DNA from leg compared to wing discs was not observed for two ubiquitously expressed genes (act5C and pyruvate dehydrogenase; data not shown), suggesting that the tissue-specificity of Tcf binding to LT is significant. Thus, consistent with our genetic experiments, these data indicate that the Wg pathway directly activates LT in leg discs by binding Tcf.
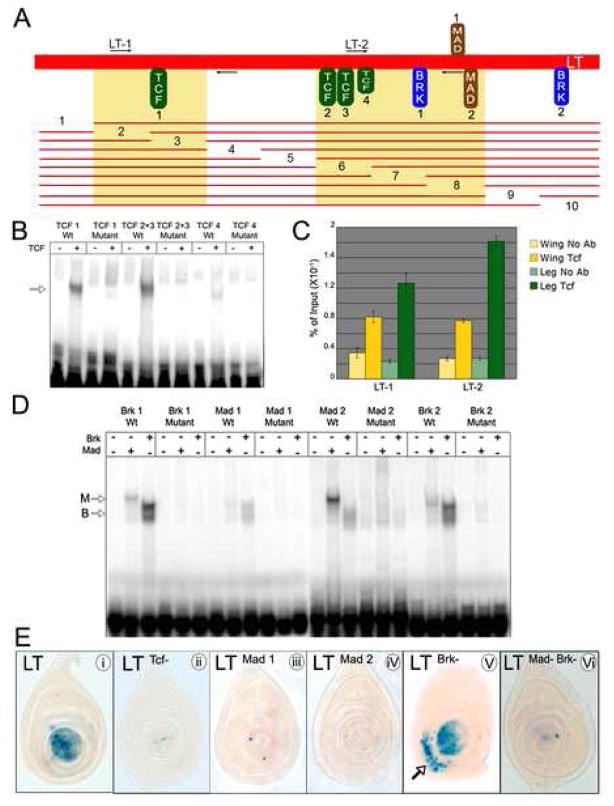
Figure 3. The regulation of LT by the Dpp and Wg pathways is direct.
(A) Diagram of LT with transcription factor binding sites. Oval size indicates the relative affinities of these binding sites in EMSAs; ovals above and below the line indicate different binding site orientations. All of these binding sites, except for Brk2, are well conserved through D. virilis. The thin red lines summarize the results of a set of ~100 bp deletions tested in reporter genes. Regions shaded in yellow indicate deletions that had reduced or no reporter activity. Except for #2 and #7, all deletions that had an effect removed a Tcf or Mad binding site. Deletions #2 and #7 indicate that other inputs besides the mapped Tcf and Mad binding sites are required for LT activity.
(B) EMSAs showing binding of Tcf to probes containing wild type or mutant binding sites (see Experimental Procedures for sequences). Arrows indicate protein-DNA complexes.
(C) ChIP experiments demonstrating specific binding of Tcf to LT in imaginal discs. Anti-Tcf antibodies pulled down LT ~twice as efficiently from leg discs as from wing plus haltere discs. Similar results were seen for two independent PCR fragments, LT-1 and LT-2 (whose positions in LT are indicated in panel A). Each column shows the averages and standard error of the mean for four independent IPs and real time PCRs.
(D) EMSAs showing binding of Mad and Brk to probes containing wild type or mutant binding sites (see Experimental Procedures for sequences). Arrows indicate protein-DNA complexes; M, Mad; B, Brk. Although Mad1 has a lower affinity for Mad than Mad2, its binding is sequence-specific.
(E) X-Gal stains of leg discs from flies containing wild type LT-lacZ (i), LTTcf−-lacZ (with all Tcf sites mutated; ii), LTMad1−-lacZ (with the Mad1 site mutated; iii), LTMad2−-lacZ (with the Mad2 site mutated; iv), LTBrk−-lacZ (with both Brk sites mutated; v), and LTMad−Brk−-lacZ (with both Mad and Brk sites mutated; vi). Mutation of the Tcf sites or either Mad site resulted in loss of activity. Mutation of both Brk sites resulted in the ventral expansion of expression (arrow). The LTBrk−-lacZ disc shown here has an intermediate amount of derepression; other transformant lines show stronger and more uniform ventral expression of lacZ. Mutation of both Mad and Brk sites resulted in no expression
We discovered four candidate binding sites for the transcriptional effectors of the Dpp pathway, Mad and Brk (Fig. 3A). Previous studies demonstrated that Mad and Brk bind to similar DNA sequences (Kirkpatrick et al., 2001). Consistently, all four of the sites we identified in LT bound to both Mad and Brk, although the relative affinities of these two factors differed from site to site (Fig. 3A and D). As with the Tcf sites, the contribution of these sites to LT activity was assessed using a lacZ reporter gene assay in transgenic flies. Two of the sites, which we named Mad1 and Mad2, were essential for LT activity (Fig. 3A and E). Mutation of either of these sites in isolation or in combination (LTMad−-lacZ) resulted in the loss of LT activity (Fig. 3E and data not shown). In contrast, mutation of the other two sites, which we named Brk1 and Brk2, resulted in the ventral expansion of LT activity (LTBrk−-lacZ) (Fig. 3E). These results are consistent with the Brk expression pattern and the derepression of LT-lacZ seen in brk− clones described above (Fig. 1E and 2E). Mutation of either Brk1 or Brk2 on their own had no effect (data not shown). We also found that LT reporter genes with both Brk and both Mad sites mutated (LTMad−,Brk−-lacZ) were not expressed or, in some cases, had very weak expression (Fig. 3E). These data suggest that LT directly integrates input from the Dpp pathway in two ways. First, Dpp directly activates LT, by binding Mad at two ‘activator’ sites, Mad1 and Mad2. Second, in ventral and lateral cells, LT activity is directly repressed due to Brk binding at two ‘repressor’ sites, Brk1 and Brk2. The requirement for Mad input was further supported by our finding that LT reporter genes with mutant Mad binding sites were not expressed in brk− clones (Supp. Fig. 5C and D). We note that the ventral expression of LTBrk−-lacZ argues that there is activated Mad in the ventral leg disc. Consistently, although Dpp signal transduction was strongest in the dorsal disc, weaker pathway activation, visualized by anti-PMad immunostaining, was observed in the ventral region of third instar leg discs (data not shown). Moreover, ventral LTBrk−-lacZ expression required Dpp signaling as it was lost in tkv− clones (Supp. Fig. 5E).
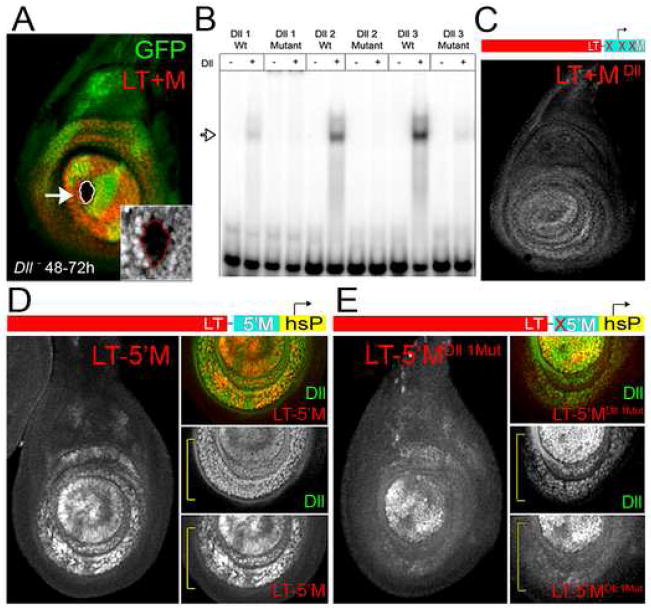
The Dll promoter region maintains LT-initiated expression
We have described a Dll regulatory element, LT, that accurately recapitulates Dll expression, and its dependency on Wg and Dpp, early in leg disc development. Unlike Dll, LT continuously requires input from Wg and Dpp and, by the end of larval development, LT is only active in a small subset of Dll-expressing cells (Fig. 1B). These data suggest that LT contains the information required to respond to Wg and Dpp, but is lacking the information required to maintain Dll expression. Because promoter regions can play important roles in enhancer activities (e.g. (Calhoun et al., 2002)), we tested a 300 bp fragment that encompasses the transcription start site of the Dll gene for maintenance activity (Fig. 1A). When this element, M, was used instead of the minimal promoter from the hsp43 gene that is in our standard reporter genes, the resulting LT+M-lacZ reporter gene accurately reproduced the normal expression pattern of Dll at all stages of leg disc development. While LT was active only in the center of the mature leg disc (Fig. 4A), the LT+M composite element was active in all cells that express Dll (Fig. 4C). On its own, the M-lacZ reporter gene was expressed very weakly throughout the leg disc, with slightly higher activity in Dll-expressing cells (Fig. 4B). M contains a functional promoter because, when used with another enhancer (the dppdiscs enhancer (Masucci et al., 1990), a dppdiscs-M-lacZ reporter gene drove dpp-like expression in both wing and leg imaginal discs (data not shown).
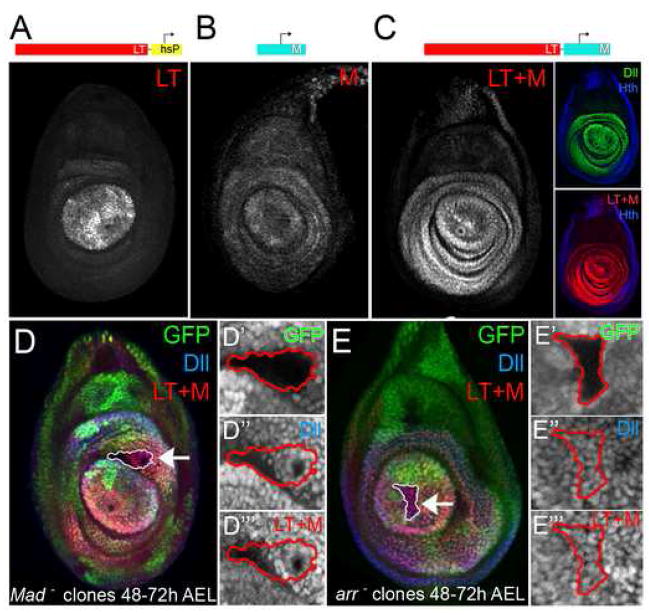
Figure 4. The Dll promoter region has maintenance activity.
(A) LT-lacZ was active in subset of Dll-expressing cells (with the hsp43 promoter, hsP).
(B) M-lacZ is expressed nearly ubiquitously in leg discs at very low levels, with slightly higher levels in the Dll domain. M-lacZ is not expressed in wing discs (not shown).
(C) LT+M-lacZ was expressed in all cells that express Dll in the leg imaginal disc, including low-level expression in the Dll-expressing trochanter ring. LT+M-lacZ was not expressed in wing discs (not shown). The inset shows the expression of Dll (green) and LT+M-lacZ (red) in combination with Hth (blue).
(D and E) Genetic tests of LT+M-lacZ maintenance.
(D-D‴) A Mad− clone induced between 48–72h AEL, marked by absence of GFP, continues to express LT+M-lacZ (red) and Dll (blue).
(E) An arr− clone induced between 48–72h AEL, marked by absence of GFP, continues to express LT+M-lacZ (red) and Dll (blue). The insets (D′,D″,D‴,E,E″,E‴) show blow-ups of the clones, outlined in red.
To test if LT+M-lacZ was, like Dll, able to maintain its expression in the absence of continuous Wg and Dpp inputs, we analyzed its expression in clones that cannot transduce these signals. As with Dll, and in contrast to LT-lacZ, the expression of LT+M-lacZ was unaffected in arr− clones generated between 48 to 72h or later (Fig. 4E). Likewise, inactivation of the Dpp pathway in Mad− clones generated at this time also had no effect on LT+M-lacZ expression (Fig. 4D). Thus, the M element provides the information to maintain LT-initiated expression, even in the absence of continuous inputs from Wg and Dpp.
The M element directly requires Dll input
One plausible mechanism for Dll maintenance is through a positive autoregulatory feedback loop (Castelli-Gair and Akam, 1995). According to this idea, Dll itself may be required for maintenance. Alternatively, Dll expression could be maintained via the Trithorax (Trx) and/or Polycomb (Pc) groups of epigenetic regulators (reviewed by (Ringrose and Paro, 2004)). We found that, when generated during the maintenance phase (i.e. after 72h), trx clones had no effect on Dll expression (data not shown), demonstrating that Dll maintenance does not require this function. In contrast, Pc− or Sex combs on midlegs (Scm−) clones resulted in a loss of Dll expression in some regions of the Dll domain (data not shown). These data suggest that PcG functions might be playing a role in Dll maintenance. However, because many genes are likely to be derepressed in the absence of these PcG functions, the loss of Dll expression observed in these clones may be indirect (see Discussion).
To test if autoregulation contributes to Dll maintenance, we generated Dll loss of function clones during the maintenance phase and examined the effect on LT+M-lacZ expression. LT+M activity was eliminated in Dll mutant clones (Fig. 5A), indicating that Dll is essential for it’s activity. To determine if the requirement for Dll is direct, we searched for candidate Dll binding sites within the M element, and tested the ability of wild type and mutant sequences to bind Dll protein in vitro. Three Dll binding sites were found in the M element (Fig. 5B). Mutating all three of these binding sites together (but not individually) in the context of the LT+M-lacZ reporter gene strongly reduced, but did not eliminate, expression (Fig. 5C). These data demonstrate that Dll is directly contributing to M’s activity, but suggest that there are additional inputs, and perhaps additional Dll binding sites, that contribute to maintenance activity.
Figure 5. Dll is required for Dll maintenance.
(A) A Dll− clone induced between 48–72h AEL, marked by absence of GFP, resulted in loss of LT+M-lacZ expression (red). The inset shows the clone outlined in red and the cell autonomous loss of LT+M-lacZ expression.
(B) EMSAs showing Dll binding to each of the three Dll sites in the M element (WT and mutant; see Experimental Procedures for sequences). The arrow indicates the Dll-induced complexes.
(C) The expression of a LT+M reporter gene with all three Dll binding sites mutated (LT+MDll−-lacZ) was only weakly expressed in Dll-expressing cells of the leg disc.
(D) Expression driven by the LT-5′M-hsP-lacZ reporter gene. The level and pattern of expression indicates that the 5′M fragment confers partial maintenance activity. The insets and brackets compare β-gal and Dll expression.
(E) When the Dll1 binding site is mutated in this reporter (LT-5′MDll1−-hsp-lacZ), expression resembles that driven by LT-lacZ (compare with Fig. 4A).
The 300 bp M element, as defined above, includes the transcription initiation site for Dll as well as 3′ and 5′ flanking sequences. To determine where within this element maintenance activity resides, we characterized additional reporter genes containing M variants. Combining LT with the 3′ half of the M element (including the Dll transcription start site; LT-3′M-lacZ) resulted in no reporter expression (data not shown). The 3′ fragment of M drove weak expression with the dppdiscs enhancer demonstrating that it contains a functional promoter (data not shown). These data suggest that the 5′ fragment of M is essential for LT-stimulated maintenance of expression. To test for the sufficiency of 5′M, we fused it to the minimal promoter from the hsp43 gene which, on its own, does not support maintenance (see above). Combining this chimeric fragment with LT (LT-5′M-hsp-lacZ) resulted in reporter gene expression that was similar to, though less uniform than, that driven by LT+M-lacZ (Fig. 5D), suggesting that 5′M provides partial maintenance activity. Mutation of the sole Dll binding site in 5′M reverted the expression pattern to one that is very similar to that driven by LT-lacZ (Fig 5E, compare it to Fig. 5D and 4A), suggesting that this Dll binding site is important for maintenance. Taken together, these data suggest that Dll directly regulates its own expression through binding sites located close to its own promoter, and this binding contributes to the maintenance activity displayed by the M element.
LT is also required for maintenance
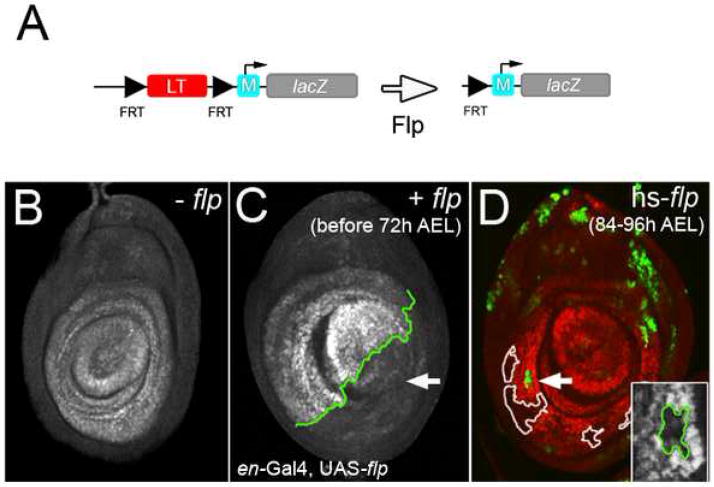
We have identified two cis-regulatory elements that together recapitulate the Wg- and Dpp-dependent and maintenance phases of Dll expression during development of the Drosophila leg. Significantly, the LT and M elements synergize with each other to produce accurate and robust expression; neither element, on their own, is capable of generating a strong Dll-like expression pattern. One question that emerges from these experiments is how LT synergizes with M to elicit maintenance. One possibility is that LT transiently interacts with M and changes its properties so that it can function as a robust autoregulatory element. Alternatively, LT may have to continuously work with M to confer maintenance activity. To distinguish between these scenarios, we created a LT+M reporter gene in which LT was flanked by FRT sequences (>LT>M-lacZ), allowing us to delete LT at various times during development using Flp-mediated recombination (Fig. 6A) (Struhl and Basler, 1993). As expected, deletion of LT during the Wg- and Dpp-dependent stage (prior to 48h AEL) resulted in no reporter expression (Fig. 6C). Interestingly, deletion of LT during the maintenance stage (after 72h AEL) also caused loss of reporter gene expression (Fig. 6D). These data argue suggest that LT is continuously required for maintenance and, therefore, LT+M-lacZ expression.
Figure 6. LT is required for maintenance.
(A) Diagram of a LT+M reporter gene in which LT is flanked by FRT sites (black triangles). After expression of Flp, LT is deleted, leaving a single FRT site and the M element.
(B) In the absence of Flp, >LT>M-lacZ generated a Dll-like expression pattern. This disc came from a larva of the same genotype as the one shown in (D) (hs-flp122; >LT>M-lacZ), but was not given a heat shock.
(C) Deletion of the LT enhancer in the posterior compartment (green) early in development (prior to maintenance) using en-Gal4, UAS-flp resulted in the loss of lacZ expression in that compartment, while leaving expression in the anterior compartment intact.
(D) Heat shock-induced expression of Flp during the maintenance stage of Dll expression resulted in the loss of reporter expression within the Dll domain. Due to the design of this experiment (see Experimental Procedures) only a subset of these heat-shock induced events were marked by GFP+; other, unmarked events are outlined. The inset shows a blow-up of the GFP-marked clone. In this experiment, Flp was provided 90 +/−6 h AEL via a 8 minute heat shock, significantly after maintenance begins.
Discussion
Molecular logic of Dll expression during leg development
We have provided evidence that Dll expression during Drosophila leg development is controlled by separate, synergistically-interacting cis-regulatory elements. The first element, LT, activates transcription only in response to high levels of Wg and Dpp signaling. The second element, M, includes the Dll promoter and has the ability to activate transcription in a Wg and Dpp-independent manner, but only when in cis to LT. Together, these results fit well with previous genetic experiments showing that the Wg and Dpp inputs into Dll are only required transiently, prior to ~60h AEL (Galindo et al., 2002; Lecuit and Cohen, 1997). Based on our data, we hypothesize that LT, and perhaps other elements with similar properties, is responsible for activating the Wg- and Dpp-dependent phase of Dll expression. Further, our data suggest that the combination of LT+M executes the Wg- and Dpp-independent phase of Dll expression. The existence of a two-component cis-regulatory system for Dll expression has several interesting implications, and provides a mechanistic understanding of how Wg, Dpp, and Dll inputs are integrated into Dll expression.
Signal integration into Dll
The requirement for multiple inputs for gene activation is a common theme in transcriptional regulation (reviewed by (Arnosti, 2003; Barolo and Posakony, 2002; Mann and Carroll, 2002)). Enhancer elements can be thought of as “logic integrators” that are only active in the presence of the correct activators and in the absence of repressors (Istrail and Davidson, 2005). The LT element defined here behaves as such a logic integrator. To be active, at least three conditions must be met. First, LT must be bound to a transcriptionally active form of Tcf, a condition which indicates high levels of Wg signaling. Second, LT must be bound to a transcriptionally active form of Mad, and, third, LT must not be bound to Brk. The second and third of these three conditions both indicate high levels of Dpp signaling. This combination of inputs ensures that LT is only triggered where Wg and Dpp signaling are both active. In addition, we hypothesize that there must be another input that restricts LT’s activity to the ventral discs (e.g., it is not active in other tissues where Wg and Dpp signaling intersect such as the wing disc). Such a ventral-specific input could be Dll, itself, which is expressed before LT is active via the Dll304 enhancer (Castelli-Gair and Akam, 1995), and/or another ventral-specific factor such as buttonhead (btd), which is also required for Dll expression (Estella et al., 2003). Consistent with this idea, LT-lacZ is lost in Dll− clones and in Dll hypomorphic discs, suggesting that Dll input, in addition to Wg and Dpp, is required for its activity (data not shown).
As noted above, Dpp signaling uses two mechanisms (Mad binding and absence of Brk) to control LT’s activity. Because Brk, a transcriptional repressor, binds directly to LT, it restricts the domain in which Wg signaling can activate this element. This conclusion is best supported by the expression pattern of the LT reporter gene in which the Brk binding sites were mutated. Specifically, the expression of this reporter (LTBrk−-lacZ) was expanded ventrally, indicating its potential to be activated more broadly by Wg signaling in the absence of this repressor. Thus, we suggest that the primary role of Brk is to provide spatial information to LT activation. The absence of Brk, however, is apparently not sufficient for LT activation; Mad input into LT appears also to be essential. Several experiments support this conclusion. Most informatively, LT-lacZ was not expressed in Mad−; brk− clones and LT-lacZ reporter genes with either Mad site mutated were not expressed in brk− clones. Thus, even in the absence of Brk, LT requires Mad input. In contrast to providing spatial information, we suggest that the Mad input into LT is important for boosting the level of its activation, together with Tcf, by providing an additional potent transcriptional activator. Further, LT is unlikely to be the only Dll cis regulatory element that integrates Wg plus Dpp signaling during leg development. Although LT was the only fragment within the 14 kb of 5′ DNA that drove strong expression in the leg disc in a standard reporter gene assay, thus allowing the dissection of Wg and Dpp signal integration, we identified a second fragment that was able to synergize with M to produce a Dll-like expression pattern (Supp. Fig. 6). In summary, these data suggest that during the Wg- and Dpp-dependent stage, Dll expression is regulated by the direct binding of Tcf, Mad, and Brk to LT and, perhaps, additional regulatory elements (Fig. 7).
Figure 7. The trigger-maintenance model.
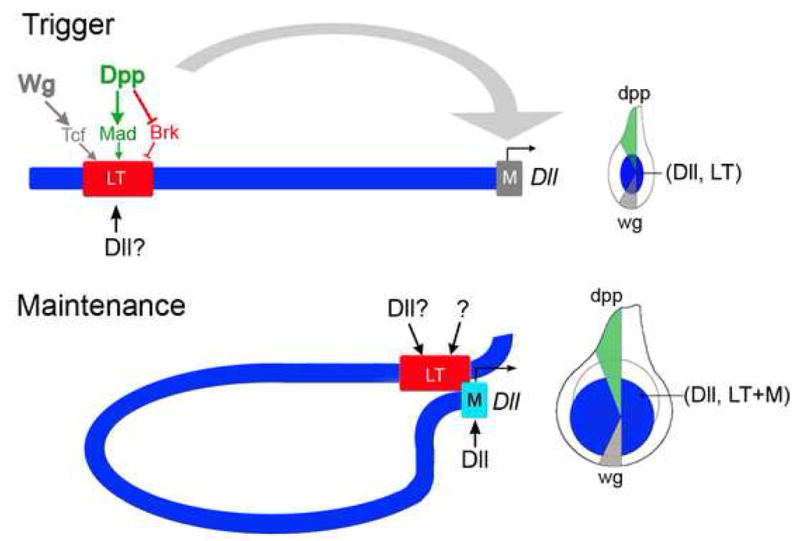
LT drives Dll expression early in larval development by directly integrating inputs from the Wg and Dpp signaling pathways. Tcf and Mad bind LT to activate, while Brk binds LT to repress, resulting in LT activity in the center of the young leg disc. Dll is also required for LT activity, although it is not known if this input is direct. We also suggest that other elements within the Dll locus may act redundantly with LT to integrate the Wg and Dpp signals (not indicated). As the disc grows, Dll becomes independent of Wg and Dpp signaling. During the maintenance phase, the composite LT+M element behaves as an autoregulatory element as it is directly activated by Dll binding to sequences close to the Dll promoter (M). Dll input into LT may also contribute to maintenance, as well as other currently unknown factors. Consistent with this model, a lineage tracing experiment using LT demonstrates that all Dll-expressing cells in a third instar leg disc are derived from LT-expressing cells (McKay et al, in preparation).
Models of maintenance
As is the case for Dll, there are examples of other genes that have separable initiation and maintenance phases of expression. For many of these examples, expression is maintained by the trxG and PcG of epigenetic regulators (reviewed by (Breiling et al., 2007; Brock and Fisher, 2005)). There are also examples of genes that require enhancer-promoter communication for maintenance. For example, a regulatory element from the Hoxb4 gene requires sequences from its own promoter for stable expression in the mouse hindbrain (Gilthorpe et al., 2002). In this case, a key input into the promoter proximal sequences is the PcG protein, YY1. We find that Dll expression is unaffected in trx mutant clones, but is lost in a subset of Pc− and Scm− clones, raising the possibility that PcG functions play a role in maintenance. However, PcG functions are more typically associated with maintaining genes in a repressed state, not an expressed state. Moreover, because of PcG’s widespread role in gene silencing, many genes are likely to be derepressed in these clones. In fact, the Hox gene Abd-B is derepressed in these clones, and Abd-B has the ability to repress Dll (our unpublished observations and (Estrada and Sanchez-Herrero, 2001)). Thus, on balance, it seems more likely that the loss of Dll expression observed in some Pc− clones is an indirect affect. In contrast, our results strongly argue that positive autoregulation, by direct binding of Dll to the M element, plays an important role in Dll maintenance (Fig. 7).
One conclusion we can draw from our observation that both LT and M are required for maintenance is that LT requires the Dll promoter to be fully active. Such promoter-specific enhancer activation has been observed previously, and is generally thought to be important for remote enhancers to stimulate transcription from the correct promoter in gene-dense regions of the genome (e.g. (Butler and Kadonaga, 2002; Calhoun et al., 2002; Li and Noll, 1994; Merli et al., 1996)). The LT+M synergy described here is distinct from these other examples. In this case, although enhancer-promoter compatibility may be part of the reason that LT works better with M (and over large distances), our results show that the combination of the two has properties that are not exhibited by either element on its own. Specifically, while M-lacZ is very weakly expressed in leg discs, and LT-lacZ requires continuous Wg and Dpp inputs, the combination of LT+M allows Dll autoregulation to occur in a Wg- and Dpp-independent manner. Moreover, LT+M is not simply a Dll autoregulatory element: even though Dll is expressed in the wing disc, transcriptional activation by LT+M remains restricted to the ventral imaginal discs. This observation implies that the Dll-input into LT+M can only occur in cells where LT was activated, which itself only happens in ventral discs. Thus, LT+M is not only a two-component Dll-autoregulatory element, but is an autoregulatory element that requires the prior Wg- and Dpp-activation of LT.
These observations lead us to suggest two classes of models by which maintenance may occur. In one, an activated LT element changes the chromatin structure of M, for example, by changing the position of a repressive nucleosome, so that it can function as an autoregulatory element. According to this model, the continued presence of LT is required to maintain this chromatin structure. A second model that would also accommodate our data is that the combination of LT plus M is required to increase the efficacy of transcriptional activation by, for example, providing additional Dll (or other activator) binding. According to this scenario, LT activation by Wg and Dpp triggers the initial interaction between the LT and M elements, which would then be stabilized in a Wg and Dpp-independent manner. These models are not mutually exclusive, and both can be tested by analyzing the chromatin status at the M and LT elements.
Our results also raise the question of what purpose this two step, ‘trigger-maintenance’ mechanism may serve. One possibility is that, by having only a transient requirement for Wg and Dpp, these morphogens are available for carrying out completely different tasks, without affecting Dll expression. In support of this idea, in addition to working together to create the PD axis, Wg and Dpp function independently to instruct ventral and dorsal leg fates, respectively (Morimura et al., 1996; Struhl and Basler, 1993 and Theisen et al., 1996). Some of these late Wg and Dpp patterning functions may also require Dll input. The ‘trigger-maintenance’ logic described here in principle allows Wg and Dpp to execute functions in collaboration with their own downstream target, Dll.
It is also noteworthy that the transient nature of the Wg and Dpp inputs into Dll is not the typical way these morphogens regulate their target genes in other tissues. In the Drosophila wing, for example, Dpp and Wg are required to continuously activate their targets, such as vestigial, optormotor blind, and spalt (de Celis et al., 1996; Grimm and Pflugfelder, 1996 and Kim et al., 1996). One signficant difference between the regulation of wing and leg target genes by these morphogens is that in the wing Wg and Dpp generally act independently, whereas in the leg they act combinatorially to activate PD genes. Specifically, although they are expressed in ventral and dorsal sectors, respectively, Wg and Dpp activate Dll and dac in circular or nearly circular domains whose centers are located where the Dpp and Wg expression domains touch, in the middle of the leg disc. The trigger-maintenance mechanism defined here avoids the need for target genes such as Dll to continuously integrate Wg and Dpp inputs as the disc grows in size, and provides a mechanism to generate circular domains of gene expression using dorsal and ventral morphogen inputs.
Experimental Procedures
Plasmids and transgenes
Our standard reporter genes were built from the hs43-nuc-lacZ vector, which contains the minimal (TATA box) promoter from the hsp43 gene. The hsp43 promoter was removed for constructs containing the M element. The LT, A, B, C, D, E, F, M fragments were selected based on sequence conservation to other Drosophilids (Vista Genome Browser), and cloned by PCR (details are available upon request). The 208 and 179 fragments were obtained by Eco R1 and Eco R1 and Bam H1 digestion, respectively (Vachon et al., 1992). LT is essentially equivalent to the Dll215 enhancer, although no larval expression was reported for this enhancer (Vachon et al., 1992). LT-Gal4 was generated by cloning the LT enhancer into hs43-Gal4. Deletions and mutations were introduced in the LT and M elements using PCR and the QuikChange Site Directed Mutagenesis Kit (Stratagene). The UAS-Tcf RNAi was generated by cloning the 3′ end of Drosophila pangolin into the pWIZ vector (Lee and Carthew, 2003); this transgene was used in combination with a UAS-Tcf RNAi that was a gift from B. Dickson for maximal effect.
For reporter genes, multiple transformants were surveyed to select lines displaying representative expression patterns. Notably, M-lacZ was very sensitive to position effects; however, most lines consistently had very low-level ubiquitous expression, with slightly higher levels in the Dll domain. The sensitivity to position effect was eliminated in the presence of LT.
Immunostaining
Imaginal discs were prepared and stained using standard procedures. The primary antibodies used were: rabbit and mouse anti-β-Gal (Cappell and Promega), mouse anti-Wg (DSHB), guinea pig anti-P-Mad (gift of E. Laufer and T. Jessell), guinea pig anti-Dll (generated by us against full length protein), rabbit anti-Hth generated against full length protein.
Protein Purification and EMSAs
GST-Mad MH1+L (Xu et al., 1998), GST-dTCF HMG (Lee and Frasch, 2000), GST-Brk 1–100 (gift of C. Rushlow) were produced and purified by standard procedures (Amersham-Pharmacia). The full-length Dll cDNA was cloned in frame into pET14b (Novagen). His-Dll was produced and purified by standard procedures (Qiagen). Protein concentrations were measured by Bradford assay and confirmed by SDS-PAGE and Coomassie blue analysis. EMSAs were performed as previously described (Gebelein et al., 2004). The amount of protein used in each EMSA was 25pmol for Brk, 60pmol for Mad, 40pmol for dTcf, and 15pmol for Dll. The sequences for transcription factor binding sites are located in the supplemental data.
Chromatin Immunoprecipitations
ChIP assays were based on a previously described protocol (Papp and Muller, 2006), with alterations described in the supplemental materials.
LT Flp-out experiment
The >LT>M-lacZ reporter (FRT sites are indicated by ‘>’) was generated by cloning LT into plasmid J33R (Struhl and Basler, 1993); >LT> was subsequently cloned into M-lacZ. To delete LT prior to the maintenance phase (before 48h AEL) we drove Flp in the posterior compartment by crossing >LT>M-lacZ-containing flies to en-Gal4, UAS-flp, UAS-GFP. To delete LT during the maintenance phase we crossed >LT>M-lacZ flies to y w hs FLP122; tub>y+>Gal4 UAS-GFP; UAS-Flp and heat shocked at 90 +/−6h AEL. In this experiment, some of the clones that lose LT+M-lacZ expression will be positively marked by GFP while others will be unmarked.
Fly genetics
brkXA is a P (lacZ) insertion and is larva lethal (Campbell and Tomlinson, 1999). Mad1–2 is a strong hypomorph (Wiersdorff et al., 1996) while brkM68 (Jazwinska et al., 1999), PcXT109 (Zirin and Mann, 2004), trxE2 (Klymenko and Muller, 2004), ScmD2(Klymenko and Muller, 2004), tkva12 (Nellen et al., 1994) which were used in the clonal analysis, are considered as nulls. Other lines used were: en-Gal4, UAS-flp, UAS-GFP (gift from Laura Johnston), dpp-Gal4/UAS-GFP (Staehling-Hampton et al., 1994).
For gain of function experiments we used the strain y w hs FLP122; tub>y+>Gal4 UAS-GFP and the following UAS transgenes:
UAS-tkvQD (Abu-Shaar and Mann, 1998), UAS-arm (delta N) (Pai et al., 1997); UAS-TCF-RNAi (The Vienna Drosophila RNAi Center; this line is reported to have no off-target effects). Flip-out clones were originated by heat-shocking the larvae for 10 minutes at 37 C.
For loss of function clones we used the following genotypes:
y f36a brkM68 FRT 19A/ubi-GFP FRT19A; hs FLP
y w hs FLP122; Mad1–2 or tkva12 FRT 40A/ubi-GFP FRT 40A
y w hs FLP122; FRT 42D arr2/FRT 42D ubi-GFP
y w hs FLP122; FRT 42D Dllsa1/FRT 42D ubi-GFP
y w hs FLP122; PcXT109 FRT2A/ubiGFP y+ FRT2A
y w hs FLP122; FRT 82B ubiGFP/FRT 82B trxE2
y w hs FLP122; FRT 82B ubiGFP/FRT 82B ScmD2
Double mutant clones for brk and Mad we used the following genotypes:.
y f36a brkM68 FRT 19A/y w hs FLP122 ubi-GFP FRT19A; Mad1–2 FRT 40A/ubi-GFP FRT 40A
Supplementary Material
Acknowledgments
We thank G. Campbell, M. Frasch, T. Jessell, E. Laufer, K. Cadigan, D. Parker, C. Rushlow, G. Struhl and the Bloomington Stock Center for flies and reagents, the Developmental Studies Hybridoma Bank at The University of Iowa for antibodies, and members of the Mann and Johnston labs for discussions and suggestions. We also thank B. Noro, D. Thanos, H. Herranz, M. Giorgianni, and B. Gebelein for comments on the manuscript, Neal Padte for early contributions to this project, and W. Zhang and M. Baek for technical assistance. This work was supported by grants from the NIH (GM058575) and March of Dimes to R.S.M. D.J.M was partially supported by the Stem Cells and Cell Lineage Specification training grant (HD055165).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- Arnosti DN. Analysis and function of transcriptional regulatory elements: insights from Drosophila. Annu Rev Entomol. 2003;48:579–602. doi: 10.1146/annurev.ento.48.091801.112749. [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Breiling A, Sessa L, Orlando V. Biology of polycomb and trithorax group proteins. Int Rev Cytol. 2007;258:83–136. doi: 10.1016/S0074-7696(07)58002-2. [DOI] [PubMed] [Google Scholar]
- Brock HW, Fisher CL. Maintenance of gene expression patterns. Dev Dyn. 2005;232:633–655. doi: 10.1002/dvdy.20298. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- Campbell G, Weaver T, Tomlinson A. Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell. 1993;74:1113–1123. doi: 10.1016/0092-8674(93)90732-6. [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J, Akam M. How the Hox gene Ultrabithorax specifies two different segments: the significance of spatial and temporal regulation within metameres. Development. 1995;121:2973–2982. doi: 10.1242/dev.121.9.2973. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Cohen B, Simcox AA, Cohen SM. Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development. 1993;117:597–608. doi: 10.1242/dev.117.2.597. [DOI] [PubMed] [Google Scholar]
- Cohen SM. Specification of limb development in the Drosophila embryo by positional cues from segmentation genes. Nature. 1990;343:173–177. doi: 10.1038/343173a0. [DOI] [PubMed] [Google Scholar]
- Cohen SM. Imaginal Disc Development. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY; CSH Lab Press: 1993. pp. 747–842. [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen B, Cohen SM. Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature. 1994;372:175–179. doi: 10.1038/372175a0. [DOI] [PubMed] [Google Scholar]
- Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–5941. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- Estrada B, Sanchez-Herrero E. The Hox gene Abdominal-B antagonizes appendage development in the genital disc of Drosophila. Development. 2001;128:331–339. doi: 10.1242/dev.128.3.331. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Bishop SA, Greig S, Couso JP. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science. 2002;297:256–259. doi: 10.1126/science.1072311. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Gilthorpe J, Vandromme M, Brend T, Gutman A, Summerbell D, Totty N, Rigby PW. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development. 2002;129:3887–3899. doi: 10.1242/dev.129.16.3887. [DOI] [PubMed] [Google Scholar]
- Goto S, Hayashi S. Specification of the embryonic limb primordium by graded activity of Decapentaplegic. Development. 1997;124:125–132. doi: 10.1242/dev.124.1.125. [DOI] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder GO. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci U S A. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Schubiger G. Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development. 1996;122:3519–3529. doi: 10.1242/dev.122.11.3519. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick H, Johnson K, Laughon A. Repression of dpp targets by binding of brinker to mad sites. J Biol Chem. 2001;276:18216–18222. doi: 10.1074/jbc.M101365200. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Cohen SM. Proximal-distal axis formation in the Drosophila leg. Nature. 1997;388:139–145. doi: 10.1038/40563. [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. Embo J. 2007;26:2284–2294. doi: 10.1038/sj.emboj.7601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. Embo J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Miltenberger RJ, Hoffmann FM. Pattern-specific expression of the Drosophila decapentaplegic gene in imaginal disks is regulated by 3′ cis-regulatory elements. Genes Dev. 1990;4:2011–2023. doi: 10.1101/gad.4.11.2011. [DOI] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- Morimura S, Maves L, Chen Y, Hoffmann FM. decapentaplegic overexpression affects Drosophila wing and leg imaginal disc development and wingless expression. Developmental Biology. 1996;177:136–151. doi: 10.1006/dbio.1996.0151. [DOI] [PubMed] [Google Scholar]
- Nellen D, Affolter M, Basler K. Receptor serine/threonine Kinases implicated in the control of Drsophila body pattern by decapentaplegic. Cell. 1994;78:225–237. doi: 10.1016/0092-8674(94)90293-3. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994;5:585–593. [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Theisen H, Haerry TE, O’Connor MB, Marsh JL. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen S. Homeotic Genes of the Bithorax Complex Repress Limb Development in the Abdomen of the Drosophila Embryo through the Target Gene Distal-less. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- Wiersdorff V, Lecuit T, Cohen SM, Mlodzik M. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development. 1996;122:2153–2162. doi: 10.1242/dev.122.7.2153. [DOI] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin JD, Mann RS. Differing strategies for the establishment and maintenance of teashirt and homothorax repression in the Drosophila wing. Development. 2004;131:5683–5693. doi: 10.1242/dev.01450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.