SUMMARY
The DNA double strand break (DSB) repair protein DNA-PKcs and the signal transducer ATM are both activated by DNA breaks and phosphorylate similar substrates in vitro, yet appear to have distinct functions in vivo. Here we show that ATM and DNA-PKcs have overlapping functions in lymphocytes. Ablation of both kinase activities in cells undergoing immunoglobulin class switch recombination leads to a compound defect in switching, and a synergistic increase in chromosomal fragmentation, DNA insertions and translocations due to aberrant processing of DSBs. These abnormalities are attributed to a compound deficiency in phosphorylation of key proteins required for DNA repair, class switching and cell death. Notably, both kinases are required for normal levels of p53 phosphorylation in B and T cells and p53 dependent apoptosis. Our experiments reveal a DNA-PKcs-dependent pathway that regulates DNA repair and activation of p53 in the absence of ATM.
INTRODUCTION
ATM and ATR are signal transducing phosphatidyl inositol 3-kinase-like kinases (PIKKs) that are central to the DSB response (Rouse and Jackson, 2002). At least 700 proteins are phosphorylated on ATM/ATR consensus sites in response to irradiation (IR) (Matsuoka et al., 2007). A third member of the PIKK family, DNA-PKcs, is itself a target of ATM/ATR (Chen et al., 2007; Matsuoka et al., 2007), and is thought to play a more restricted role in DSB repair (Kurimasa et al., 1999). DNA-PKcs is essential for nonhomologous end-joining (NHEJ), a pathway that promotes cell survival in response to irradiation (Smith and Jackson, 1999) and repairs programmed DSBs created during V(D)J recombination and class switch recombination (CSR) in lymphocytes (Dudley et al., 2005; Jankovic et al., 2007; Smith and Jackson, 1999). Although PIKKs recognize similar target consensus sites in vitro, the physiological substrate(s) of DNA-PKcs that facilitates NHEJ remain unknown.
ATM and DNA-PKcs are both recruited to and activated by DSBs. ATM is targeted to DSBs through an interaction with the DSB sensor Mre11/Rad50/Nbs1 complex (Lee and Paull, 2005), whereas DNA-PKcs recruitment is dependent on prior loading of the Ku70/80 heterodimer on DNA ends(Dvir et al., 1992; Gottlieb and Jackson, 1993). In contrast, the molecular species recognized by ATR is single stranded DNA (ssDNA) coated by replication protein A (RPA), a ssDNA binding protein (Zou and Elledge, 2003). Maximal ATR activation is dependent on ATM and on Mre11-dependent nucleolytic processing of DSBs into ssDNA (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Yoo et al., 2007).
While most DSB signal transduction pathways are thought to be governed cooperatively by ATM and ATR, recent studies raise the possibility of cross-talk between DNA-PKcs and ATM in DNA damage signaling. For example, a DNA-PKcs dependent component to the G2/M checkpoint has been reported in epithelial cells (Arlander et al., 2008; Liu et al., 2008), and H2AX, RPA and DNA-PKcs phosphorylation is co-dependent on ATM and DNA-PKcs in irradiated fibroblasts and tumor cell lines (Chen et al., 2007; Stiff et al., 2004; Wang et al., 2001). However, combined inactivation of ATM and DNA-PKcs does not yield an additive DSB joining defect in fibroblasts (Riballo et al., 2004), nor increased radiation sensitivity in cancer cell lines (Cowell et al., 2005) compared to loss of either kinase alone.
Several lines of evidence indicate that ATM and DNA-PKcs play distinct roles in the DSB response in vivo. First, compared with the very broad spectrum of ATM substrates, the targets of DNA-PKcs in vivo remain undefined. Second, the phenotypes associated with ATM and DNA-PKcs deficiencies are distinct. ATM knockout mice are sterile due to a block in meiosis, lymphopenic and predisposed to lymphomas, whereas DNA-PKcs knockout mice are fertile but exhibit severe combined immunodeficiency (SCID) without increased incidence of tumorigenesis. Third, combined deficiency in DNA-PKcs and ATM results in synthetic lethality early in embryogenesis (Gurley and Kemp, 2001; Sekiguchi et al., 2001). Finally, whereas DNA damage induced cell cycle and apoptotic checkpoints are severely compromised in cells lacking ATM (Shiloh, 2003), DNA-PKcs is dispensable for DSB signaling, p53 activation and cell cycle arrest (Burma et al., 1999; Jhappan et al., 2000; Jimenez et al., 1999; Rathmell et al., 1997). Thus, the majority of the evidence indicates that although ATM and DNA-PKcs are both activated by IR induced DSBs they cannot substitute for each other in the DNA damage response in vivo, potentially because of distinct physiological targets.
In addition to IR induced DSBs, physiological DSBs are processed during class switch recombination in mature B cells in a reaction that alters the effector properties of antibodies (Barreto et al., 2005; Chaudhuri et al., 2007; Di Noia and Neuberger, 2007; Honjo et al., 2005; Li et al., 2007; Stavnezer et al., 2008). CSR is initiated by activation-induced cytidine deaminase (AID) (Muramatsu et al., 2000; Muramatsu et al., 1999; Revy et al., 2000), an enzyme that deaminates cytosine residues in ssDNA, leading to DSBs in switch regions which lie upstream of each of the constant region genes (Chaudhuri et al., 2003; Dickerson et al., 2003; Petersen-Mahrt et al., 2002; Pham et al., 2003; Ramiro et al., 2003). AID dependent DNA breaks trigger a DNA damage response, evidenced by the recruitment of DNA damage response proteins Nbs1, MDC1, ATM, γ-H2AX, and 53BP1 to IgH chromatin in the G1 phase of the cell cycle (Jankovic et al., 2007). Each of these factors is required for normal resolution of the CSR reaction (Difilippantonio et al., 2008; Franco et al., 2006; Jankovic et al., 2007; Ramiro et al., 2006; Reina-San-Martin et al., 2003). ATM and 53BP1 have been proposed to play a role in switch region synapsis during CSR (Lumsden 2004; Reina-San-Martin 2004; Manis, 2004 #25; Ward, 2004 #27; Reina-San-Martin, 2007 #78}, while DNA-PKcs is implicated in joining of the subset of AID induced DSBs (Franco et al., 2008).
Here we report that DNA-PKcs and ATM coordinately control programmed DSB repair during class switch recombination and cell death in lymphocytes suggesting a broader role for DNA-PKcs beyond end-joining in the maintenance of genomic stability.
RESULTS
DNA-PKcs catalytic activity is essential for CSR in the absence of ATM
To investigate the relationship between DNA-PKcs and ATM we used a well characterized inhibitor NU7026 (PKi) to interfere with DNA-PKcs kinase activity in B cells (Leahy et al., 2004; Veuger et al., 2003). In agreement with studies in fibroblast cell lines, we found compromised IR induced H2AX phosphorylation in ATM-/- but not in wild type B cells that were pre-incubated with PKi (Figure S1)(Riballo et al., 2004; Stiff et al., 2004). Potential off-target effects of PKi on other PIKKs were not evident based on the robust IR-and replication stress-induced substrate phosphorylations in PKi treated cells (Figures S2 and S3).
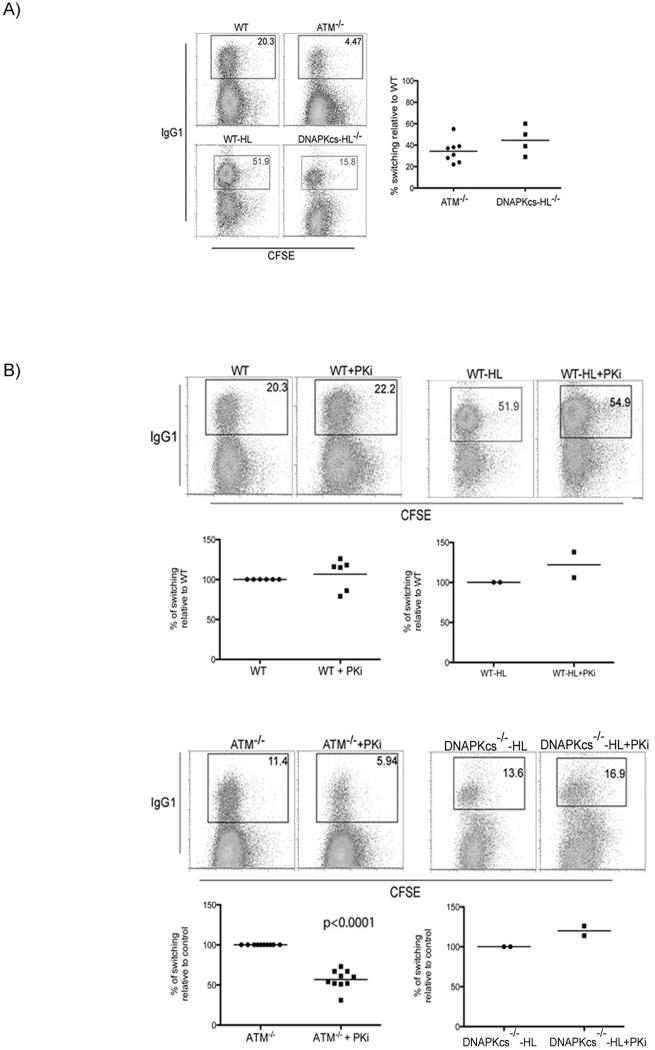
ATM-/- B cells show decreased CSR but normal levels of cell division in response to LPS and IL-4 (Lumsden et al., 2004; Reina-San-Martin et al., 2004) (Figure 1A). B cells fail to develop in DNA-PKcs-/- due to absence of V(D)J recombination, but can be reconstituted by introduction of pre-rearranged Ig heavy and light chain transgenes (DNA-PKcs-/-HL). In our experiments, DNA-PKcs-/-HL B cells showed defects in CSR to IgG1 and IgG3 ranging from 30-70% of controls (Cook et al., 2003; Franco et al., 2008; Manis et al., 2002) (Figure 1A; Figure S4).
Figure 1. Contribution of DNA-PKcs holoenzyme and DNA-PKcs kinase activity to CSR.
(A) Flow cytometric analysis of CSR to IgG1 in ATM-/-, wild type (WT, littermate control for ATM), DNA-PKcs-/--HL and WT-HL (littermate control for DNA-PKcs-/--HL). DNA-PKcs-/--HL and WT-HL are matched littermates expressing immunoglobulin heavy VB1-8 and light chain V3-83 genes that rescue B cell development in DNA-PKcs-/-mice (Manis et al., 2002). B cells were labeled with CFSE to assess cell division and stimulated with LPS+IL4 for 3 days. Graphs show switching relative to matched wild type sample, each dot represents an individual experiment and the bar represents the average. (B) Flow cytometric analysis of CSR as in (A) in the presence or absence of PKi NU7026. Graphs as in (A) but show switching relative to the matched sample in the absence of Pki as indicated.
To examine the role of DNA-PKcs in CSR, B cells were labeled with CFSE [5-(and 6-) carboxyfluorescein diacetate succinyl ester] to track cell division by dye dilution and stimulated with LPS and IL-4 to induce switching to IgG1 in the presence or absence of PKi. We found a small increase in CSR to IgG1 in PKi treated wild type and DNA-PKcs-/-HL B cells but no change in cell division (Figure 1B; Figure S5). Thus, while loss of the DNA-PKcs protein impairs CSR, inhibition of DNA-PKcs activity does not.
To examine the possibility that DNA-PKcs activity might be redundant with ATM during CSR we examined switching to IgG1 and IgG3 by ATM-/- B cells in the presence or absence of PKi. In contrast to wild type B cells, reducing DNA-PKcs activity in ATM-/- B cells resulted in a decrease in CSR while cell division remained intact (Figure 1B; Figure S5). This decrease in CSR was not due to altered AID protein levels or switch region accessibility as measured by germline transcription of switch regions (Figure S5). Thus, ATM and DNA-PKcs do not appear to affect events upstream of AID-mediated switch lesion formation. We conclude that ATM and DNA-PKcs have overlapping functions during CSR.
Loss of ATM and DNA-PKcs activity results in DNA insertions into CSR junctions
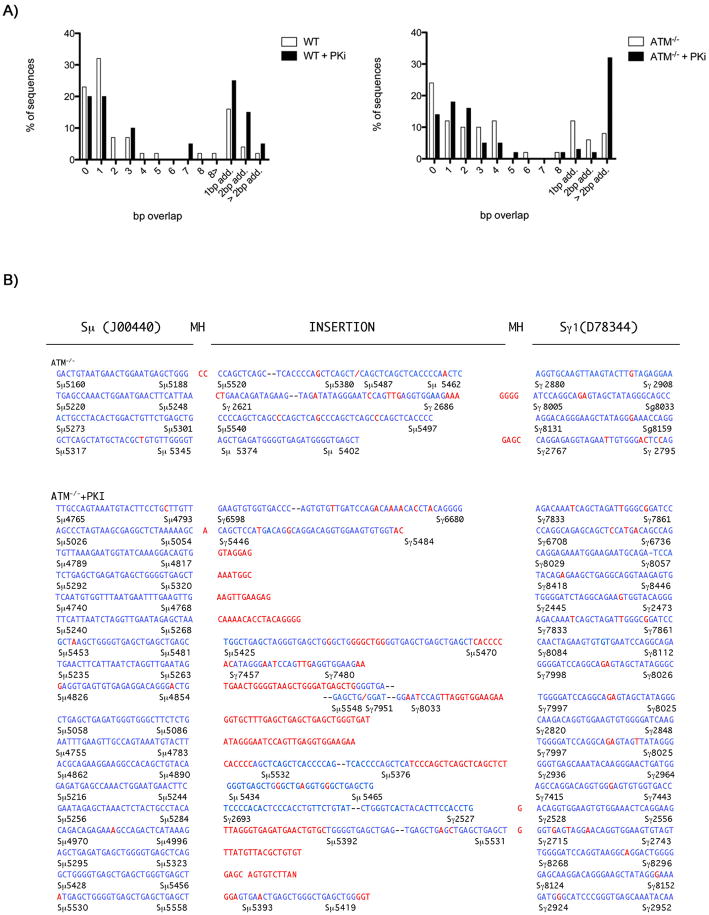
To determine whether loss of both PIKK activities impacts on DNA joining at immunoglobulin switch (S) regions we cloned and sequenced Sμ-Sγ1 junctions from IgG1 positive B cells that have completed CSR (Figure 2A). Consistent with previous analyses, we found no statistically significant alterations in the switch junctions in ATM deficient cells (Reina-San-Martin et al., 2004). However, loss of ATM resulted in a small increase in junctions showing three or more nucleotide insertions (Figure 2A). Such junctions are rare in wild type cells and the addition of PKi to wild type cells resulted in a small increase in these events (Figure 2A). Strikingly, addition of PKi to ATM-/- B cells increased the frequency of insertions to 32% of the total CSR junctions analyzed (Figure 2A). Many of these insertions were large (~100-200bp), and came from non-contiguous regions of switch DNA, consistent with the idea that AID introduces multiple lesions in switch regions (Figure 2B)(Dudley et al., 2005; Reina-San-Martin et al., 2003). We conclude that DNA-PKcs and ATM have a redundant function in switch break repair, and are required to prevent intra-molecular insertions at the switch junctions. A similar function has been ascribed to 53BP1 (Difilippantonio et al., 2008; Manis et al., 2004; Reina-San-Martin et al., 2007).
Figure 2. Inhibition of DNA-PKcs kinase activity increases the frequency of CSR junctions with insertions.
(A) Wild type and ATM-/- B cells were either untreated or pretreated with PKi NU7026, stimulated with LPS+IL4 for 72 hours, and IgG1+ cells were sorted for further analysis. Genomic DNA was amplified by PCR, and Sμ/Sγ1 junctions were sequenced. Percentage of junctions with indicated nucleotide overlap or insertions is indicated. (B) Sμ/Sγ1 switch junctions with insertions greater than 2bp from ATM-/- B cells in the presence or absence of PKi NU7026. The numbers below each sequence correspond to Sμ (Genbank J00440) and Sγ1 (Genbank D78344) germline sequences. Blue letters indicate sequences homologous to germline, red letters indicate mutations, insertions with no known homology or microhomology at junctions, -- indicates contiguous sequences that are not shown, / indicates a break in the sequence. Number of sequences analyzed: ATM-/-=50; ATM-/-+PKi=56; WT=44; WT+PKi=20.
Synergistic increase in genomic instability
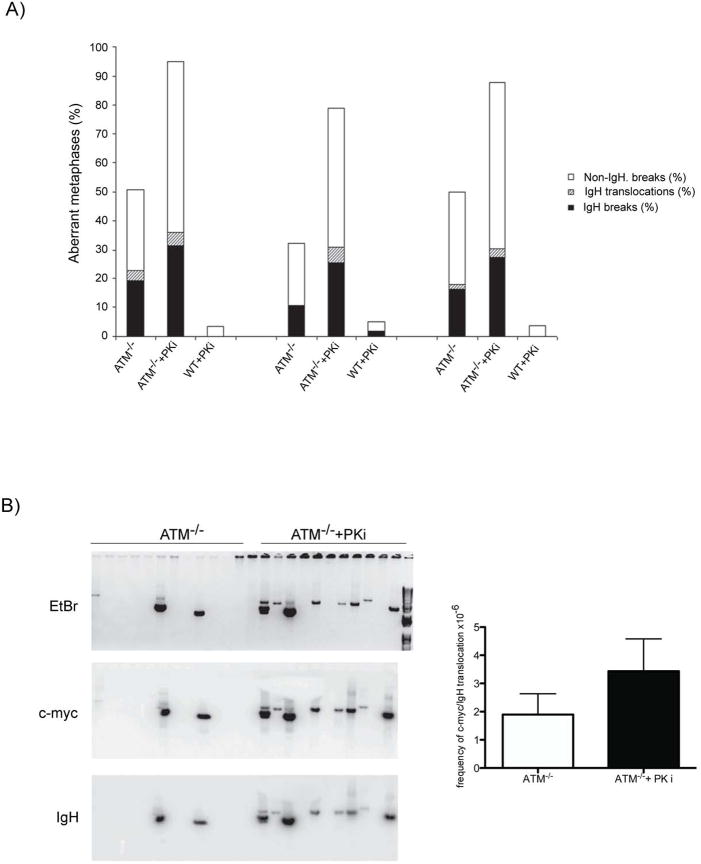
DSBs are essential intermediates during antigen receptor gene diversification reactions, but they are also substrates for chromosomal translocations. As a result of their end-joining defects, DNA-PKcs-/- and ATM-/- B cells stimulated to undergo CSR accumulate IgH -specific chromosome breaks and general instability outside of the IgH locus (Callen et al., 2007; Franco et al., 2006; Franco et al., 2008; Ramiro et al., 2006). Similar defects are also found in wild type B cells treated with the small molecule inhibitor of ATM (KU55933) (Callen et al., 2007). To determine whether ATM and DNA-PKcs are redundant in maintaining genomic integrity during CSR we compared the level of genomic instability in wild type or ATM-/- B cells cultured in the presence or absence of PKi. IgH associated breaks, translocations, and general instability outside of the IgH locus were monitored by fluorescent in situ hybridization (FISH) using chromosome 12, telomere, and IgHCα-specific probes (Callen et al., 2007; Franco et al., 2006) (Figure 3A). In three independent experiments, wild type B cells showed low levels of general chromosome instability, and little or no IgH instability in the presence or absence of PKi (Figure 3A). As previously reported, 10-20% of ATM-/- B cells harbored IgH specific breaks, 0.5-3.5% carried IgH translocations and 20-30% had non-IgH associated instability (Figure 3A) (Callen et al., 2007). In marked contrast to wild type, inhibition of DNA-PKcs in ATM-/- B cells led to a synergistic increase in IgH breaks, translocations, and general genomic instability (Figure 3A). We conclude that DNA-PKcs and ATM are redundant for the switch reaction and for maintaining general and IgH chromosome stability in B cells undergoing CSR.
Figure 3. Loss of DNA-PKcs kinase activity results in a synergistic increase in genomic instability in ATM-/- B cells.
(A) Frequency of IgH locus breaks (black), IgH locus translocations (grey, striped), and non-IgH breaks (white) in 3 independent cultures of ATM-/- and wild type B cells mock or pre-treated with PKi NU7026. Metaphase spreads (>80 analyzed for each experiment) were prepared 72 hours after B cell stimulation with LPS and IL4. (B) c-myc/IgH translocations. Representative agarose gel (top) and Southern blots with c-myc (middle) and IgH (bottom) probes. Frequency of c-myc/IgH translocation per 106 cells is plotted on the right p=0.0002 (ATM-/- vs. ATM-/- pretreated with PKi NU7026).
Increase in c-myc/IgH translocations
While both ATM and DNA-PKcs have been implicated in the repair of a subset of AID-induced DSBs (Franco et al., 2006; Franco et al., 2008; Ramiro et al., 2006; Reina-San-Martin et al., 2004), ATM and p53 have an additional apoptotic function: protecting against CSR-associated tumor promoting translocations between c-myc and IgH (Ramiro et al., 2006). In contrast, DNA-PKcs-/- B cells have an intact p53 dependent apoptotic checkpoint, evidenced by the finding that LPS stimulated DNA-PKcs-/- B cells that suffer DNA damage are eliminated by p53 (Franco et al., 2008).
To determine whether ATM and DNA-PKcs also serve a redundant function in protecting against c-myc/IgH translocation, we assayed for the frequency of these events in ATM-/- B cells treated with PKi (Ramiro et al., 2006; Ramiro et al., 2004) (Figure 3B). Consistent with the effects on CSR, we found a significant increase in the frequency of c-myc/IgH translocations in ATM-/- B cells treated with PKi compared to ATM-/- B cells alone (Figure 3B). Therefore, DNA-PKcs kinase activity does indeed protect against c-myc/IgH translocation in the absence of ATM.
Impaired phosphorylation of DNA repair and checkpoint proteins
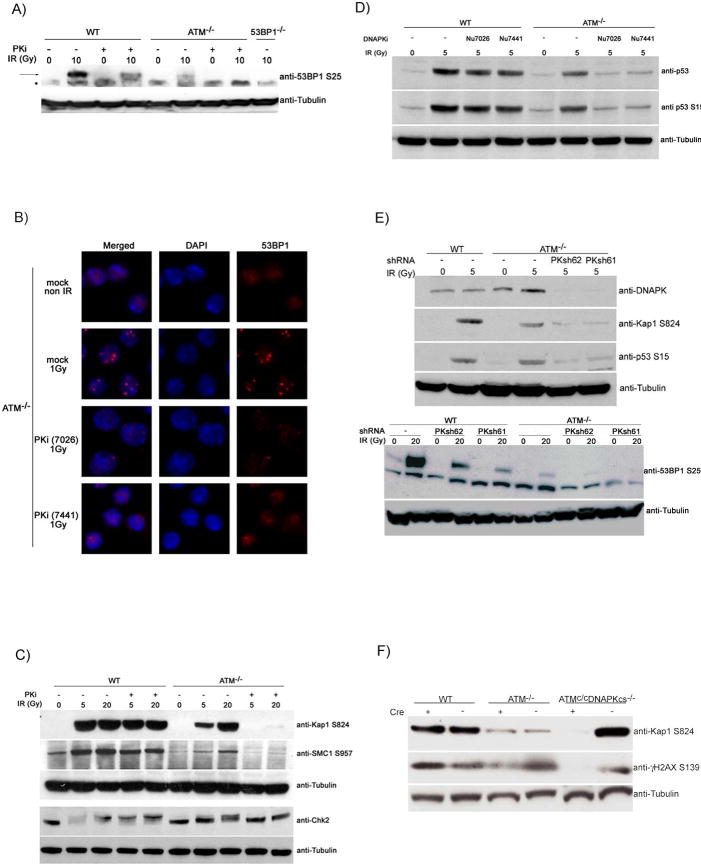
Combined inhibition of ATM and DNA-PKcs resembles deficiency in 53BP1(Manis et al., 2004; Ramiro et al., 2006; Reina-San-Martin et al., 2007; Ward et al., 2004) in that it results in dramatically decreased CSR, switch region insertions, and IgH instability (Figures 1-3). To determine whether combined PIKK inhibition alters 53BP1 responses we measured its phosphorylation and focus formation in response to irradiation (Figures 4A and 4B). Inhibition of DNA-PKcs or ATM deficiency resulted in decreased 53BP1 Ser25 phosphorylation, and the combination led to the complete absence of phosphorylation (Figure 4A). In addition, PKi abrogated 53BP1 focus formation in ATM-/- but not in wild type B cells exposed to IR (Figure 4B). Thus, ATM and DNA-PKcs phosphorylate 53BP1 and regulate 53BP1 focus formation in a redundant manner.
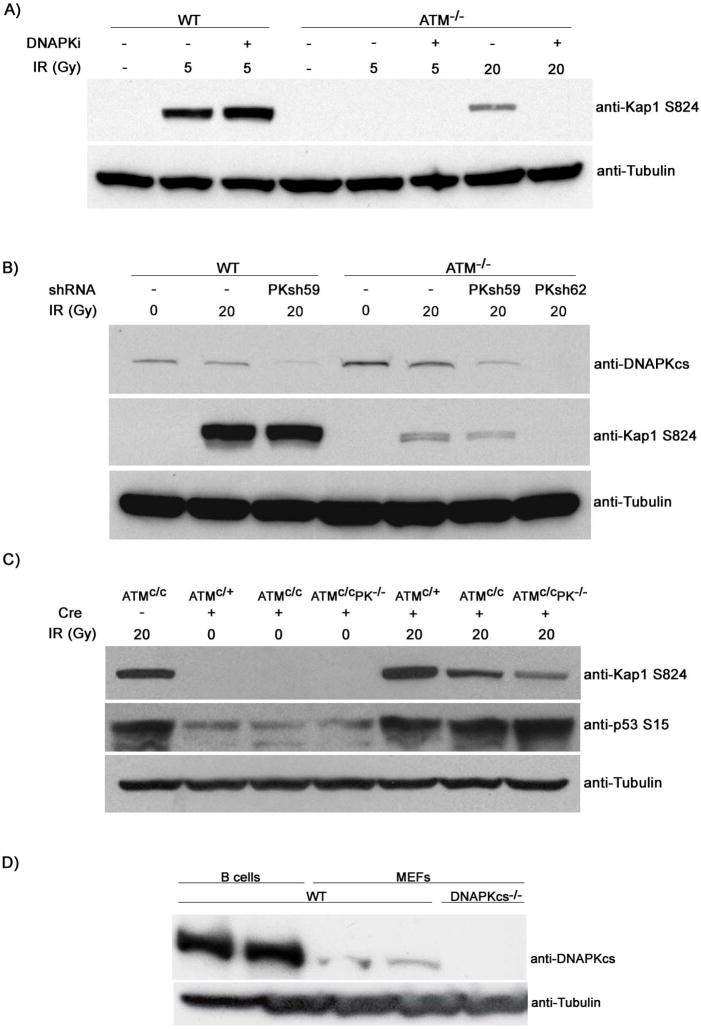
Figure 4. ATM and DNA-PKcs collaborate to phosphorylate 53BP1, Kap-1, p53, SMC1 and Chk2 in lymphocytes.
(A) Wild type, ATM-/- and 53BP1-/- B cells were cultured with LPS and IL4 with or without PKi NU7026 for 48h, and harvested 1 hour after 0 or 10 Gy irradiation as indicated. Western blots show 53BP1 phosphorylation (arrow) and tubulin loading controls. * indicates non-specific band. (B) 53BP1 focus formation in ATM-/- B cells 6 hours after treatment with PKi (NU7026) or another potent and selective inhibitor of DNA-PKcs (NU7441) (Leahy et al., 2004) and 1Gy irradiation. (C) Western blots for Kap-1, SMC1, and Chk2 phosphorylation and tubulin loading controls in irradiated B cells treated as in (A). (D) Western blot shows p53 protein levels and p53 phosphorylation on Ser15, 4 hours after 0 or 5 Gy treatment. Cells were pretreated with PKi (NU7026) or an independent selective inhibitor of DNA-PKcs (NU7441). (E) DNAPKcs was depleted in cultured WT and ATM-/- B cells using short hairpin RNAs. Five days after infection, cells were untreated or irradiated, sorted and blotted for DNA-PKcs, phosphorylated Kap-1, phosphorylated p53 (upper panel) or phosphorylated 53BP1 (lower panel). (F) B cells from WT, ATM-/- and conditional ATMc/cDNAPK-/- mice were infected with a retrovirus expressing Cre-GFP (+) or GFP (-) for 5 days. Cells were sorted, irradiated and levels of Kap-1 and H2AX phosphorylation were assessed by Western blotting.
To determine whether ATM and DNA-PKcs collaborate in phosphorylating additional downstream substrates, we examined the effects of combined loss of the two PIKKs on the classical ATM substrates Kap-1, Chk2, Chk1, p53 and SMC1 in irradiated B cells and thymocytes. We found that IR induced phosphorylation of Kap-1, SMC1, Chk2, p53 and Chk1 was further compromised in ATM-/- B cells or thymocytes treated with PKi (Figures 4C-D; Figure S6). These data suggest that ATM and DNA-PKcs are generally redundant for substrate phosphorylation in mouse lymphocytes.
To further confirm these findings, we targeted DNA-PKcs in primary B cells with short hairpin RNAs (shRNAs). Western blot analysis revealed reduction of DNA-PKcs after infection with two independent shRNAs, PKsh61 and PKsh62 (Figure 4E). Consistent with the results with PKi, downregulation of DNA-PKcs in ATM deficient cells led to further reduction in IR induced phosphorylation of Kap-1, p53 and 53BP1 (Figure 4E). Finally, we used a genetic approach to produce B cells deficient in ATM and DNA-PKcs. DNA-PKcs-/-HL and ATM conditional knock out mice (Zha et al., 2008)(Zha et al, in preparation) were bred to generate DNA-PKcs-/-HL/ATMc/c B cells. To delete ATM activity, mutant B cells were infected with a virus expressing cre recombinase or vector control expressing GFP as a marker. GFP+ cells were sorted, and efficient deletion of ATM was confirmed by PCR (Figure S7). The sorted cells were irradiated and analyzed by Western blotting. Whereas ATM or DNA-PKcs deficient cells are capable of phosphorylating Kap1-1 and H2AX in response to IR (Figure 4F), deletion of ATM in DNA-PKcs knockout B cells led to a near complete ablation of in Kap-1 and H2AX phosphorylation (Figure 4F). Thus, combined genetic depletion of ATM and DNA-PKcs confirms that the two kinases are redundant in lymphocytes.
To determine whether the functional overlap in lymphocytes extends to other cell types we analyzed Kap-1 phosphorylation in mouse embryo fibroblasts (MEFs). DNA-PKcs activity was depleted from ATM-/- immortalized MEFs either by treatment with PKi (Figure 5A), or by PKcs shRNA (Figure 5B), or by adenoviral CRE infection of ATMc/cDNA-PKcs-/- primary MEFs (Figure 5C). In all cases, combined deficiency in ATM and DNA-PKcs led to a reduction in Kap-1 phosphorylation relative to that of ATM deficiency alone (Figures 5A-C). However, in contrast to lymphocytes, DNA-PKcs did not affect the activation of p53 in primary ATM-/- MEFs treated with 20 Gy irradiation (Figure 5C). Consistent with this result, it has been reported that ATR cooperates with ATM in the IR induced phosphorylation of p53 in primary MEFs (Brown and Baltimore, 2003). Differences in the degree of redundancy between ATM, DNA-PKcs and ATR may reflect differences in the proportion of cycling cells (see discussion) or differences in the levels of DNA-PKcs in different cell types (Figure 5D).
Figure 5. ATM and DNA-PKcs collaborate in IR induced phosphorylation of Kap-1 but not in p53 activation in MEFs.
(A) Immortalized WT and ATM-/- MEFs were preincubated for 1hour with or without PKi and levels of phosphorylated Kap-1 were determined 1hour after 5 or 20Gy irradiation. (B) Levels of DNAPKcs and IR induced phosphorylated Kap-1 in immortalized WT and ATM-/- MEFs stably expressing lentiviruses harboring PKcs shRNA. Down regulation of DNA-PKcs in ATM-/- MEFs was more efficient using PKsh62 compared to PKsh59. (C) Passage 2 primary MEFs derived from ATMc/+, ATMc/c or ATMc/cDNAPKcs-/- mice were infected with an adenovirus expressing Cre-GFP (+), irradiated with 20Gy and levels of Kap-1 and p53 phosphorylation were analyzed by Western blotting 1 hour post irradiation. (D) Decreased levels of DNA-PKcs in WT MEFs relative to B cells as determined by Western blotting.
Collaboration between ATM and DNA-PKcs in p53 dependent apoptosis
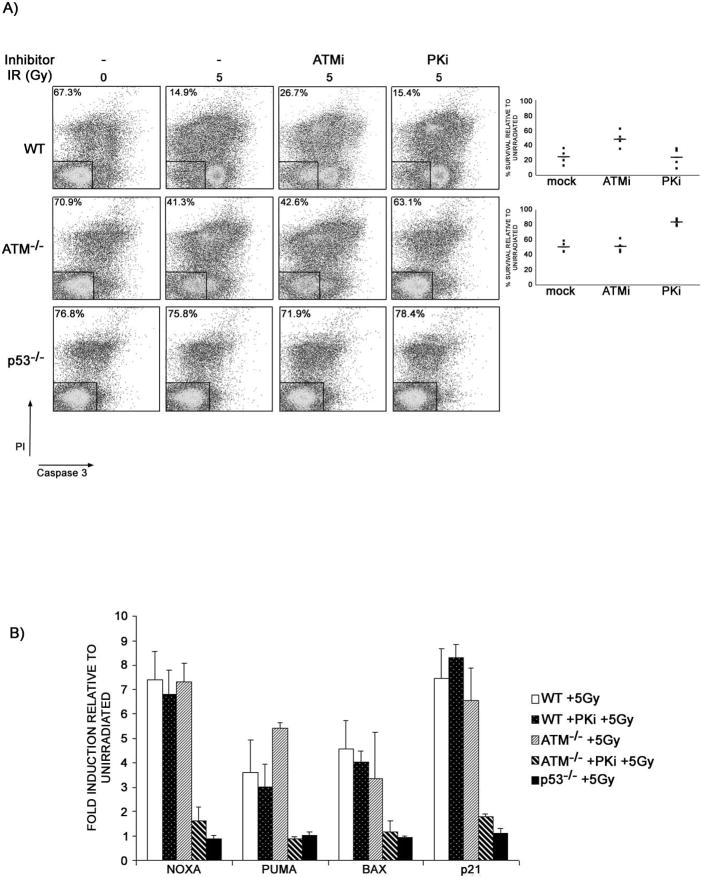
DNA-PKcs is not known to phosphorylate p53 in vivo (Jimenez et al., 1999); on the other hand, it does contribute to IR induced p53 activation in lymphocytes lacking ATM (Figure 4D; Figure S6). Therefore we speculated that DNA-PKcs might be an important inducer of apoptosis in irradiated lymphocytes lacking ATM. To test this hypothesis, we pre-treated ATM-/- and wild type thymocytes with PKi, and measured cellular viability by flow cytometry 24 hours after irradiation (Figure 6A). We found that PKi protected irradiated ATM-/- cells to levels approaching p53-/-, whereas viability of wild type thymocytes was unaffected by PKi (Figure 6A; Figure S8). We conclude that DNA-PKcs is essential for triggering p53 dependent apoptosis in response to irradiation in ATM-/- thymocytes.
Figure 6. ATM and DNA-PKcs collaborate in IR induced p53 dependent apoptosis.
(A) Wild type, ATM-/-, and p53-/- thymocytes were mock or pre-treated with ATMi (KU55933) or PKi (NU7026), irradiated with 5 Gy and the percentage of live cells was measured by flow-cytometry 24 hours post-IR using a caspase-3-substrate reagent (OncoImmunin Inc.). Percentage of live cells (Propidium Iodide (PI) and Caspase 3 negative) is indicated. Graph shows results of individual experiments (dots) normalized to unirradiated drug-treated samples of the same genotype. Bars indicate the average. (B) Quantitative RT-PCR analysis of p53-dependent induction of NOXA, PUMA, BAX and p21 mRNAs 4 hours after treatment with 5Gy plotted relative to un-irradiated controls of the same genotype. An average of two experiments performed in triplicate is shown. The experiments were repeated 6 times with similar results.
Irradiation induced thymocyte apoptosis is regulated in part through p53 dependent induction of the pro-apoptotic genes NOXA, PUMA, and BAX. To gain insight into the basis for the apoptotic defect in ATM-/- thymocytes we examined the expression of these genes by real time quantitative RT-PCR four hours after treatment with 5 Gy of irradiation. The irradiation induced increase in messenger RNA for these genes was not significantly different between ATM-/- and wild type thymocytes (Figure 6B). However, pre-treatment of ATM-/- thymocytes with PKi completely abrogated the induction of pro-apoptotic genes as well as the induction of the p21 cell cycle inhibitor, all of which accumulate in a p53-dependent manner (Figure 6B). Thus, DNA-PKcs is required for ATM-independent transcriptional activation of p53 dependent genes in lymphocytes.
DISCUSSION
The ATM and ATR signaling network regulates replication fork stability, transcription, checkpoint signaling, and DNA repair. In contrast, DNA-PKcs is thought to have a more restricted role in the re-ligation of broken ends by NHEJ (Smith and Jackson, 1999). Our studies reveal a broader function for the DNA-PKcs dependent pathway in the DNA damage response in lymphocytes.
Non-catalytic role of DNA-PKcs in CSR
DNA-PKcs catalytic activity is reported to be essential for V(D)J recombination (Kurimasa et al., 1999) and SCID thymocytes lacking DNA-PKcs activity exhibit an increase in p53 activation and apoptotic cell death (Guidos et al., 1996). The role of DNA-PKcs in CSR is the subject of debate with some investigators reporting near normal switching in SCID mice (Bosma et al., 2002) and in DNA-PKcs-/- mice (Kiefer et al., 2007), while others have demonstrated variable levels of impairment in CSR (Cook et al., 2003; Manis et al., 2002) with the most profound defects found in null mutant mice (Manis et al., 2002). Our experiments confirm the existence of partial CSR defects to both IgG1 and IgG3 in mice lacking DNA-PKcs protein. In contrast, CSR or end-joining of IgH switch region breaks was not impaired in wild type cells depleted of DNA-PKcs catalytic activity. Our results suggest that in ATM sufficient cells DNA-PKcs has functions other than catalysis in promoting CSR. For example, DNA-PKcs may play a structural role acting as a scaffold to align, synapse or protect CSR breaks (DeFazio et al., 2002).
Impaired CSR in the absence of DNA-PKcs and ATM resembles 53BP1 deficiency
While DNA-PKcs enzymatic activity is dispensable for end-joining during CSR in normal cells, similar reduction of DNA-PKcs activity in ATM-/- B cells results in decreased class switching, accumulation of intra-molecular insertions at switch junctions, and a synergistic increase in IgH and general chromosomal instability. Our study suggests that these observations can be explained by a previously unappreciated compound defect in phosphorylation of DNA repair and damage signaling proteins in doubly deficient lymphocytes as discussed below.
ATM plays a central role in DSB repair by phosphorylating numerous substrates including focus forming factors Nbs1, MDC1, H2AX, and 53BP1 and NHEJ proteins that also participate in CSR break repair (Jankovic et al., 2007). Among these factors, the most profound defect in CSR is found in 53BP1 deficient B cells, which show a nearly complete block in the reaction (Manis et al., 2004; Reina-San-Martin et al., 2007; Ward et al., 2004). 53BP1-/- cells undergo a perturbed form of the CSR reaction evidenced by an increase in short range intra-switch region joints and a concomitant decrease in long range junctions that contain insertions of non-contiguous switch region DNA (Reina-San-Martin et al., 2007). Similar but less pronounced defects in CSR are also found in ATM, H2AX, Nbs1 and DNA-PKcs deficient B cells. However, in contrast to 53BP1 deficiency, the CSR junctions in these mice are nearly normal (Celeste et al., 2002; Kracker et al., 2005; Lumsden et al., 2004; Manis et al., 2002; Reina-San-Martin et al., 2004; Reina-San-Martin et al., 2003; Reina-San-Martin et al., 2005). Our finding that inhibition of DNA-PKcs activity in ATM-/- abrogates 53BP1 phosphorylation and focus formation, and results in CSR junctions that resemble those found in 53BP1-/- B cells, suggests that the two kinases can substitute for each other in activating key DNA damage response factors required for the repair of DSBs. This idea is further supported by cooperative phosphorylation of Kap-1 and SMC1, both of which have been implicated in chromosomal repair and cell survival after DNA damage (Kitagawa et al., 2004; Ziv et al., 2006).
Synergistic increase in genomic instability
In addition to decreased CSR, loss of ATM leads to genomic instability and an increase in AID mediated c-myc/IgH translocations (Callen et al., 2007; Ramiro et al., 2006). These aberrations are further increased by loss of DNA-PKcs catalytic activity. B cells containing chromosomal breaks or oncogenic translocations are normally eliminated via a p53 dependent pathway, which is thought to be activated in part by ATM (Ramiro et al., 2006). Our experiments show that ATM and DNA-PKcs are redundant with respect to activation of p53 and the Chk2 pathway. The combined deficiency of the two PIKKs results in a severe defect in DNA repair and activation of apoptosis, which we propose contributes to the synergistic increase in chromosomal aberrations in B cells lacking ATM and DNA-PKcs.
Upon DNA damage, the p53 tumor suppressor undergoes ATM- and Chk2-dependent phosphorylation, leading to increased transcription of target genes that promote apoptosis. It has been suggested that ATR may contribute to p53 mediated apoptosis because there is a partial defect in p53 stabilization in irradiated ATM and Chk2 deficient mice (Hirao et al., 2002; Takai et al., 2002); furthermore, there is a requirement for phosphorylation of SQ/TQ sites in Chk2 to induce ATM independent p53 activation (Hirao et al., 2002); and finally p53 activation in primary MEFs is co-dependent on ATM and ATR (Brown and Baltimore, 2003). In contrast, our results strongly suggest that ATM and DNA-PKcs collaborate to promote p53 activation in irradiated thymocytes and B cells undergoing CSR. This is consistent with the findings that ATR signaling is subject to cell cycle control, occurring primarily in S/G2 (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Yoo et al., 2007), whereas the vast majority of irradiation induced breaks in thymocytes, and physiological AID dependent breaks in B cells, are found in the G1 phase of the cell cycle (Petersen et al., 2001; Schrader et al., 2007). Moreover, we have found that even the residual IR induced ATR activation in thymocytes is co-dependent on ATM and DNA-PKcs, as evidenced by the abrogation of ATR dependent Chk1 phosphorylation in cells with compromised ATM and DNA-PKcs function (Figure S6B). A larger population of S phase cells in MEFs could dictate a larger role for ATR in the absence of ATM (Cimprich and Cortez, 2008).
If ATM and DNA-PKcs are both stimulated by the same lesion (a DSB), recruited with rapid kinetics, and have similar substrate specificity, why are CSR, chromosomal integrity, apoptosis, and substrate phosphorylation more severely compromised by the loss of ATM than of DNA-PKcs, and why are the phenotypes of ATM-/- and DNA-PKcs-/- mice so different? DSBs recruit and activate DNA-PKcs and also induce rapid ATM-dependent DNA-PKcs phosphorylation, which paradoxically contributes to the dissociation of DNA-PKcs from DNA ends (Chen et al., 2007; Merkle et al., 2002; Uematsu et al., 2007). As suggested by the defects in CSR, DNA-PKcs may play an important structural role in repair (Figure 1; Figure S4). Therefore, we propose that if a lesion fails to be repaired in a timely manner by a DNA-PKcs-dependent end-joining mechanism, ATM-mediated DNA-PKcs phosphorylation would induce dissociation of DNA-PKcs from DNA ends, while at the same time triggering cell cycle arrest, apoptosis, or delayed repair (Figure S9). In this model, DNA-PKcs would only maintain its presence at a DSB and efficiently phosphorylate DNA repair and checkpoint substrates in the absence of ATM. This switch from functioning as a structural component of the NHEJ pathway in ATM sufficient cells to a back-up signaling molecule in ATM deficient cells would explain why DNA-PKcs can have paradoxically opposite roles in the DNA damage response. For example, loss of DNA-PKcs promotes a p53-dependent checkpoint in ATM sufficient thymocytes (Guidos et al., 1996), but severely impairs p53-dependent apoptosis in ATM-/- thymocytes (Figure 6).
A non-exclusive explanation for why ATM might be the “primary” DSB kinase is that ATM signaling is associated with massive signal amplification. A small number of breaks are capable of activating most of the cellular pool of ATM (Bakkenist and Kastan, 2003), whereas full DNA-PKcs activation, measured by auto-phosphorylation, seems to require higher levels of DNA damage (Douglas et al., 2007; Uematsu et al., 2007). ATM amplification is promoted by interaction with Mre11/Rad50/Nbs1 on chromatin flanking the DSB site, whereas DNA-PKcs activation seems to be limited to the DNA end bound by Ku (You et al., 2007) (Figure S9). Taken together, the combined effect of ATM mediated rapid dissociation of DNA-PKcs from the DNA break as well as amplification of ATM signaling may explain why very few in vivo substrates for DNA-PKcs have hitherto been discovered. Consistent with both hypotheses, aberrant ATM recruitment and or amplification in Nijmegen breakage syndrome (NBS) cells and cells expressing a C-terminally truncated Nbs1 protein results in a greater dependence on DNA-PKcs-phosphorylation of H2AX (Falck et al., 2005) and Chk2 (Figure S10) relative to normal cells.
Ataxia telangiectasia (AT) is a pleotropic disease caused by dysfunction of the ATM kinase. Clinically, the most devastating feature of AT is loss of cerebellar neurons, which is believed to be due to defects in the DNA damage response. Neurons in ATM-/-mice are resistant to DNA damage induced apoptosis and there is little neuronal degeneration in mice, whereas other tissues, such as the intestines, are hypersensitive, and other cell types, such as lymphocytes, show intermediate radiation sensitivity (Herzog et al., 1998). We speculate that these variations in cellular responses to DNA damage may reflect tissue and species specific differences in the degree to which PIKK family members compensate for lack of ATM. This may reflect variations in the cell cycle status, and the abundance of DNA-PKcs in different tissues.
EXPERIMENTAL PROCEDURES
Mice
ATM-/- and 53BP1-/- mice were gifts from Anthony Wynshaw-Boris and Junjie Chen respectively. p53-/- mice were purchased from Taconic laboratories, and Nbs1657Δ5, Nbs1tr735, and DNA-PKcs-/-HL mice were generated as described (Difilippantonio et al., 2007; Manis et al., 2002; Zha et al., 2008). The ATMc/c allele was produced as described (Zha et al., 2008). The characterization of the ATMc/c mouse phenotype will be published elsewhere (Zha et al. in preparation).
B cell cultures, FISH detection of genomic instability and c-myc/IgH translocations
B cells were isolated, stimulated and assessed for CSR as described (Reina-San-Martin et al., 2003). For irradiation experiments, cells were mock or pre-treated with ATMi Ku55933 (5uM), PKi NU7026 (20uM) or PKi NU7441 (500nM) for 1 hour. For CSR and metaphases, cells were mock or pretreated with PKi NU7026 (20uM) for 72 hours. For FISH analysis, metaphases were hybridized with chromosome 12 painting, IgH Cα BAC, and telomere-repeat specific PNA probes as described (Callen et al., 2007). PCR and Southern blotting analysis of c-myc/IgH translocations were performed as described (Ramiro et al., 2004).
Western blotting
Western blot analysis was performed as described (McBride et al., 2006; Pellegrini et al., 2006). Primary antibodies were used at the following dilutions: anti-mouse Atm S1987p (1:400) (Pellegrini et al., 2006), anti-Smc1 S957p (1:400, Rockland Immunochemicals), anti-Smc1 (1:2,000, Novus), anti-p53 S15p (1:500, Cell Signaling), anti-p53 (1:2,000, Cell Signaling), anti-Chk1 317p (1:600, Bethyl Labs), anti-Chk2 (1:1,500, Upstate Biotechnology), anti-tubulin (1:15,000, Sigma), anti-γH2AX (1: 1,000 Upstate Biotechnology), anti-Kap-1 S824p (1:700, Bethyl Labs), anti-53BP1 S25p (0.2 μg/ml, Bethyl Laboratories) and anti-AID (1:1000) (McBride et al., 2006).
shRNA and viral infections
A series of small hairpin RNA vectors against DNAPKcs were selected from Open Biosystems, with the following sequences: Prkdc 59: GCGACATATTATGGAAGAATT; Prkdc 61:CCACCCAACAACAATATGATT; Prkdc 62: GCCATACAAATGTGGAATTAA. For experiments in MEFs, DNA fragments encoding ShRNA’s were cloned into the lentiviral vector pLKO.1. For experiments in B cells DNA fragments encoding shRNAs were cloned in LMP retroviral vector (Open Biosystems). For immortalized MEF infection, ShRNA lentiviruses were added to MEFs and 24 hours later cells were selected with puromycin. Primary MEFs were isolated from mice harboring conditionally deleted alleles for the kinase domain of ATM as described in (Zha et al., 2008). ATMc/+, ATMc/c and ATMc/c DNAPK-/-cells were directly infected with a high-titer adenovirus Cre-GFP (Gift from R. C. Mulligan, Harvard Medical School). Four days after infection cells were harvested and protein lysates were prepared. B cells were activated with LPS and IL4 were infected with retrovirus carrying DNA-PKcs specific or control shRNA as described (Ramiro et al., 2006) or with a retrovirus carrying Cre recombinase as described (Robbiani et al., 2008). GFP positive cells were sorted after 5 days, irradiated and phosphorylation was measured by western blot.
Quantitative RT-PCR
Switch region transcription was measured by RT-PCR as described (Reina-San-Martin et al., 2004). Thymocytes were pre-incubated with DNA-PKcs inhibitor for 1 hr, irradiated, and incubated in media for 4 hours before RNA was extracted and reverse transcribed using a Super ScriptIII cDNA synthesis kit (Invitrogen). Quantitative RT-PCR was performed using SYBR Green (Perkin Elmer) with the 7900HT Fast Real-time PCR system (Applied Biosystems). PCR conditions were 40 cycles of 95C for 30s followed by 1 min at 61C. All samples were run in triplicate. PUMA primers are as described in (Alvarez et al., 2006). P21 primers: F5’-AAGTGTGCCGTTGTCTCTTC-3’, R 5’-ACTTCAGGGTTTTCTCTTGC-3’ NOXA primers: F5’-GTTGAGCACACTCGTCTTCAA-3’, R5’-GTTGAGCACACTCGTCCTTCAA-3’ GAPDH primers: F5’-CCAGTATGACTCCACTCACG-3’ and R5’-GACTCCACGACATACTCAGC-3’. BAX primers are described in (Boley et al., 2002).
Supplementary Material
Acknowledgments
We thank Junjie Chen and Anthony Wynshaw-Boris, for mice; L. Stapelton for chromosome painting probes; G. Smith for small molecule inhibitors; Jeremy Daniel, Davide Robbiani, Oscar Fernandez-Capetillo, and Yair Dorsett for comments on the manuscript, Rafael Casellas for advice on ShRNA. A.N. is supported by the Intramural Research program of the NIH, National Cancer Institute, Center for Cancer Research. F.W.A. is supported by grants from NIH. F. W. A. is a Howard Hughes Institute Investigator M.C.N. is supported by grants from NIH. M.C.N. is a Howard Hughes Institute Investigator.
Footnotes
The authors declare no financial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez S, Drane P, Meiller A, Bras M, Deguin-Chambon V, Bouvard V, May E. A comprehensive study of p53 transcriptional activity in thymus and spleen of gamma irradiated mouse: high sensitivity of genes involved in the two main apoptotic pathways. International journal of radiation biology. 2006;82:761–770. doi: 10.1080/09553000600949624. [DOI] [PubMed] [Google Scholar]
- Arlander SJ, Greene BT, Innes CL, Paules RS. DNA protein kinase-dependent G2 checkpoint revealed following knockdown of ataxia-telangiectasia mutated in human mammary epithelial cells. Cancer research. 2008;68:89–97. doi: 10.1158/0008-5472.CAN-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Barreto VM, Ramiro AR, Nussenzweig MC. Activation-induced deaminase: controversies and open questions. Trends in immunology. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Boley SE, Wong VA, French JE, Recio L. p53 heterozygosity alters the mRNA expression of p53 target genes in the bone marrow in response to inhaled benzene. Toxicol Sci. 2002;66:209–215. doi: 10.1093/toxsci/66.2.209. [DOI] [PubMed] [Google Scholar]
- Bosma GC, Kim J, Urich T, Fath DM, Cotticelli MG, Ruetsch NR, Radic MZ, Bosma MJ. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. The Journal of experimental medicine. 2002;196:1483–1495. doi: 10.1084/jem.20001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes & development. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Kurimasa A, Xie G, Taya Y, Araki R, Abe M, Crissman HA, Ouyang H, Li GC, Chen DJ. DNA-dependent protein kinase-independent activation of p53 in response to DNA damage. The Journal of biological chemistry. 1999;274:17139–17143. doi: 10.1074/jbc.274.24.17139. [DOI] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science (New York, NY) 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Advances in immunology. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. The Journal of biological chemistry. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AJ, Oganesian L, Harumal P, Basten A, Brink R, Jolly CJ. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J Immunol. 2003;171:6556–6564. doi: 10.4049/jimmunol.171.12.6556. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Durkacz BW, Tilby MJ. Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochemical pharmacology. 2005;71:13–20. doi: 10.1016/j.bcp.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. The Journal of experimental medicine. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. The EMBO journal. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annual review of biochemistry. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. The Journal of experimental medicine. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Kruhlak MJ, Lee Y, Difilippantonio MJ, Feigenbaum L, Jackson SP, McKinnon PJ, Nussenzweig A. Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis. The Journal of experimental medicine. 2007;204:1003–1011. doi: 10.1084/jem.20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Molecular and cellular biology. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Advances in immunology. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Franco S, Murphy MM, Li G, Borjeson T, Boboila C, Alt FW. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. The Journal of experimental medicine. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MT, Danska JS. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes & development. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- Gurley KE, Kemp CJ. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr Biol. 2001;11:191–194. doi: 10.1016/s0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science (New York, NY) 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Molecular and cellular biology. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nature immunology. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Nussenzweig A, Nussenzweig MC. Antigen receptor diversification and chromosome translocations. Nature immunology. 2007;8:801–808. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM-and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nature cell biology. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Yusufzai TM, Anderson S, Anver MR, Merlino G. The p53 response to DNA damage in vivo is independent of DNA-dependent protein kinase. Molecular and cellular biology. 2000;20:4075–4083. doi: 10.1128/mcb.20.11.4075-4083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez GS, Bryntesson F, Torres-Arzayus MI, Priestley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo PA, Taccioli GE, et al. DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature. 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- Kiefer K, Oshinsky J, Kim J, Nakajima PB, Bosma GC, Bosma MJ. The catalytic subunit of DNA-protein kinase (DNA-PKcs) is not required for Ig class-switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2843–2848. doi: 10.1073/pnas.0611359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes & development. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracker S, Bergmann Y, Demuth I, Frappart PO, Hildebrand G, Christine R, Wang ZQ, Sperling K, Digweed M, Radbruch A. Nibrin functions in Ig class-switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Molecular and cellular biology. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorganic & medicinal chemistry letters. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science (New York, NY) 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Li Z, Luo Z, Ronai D, Kuang FL, Peled JU, Iglesias-Ussel MD, Scharff MD. Targeting AID to the Ig genes. Advances in experimental medicine and biology. 2007;596:93–109. doi: 10.1007/0-387-46530-8_9. [DOI] [PubMed] [Google Scholar]
- Liu X, Matsuda A, Plunkett W. Ataxia-telangiectasia and Rad3-related and DNA-dependent protein kinase cooperate in G2 checkpoint activation by the DNA strand-breaking nucleoside analogue 2’-C-cyano-2’-deoxy-1- -D-arabino-pentofuranosylcytosine. Molecular cancer therapeutics. 2008;7:133–142. doi: 10.1158/1535-7163.MCT-07-0416. [DOI] [PubMed] [Google Scholar]
- Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC, 3rd, Gearhart PJ, Wynshaw-Boris A, Max EE, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. The Journal of experimental medicine. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Dudley D, Kaylor L, Alt FW. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nature immunology. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science (New York, NY) 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. The Journal of biological chemistry. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. The Journal of biological chemistry. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443:222–225. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature immunology. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Rathmell WK, Kaufmann WK, Hurt JC, Byrd LL, Chu G. DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer research. 1997;57:68–74. [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. The Journal of experimental medicine. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/-B cells. European journal of immunology. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. The Journal of experimental medicine. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Molecular cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Rieina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland D, Chen HT, Corcoran A, Nussenzweig A, et al. Activation Induced Deaminase is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008 doi: 10.1016/j.cell.2008.09.062. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science (New York, NY) 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, et al. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature reviews. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & development. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and Regulation of Class Switch Recombination. Annual review of immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer research. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, Anderson CW, Appella E, Nakanishi M, Suzuki H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. The EMBO journal. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. The Journal of cell biology. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer research. 2003;63:6008–6015. [PubMed] [Google Scholar]
- Wang H, Guan J, Wang H, Perrault AR, Wang Y, Iliakis G. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer research. 2001;61:8554–8563. [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. The Journal of cell biology. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. The Journal of biological chemistry. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nature cell biology. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9302–9306. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM-and KAP-1 dependent pathway. Nature cell biology. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science (New York, NY) 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.