Summary
Genetic analysis of the drosophila antiviral response indicates that RNA interference plays a major role. This contrasts with the situation in mammals, where interferon-induced responses mediate innate antiviral host-defense. An inducible response also contributes to antiviral immunity in drosophila, and similarities in the sensing and signaling of viral infection are becoming apparent between drosophila and mammals. In particular, DExD/H box helicases appear to play a crucial role in the sensing of RNA virus infections in flies and mammals.
Introduction
Viruses are the most abundant infectious agents on earth, and the diseases caused by these intracellular pathogens represent a constant threat and an important cause of mortality worldwide. Because they replicate intracellularly, and evolve rapidly to adapt to the changing environment of their host cells, viruses are a great challenge to their hosts. Although the fruit-fly Drosophila melanogaster, a long-time favourite model of geneticists and developmental biologists, emerged in the 1990s as an interesting model to investigate innate immunity, the issue of antiviral immunity only started to be addressed in the past five years. Several RNA viruses belonging to different families (Rhabdoviridae, Dicistroviridae, Birnaviridae, Reoviridae) have been identified in drosophila and provide useful models to study antiviral immunity[1]. Based on the findings obtained for antibacterial and antifungal immunity[2], it is hoped that the study of antiviral resistance in drosophila may reveal novel evolutionary conserved molecules or pathways, leading to a better understanding of innate antiviral immunity in mammals. Another rational for investigating host-virus interactions in invertebrate hosts is that some of them, like the mosquitoes Aedes, are vectors for viruses that have major impacts on humans (e.g. Dengue).
RNA interference as an intrinsic defense against viral infection
RNAi was first identified as a potent antiviral defense mechanism in plants[3]. More recently, RNAi was also found to play an important role in the control of viral infection in drosophila. Central to the RNAi mechanism are the slicing enzymes of the Argonaute family (five members in drosophila), which mediate highly specific cleavage of target RNA molecules. The specificity of Argonaute enzymes is achieved by their association with small RNAs, which guide them to complementary sequences. Three RNAi pathways, involving different members of the Argonaute family, have been defined in drosophila: (i) the small interfering (si)RNA pathway involves Argonaute (AGO)-2, and is activated by double stranded (ds)RNA. siRNAs are produced by the RNaseIII enzyme Dicer-2, which forms a complex with the dsRNA binding protein (dsRBP) protein R2D2[3] (Figure 1); (ii) the micro (mi)RNA pathway involves AGO-1, Dicer-1 and its dsRBD co-factor R3D1, and regulates expression of drosophila genes, in particular during development; (iii) Finally, the piRNA pathway involves the three other AGO proteins encoded by the drosophila genome, namely Piwi, Aubergine and AGO3. piRNAs (Piwi-associated RNAs) are involved in the control of mobile genetic elements, including the retrovirus gypsy, in the germ-line[4]. Demonstration of the critical role of RNAi as a potent antiviral mechanism in drosophila is based on three lines of evidence: first, genetic data indicating that RNAi pathway mutants are hypersensitive to RNA virus infections, and contain increased viral load; second, identification of viral suppressors of RNAi (VSRs), which counteract the immune defense of the fly; and third, presence of siRNAs of viral origin in infected cells/flies. As discussed below, there is good experimental support in drosophila for all these lines of evidence.
Figure 1.
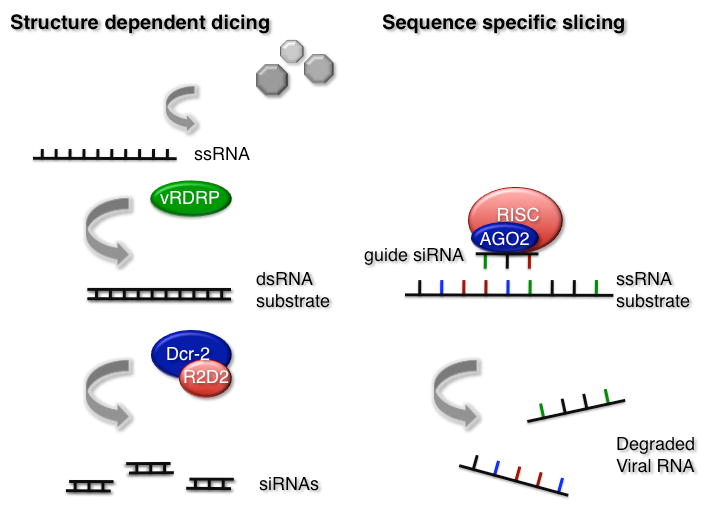
Degradation of viral nucleic acids by RNA interference. Dicer-2 senses double stranded (ds)RNA produced by the viral RNA-dependent RNA-polymerase (vRDRP) during replication of the virus. Following recognition, the RNaseIII enzyme Dicer-2, which is found in a complex with the dsRNA binding protein R2D2, processes dsRNA into duplex siRNAs (left panel). The guide strand of these siRNAs is then incorporated in the RNA induced silencing complex (RISC), while the passenger strand is degraded (not shown on the figure). The loaded RISC complex mediates sequence specific slicing of viral ssRNA by its core component AGO2 (right panel).
Analysis of the role of the siRNA pathway was greatly facilitated by the fact that flies mutant for Dicer-2, AGO2 or R2D2 are viable[5-7]. These mutant flies succumb more rapidly than wild-type controls to infection with the RNA viruses Drosophila C virus (DCV), Cricket paralysis virus (CrPV), Flock house virus (FHV) or Sindbis virus (SINV)[8-10]. Increased lethality correlates with increased viral titers in infected flies. Because DCV and CrPV (Dicistroviridae), FHV (Nodaviridae) and SINV (Alphaviridae) belong to different families of viruses, these data indicate that the siRNA pathway mediates a broad antiviral defense in flies. Interestingly, resistance to the virus Drosophila X virus (DXV; Birnaviridae) and West-Nile virus (WNV; Flaviviridae) seems to involve AGO2, but not Dicer-2[11,12]. In addition, the piRNA pathway appears to mediate resistance to these two viruses, since mutants for Piwi, Aubergine or the helicase Spindle E are more sensitive to DXV and/or WNV. Hence, antiviral RNAi may involve more than one pathway.
Viral suppressors of RNA interference
In agreement with the idea that RNAi exerts an evolutionary pressure on viruses, VSRs have been identified in insect RNA viruses[9,10,13]. Two of these, B2 from FHV and 1A from DCV interact with dsRNA, thus preventing recognition and cleavage by Dicer-2, whereas a third one (CrPV-A) does not appear to bind dsRNA. DCV-1A contains a bona fide dsRNA binding domain[9], whereas FHV-B2 contains an original fold and binds the side of dsRNA as a four α-helices bundle[14]. Both proteins bind to dsRNA with high (nanomolar) affinity, and point mutations affecting interaction with dsRNA abolish their VSR activity. Unlike DCV-1A, which only binds long dsRNAs, FHV-B2 can interact with both long dsRNA and siRNAs. Thus, FHV-B2 has the potential to interfere with both steps of RNAi, whereas DCV-1A only blocks dicing of long dsRNAs.
Cricket paralysis virus (CrPV) belongs to the same family as DCV (Dicistroviridae), and the two viruses share extensive sequence similarity. Surprisingly, they diverge in the 5′ end of their RNA genome, which encodes the VSR in both viruses[9,10]. The RNAi suppressor CrPV-A may inhibit RNA silencing by targeting protein components of the RNAi pathway, as shown for some plant viruses VSRs (reviewed in [15]). Interestingly, the existence of VSRs in insect viruses targeting key players of the siRNA pathway is indirectly supported by the rapid evolution of these host genes. Indeed, dicer-2, Ago2 and r2d2 are among the 3% fastest evolving genes in drosophila, in sharp contrast with their counterparts of the miRNA pathway dicer-1, Ago1 and r3d1[16]. Such rapid evolution may be driven by the pressure exerted by viruses, through their VSRs, on these essential host-defense genes. Thus, the identification and characterization of VSRs from insect viruses represent a promising avenue of research to dissect the antiviral RNA interference pathway.
Small RNA-mediated antiviral immunity
RNA interference provides a highly specific mechanism to degrade viral nucleic acids in cells. The mechanism involves two tightly connected steps: first, viral dsRNA is recognized by Dicer-2, which processes it into siRNAs. Second, these siRNAs are incorporated in the RNA-induced silencing complex (RISC), and guide the slicing enzyme AGO2 towards complementary sequences, which will be cleaved (Figure 1). In support of this scheme, flies carrying a transgene directing expression of FHV dsRNA are protected against a challenge by FHV, but not by DCV[8].
In good agreement with genetic data, siRNAs of viral origin can be detected in infected cells or flies[8,10,13,17]. In the case of FHV, these siRNAs are more abundant in the absence of the VSR B2, and coimmunoprecipitate with AGO2, rather than AGO1[17,18]. Furthermore, viral siRNAs cannot be detected in dicer-2 mutant embryos injected with an FHV replicon. Profiling of small RNAs produced in FHV infected cells (in the presence and absence of B2) has recently been performed using pyrosequencing, and provided useful information on the mechanism of antiviral RNAi[17-19]. Three major observations were made: (i) roughly equal amounts of (+) and (-) polarity siRNAs are observed, indicating that dsRNA generated during viral replication is used as a substrate by Dicer-2. Indeed, a large excess of siRNAs of (+) polarity would be observed if Dicer-2 was using secondary RNA structures as a substrate, because FHV-infected cells produce about 100-fold excess of RNA of (+) polarity compared to (-) polarity[20]. (ii) The siRNAs cover the whole bipartite FHV RNA genome, but there are hotspots at the extremities of both viral RNAs, suggesting that dicing occurs during the early steps of viral replication. (iii) In the absence of the VSR B2, a spectacular increase of siRNAs corresponding to the 5′ end of RNA1 (and to a lesser extent RNA2) is observed[17,19]. This confirms that the dsRNA replicative intermediates formed at the extremities of viral RNAs are major targets for Dicer-2, and suggests that these 5′ terminal siRNAs play a critical role in the abortive infection of drosophila cells by FHV when B2 is not functional. Interestingly, B2 is associated with the viral polymerase in infected cells, which may explain how it can suppress production of siRNAs from dsRNA in close proximity to or within the RNA replication complex[17] (Figure 2).
Figure 2.
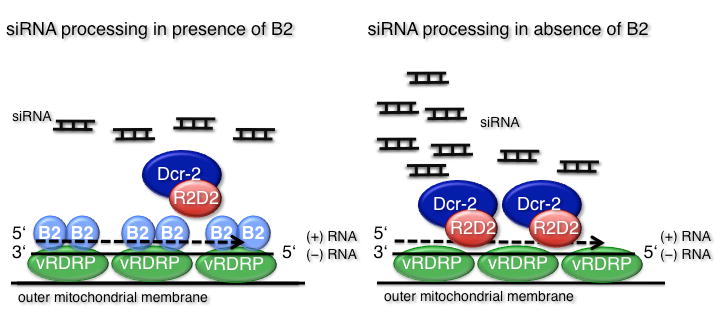
A model for the Dicer-2 mediated control of FHV infection. The viral suppressor of RNAi B2 is associated with the viral RDRP, which is anchored in the outer mitochondrial membrane. B2 binds to dsRNA replicative intermediates as they are synthesized, thus protecting them from the action of Dicer-2 (left panel). When B2 is mutated, Dicer-2 binds the nascent dsRNA replication intermediates and generates massive amounts of siRNAs corresponding to the 5′ end of the genome, leading to abortive replication and protection of the infected cells (right panel).
It will be interesting in future siRNA profiling studies to see if the conclusions reached in the case of FHV hold true for other viruses. Indeed, studies in plants indicate that dsRNA or secondary RNA structures can be targeted by the RNAi pathway, depending on the virus (reviewed in [3,15]). Of note, one region of RNA2 known to form a bulged stem loop can be processed by Dicer-2, since an excess of (+) strand siRNAs targeting this region of the viral genome is recovered from infected cells[19,21].
Inducible antiviral defense in flies
In addition to RNA interference, genome-wide microarray analysis of DCV-infected flies revealed that viral infection triggers expression of some 150 genes. There is little overlap between these genes and those induced by bacteria and fungi, indicating that flies respond differently to infection by DCV and extracellular pathogens[22-24]. Other viruses also induce a transcriptional response in drosophila [25,26].
Analysis of the promoter of the gene vir-1, which is strongly induced by DCV and FHV infection (but not bacteria or fungi), revealed that the virus response element coincides with a consensus DNA-binding site for the transcription factor D-STAT. Genetic analysis confirmed that the drosophila JAK kinase encoded by the gene hopscotch and the cytokine receptor Domeless (an homologue of the gp130 subunit of the interleukin-6 receptor) are required for induction of vir-1 and several other genes. Flies mutant for the gene hopscotch succumb rapidly to DCV infection, with high viral titers, indicating that the JAK/STAT pathway contributes to the control of the viral infection[22]. Altogether, these data indicate that DCV infection triggers production of a cytokine in infected cells, leading to activation of the JAK/STAT pathway in non-infected cells, and induction of an antiviral state[27]. An important question is now to understand the function of the induced genes, and to establish if antiviral effector mechanisms other than RNAi exist in drosophila.
Flies mutant for the DCV-induced gene Vago were recently shown to contain higher levels of virus than wild-type controls[28]. Vago therefore represents a good candidate for an antiviral molecule induced following viral infection. This 18kDa molecule contains a signal peptide, and 8 cysteine residues forming a conserved CX20CX4CX10-11CX7-9CX13-14CCX4C motif. A similar motif is found in granularin, an inducible peptide from the snail Biomphalaria glabrata, acting as an opsonin for the schistosoma parasite[29]. However, the mode of action of Vago is still unknown. Among the other induced molecules[22], most of which have no known functions, some possess interesting sequence characteristics and warrant further investigation, including a caspase (Damm), a GNBP-like molecule devoid of a signal peptide (CG12780; GNBPs are secreted proteins involved in the sensing of Gram-positive and fungal infections in drosophila[30]); or the gene CG1667, encoding an orthologue of the factor MITA/STING, which was recently shown to participate in antiviral innate immunity signaling in mammals[31,32].
Even though much remains to be learned about the function of the induced genes, it is clear at this stage that recognition of viral infection in drosophila cells leads to an inducible response and the production of antiviral molecules and cytokines. This raises the question of the nature of the viral molecular pattern recognized by the drosophila innate immune system, and of the receptor sensing this pattern.
Sensing viral infection by the DExD/H helicase Dicer-2 in drosophila
The gene Vago provides an interesting tool to investigate the sensing mechanism of the inducible antiviral response. Indeed, it is not regulated by the cytokine-activated JAK/STAT pathway[22], and is induced in cells from the fat-body, which is a major target for DCV replication. Vago is also induced by SINV but, surprisingly, not by a third RNA virus, FHV[28]. This difference can be attributed to the potent VSR B2, which suppresses induction of Vago. Because B2 was shown to interact with dsRNA, but not with single-stranded RNA, or DNA[14], this result strongly suggests that dsRNA is a viral molecular pattern recognized by the drosophila innate immune system. In agreement with the fact that B2 suppresses Dicer-2 function, expression of Vago is considerably impaired in dicer-2 mutant flies, establishing a connection between RNA interference and the inducible antiviral response[28]. Importantly however, Vago remains fully inducible in r2d2- and AGO2-mutant flies. This indicates that Dicer-2 plays a dual function in antiviral defenses in flies, acting on one hand as a critical component of the RNAi machinery, and on the other hand inducing expression of an antiviral program upon sensing viral dsRNAs (Figure 3). Dicer-2 contains an amino-terminal DExD/H box helicase domain, which is important for the inducible expression of Vago. Interestingly, this domain resembles and is phylogenetically related to the helicase domain of the mammalian RIG-I-like receptor (RLR) family[28], which mediate induction of interferon gene expression[27,33].
Figure 3.
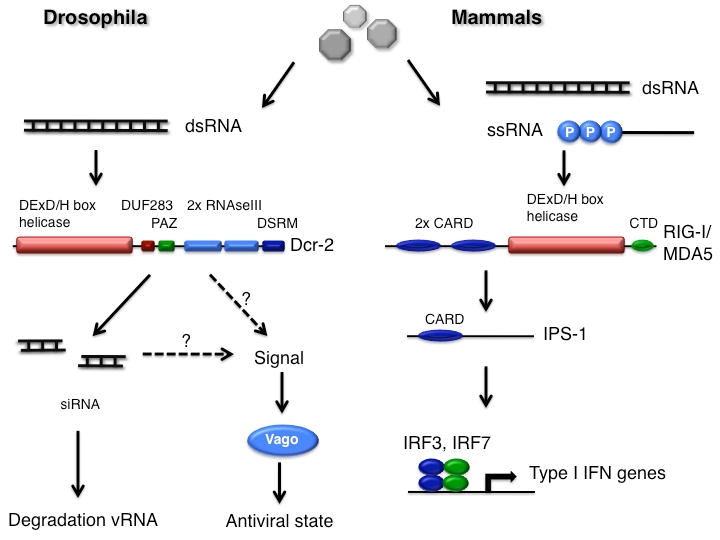
Sensing of viral nucleic acids by DExD/H box helicases in drosophila and mammals. dsRNA is detected by Dicer-2 and RLRs (RIG-I and MDA5) in flies and mammals, respectively. These molecules contain phylogenetically related DExD/H box helicase domains, which mediate a conformational change in response to interaction with viral RNAs (dsRNA or single stranded RNA with a 5′ triphosphate extremity in the case of RIG-I). In drosophila (left panel), Dicer-2 processes dsRNA into siRNAs, and triggers an unidentified signal that leads to the inducible expression of the gene Vago, which controls the viral load in the fat body of infected flies. In mammals (right panel), RLRs activate the signal transducer IPS-1 and transcription factors of the IRF family, leading to production of interferons.
Thus, a common family of DExD/H box helicases, composed of Dicer-like enzymes and RLRs, plays a critical role in the sensing of viral nucleic acids by the innate immune system in insects and vertebrate animals (Figure 3). Both for Dicers and RLRs, the exact role of the DExD/H helicase domain, which appears to function as an auto-inhibitory domain, remains mysterious[33,34]. The helicase domain does not seem to be directly involved in the sensing of dsRNA, but rather mediates a conformational change of the molecule. In the case of Dicer, this change promotes the transfer of the guide strand of the siRNA to the RISC complex, whereas in the case of RIG-I it exposes the signaling CARD domains[33,35]. In RIG-I, the helicase-mediated conformational change is induced upon binding of 5′-triphosphate RNA to a ∼130 amino-acids regulatory domain immediately following the helicase domain[36,37]. It is puzzling to note that in Dicer enzymes, the helicase domain is followed by a ∼80 amino-acids domain of unknown function, DUF283, which appears to be a divergent dsRNA binding domain[38]. Thus, the association of an RNA-binding domain on the C-terminal side of the DExD/H helicase domain may be important for the induction of the helicase activity by viral nucleic acids.
Apart from the helicase domain, Dicer-2 also contains two RNaseIII domains[5]. It will be interesting to test the importance of these domains in the induction of Vago. Indeed, recent work from S. Akira and colleagues show that RLRs can discriminate between dsRNAs of different sizes, and that a long dsRNA activating MDA5 can be processed into a shorter dsRNA (≤ 1kb) activating RIG-I upon in vitro incubation with Escherichia coli RNaseIII (or recombinant human Dicer)[39]. The recent identification of mutant alleles affecting the catalytic site of the RNaseIII domains of Dicer-2 now makes it possible to rapidly test the role of this domain in the drosophila inducible antiviral response[40].
Conclusions and future directions
In summary, even though antiviral defenses in drosophila and mammals rely on different mechanisms (RNAi versus interferon-induced responses), similarities exist both for signaling (involvement of cytokines regulating the JAK/STAT pathway) and sensing of nucleic acids (DExD/H box helicases). Based on findings in other invertebrates, plants, and even prokaryotes, RNAi is an ancestral response to viral infections[3,41,42]. The reason why it was lost in vertebrates and replaced by the inducible interferon system is not known, but it makes evolutionary sense to find a family of DExD/H box helicases related to Dicer at the origin of the interferon pathway. Importantly, the fact that vir-1 remains fully inducible by DCV in dicer-2 mutant and in B2-expressing flies[28] indicates that other sensors participate in the detection of viral infection in drosophila.
In spite of the progress made recently, important questions remain. These include the in vivo contribution of virus-induced stress (e.g. apoptosis[43]) and altered physiology (e.g. role of KATP channels in the heart[44]) in the resistance to viral infection, or the molecular characterization of the protective antiviral effect conferred by Wolbachia endosymbionts[45,46]. Models of natural viral infection (e. g. per os[47]) also need to be developed to confirm/extend the findings made with the injection model, and set the stage for future studies with insect-borne viruses in mosquitoes[48]. Another fascinating issue, both in flies and mammals, relates to the cell biology of the interaction between DExD/H box helicases and viral nucleic acids, namely, how the cellular sensors access to viral replication complexes, which are secluded into modified cytoplasmic membranes[20,49]. Finally, an important question pertains to the priming of uninfected tissues for a rapid and efficient antiviral response. In plants and the nematode C. elegans, this is achieved by the amplification and spread of antiviral silencing, mediated by cellular RNA-dependent RNA polymerases (reviewed in[3]). These enzymes are not encoded by the drosophila genome, and RNAi is thought to be a cell-autonomous process, suggesting that cytokines mediate amplification of the antiviral response[22,50]. However, the fact that drosophila cells can specifically bind and internalize dsRNA leaves open the possibility that viral replication intermediates released from infected cells may prime RNAi in uninfected cells[51,52].
Acknowledgments
We thank members of the group and Jules Hoffmann for stimulating discussions, and Safia Deddouche and Steffi Müller for critical reading of the manuscript. This work was supported by NIH (PO1 AI070167), ANR (Microbiologie-Maladies émergentes) and CNRS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huszar T, Imler JL. Drosophila viruses and the study of antiviral host-defense. Advances in Virus Research. 2009;72 doi: 10.1016/S0065-3527(08)00406-5. in press. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann J. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- •• 3.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; A very thorough review on RNAi as an antiviral defense in plants and animals
- 4.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 7.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 9.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chotkowski HL, Ciota AT, Jia Y, Puig-Basagoiti F, Kramer LD, Shi PY, Glaser RL. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology. 2008;377:197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 14.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005 doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 15.Mlotshwa S, Pruss GJ, Vance V. Small RNAs in viral infection and host defense. Trends Plant Sci. 2008;13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- • 16.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]; This article demonstrates that genes of the siRNA pathway are under selective pressure and evolve rapidly.
- •• 17.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first in-depth profiling study of viral siRNAs produced in an animal model.
- 18.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 19.van Rij RP, Berezikov E. Small RNAs and the control of transposons and viruses in drosophila. Trends in Microbiology. 2009 doi: 10.1016/j.tim.2009.01.003. in press. [DOI] [PubMed] [Google Scholar]; A very good review on the control of foreign nucleic acids by RNAi, together with an analysis of the profile of virus-derived siRNAs identified in ref. 18.
- •• 20.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]; An elegant and detailed analysis of the cell biology aspects of FHV replication.
- 21.Zhong W, Dasgupta R, Rueckert R. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc Natl Acad Sci U S A. 1992;89:11146–11150. doi: 10.1073/pnas.89.23.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 23.Hedges LM, Johnson KN. Induction of host defence responses by Drosophila C virus. J Gen Virol. 2008;89:1497–1501. doi: 10.1099/vir.0.83684-0. [DOI] [PubMed] [Google Scholar]
- 24.Sabatier L, Jouanguy E, Dostert C, Zachary D, Dimarcq JL, Bulet P, Imler JL. Pherokine-2 and -3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur J Biochem. 2003;270:3398–3407. doi: 10.1046/j.1432-1033.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsai CW, McGraw EA, Ammar ED, Dietzgen RG, Hogenhout SA. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl Environ Microbiol. 2008;74:3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira S. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- •• 28.Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]; This article demonstrates that Dicer-2 is involved in the induction of an antiviral response in drosophila, and that its DExD/H box helicase is phylogenetically related to that of mammalian RIG-I-like receptors.
- 29.Smit AB, De Jong-Brink M, Li KW, Sassen MM, Spijker S, Van Elk R, Buijs S, Van Minnen J, Van Kesteren RE. Granularin, a novel molluscan opsonin comprising a single vWF type C domain is up-regulated during parasitation. Faseb J. 2004;18:845–847. doi: 10.1096/fj.03-0590fje. [DOI] [PubMed] [Google Scholar]
- 30.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;456:274. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- • 34.Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, Collingwood MA, Behlke MA, Rossi JJ. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 34-37 indicate that the DExD/H box helicase domain in Dicer and RLRs mediates a conformzational change leading to activation of effector molecules.
- • 35.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 36.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- • 37.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Dlakic M. DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold. Bioinformatics. 2006;22:2711–2714. doi: 10.1093/bioinformatics/btl468. [DOI] [PubMed] [Google Scholar]
- •• 39.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demontrates that a long dsRNA activating MDA5 can be processed by RNaseIII into a shorter dsRNA activating RIG-I.
- 40.Lim do H, Kim J, Kim S, Carthew RW, Lee YS. Functional analysis of dicer-2 missense mutations in the siRNA pathway of Drosophila. Biochem Biophys Res Commun. 2008;371:525–530. doi: 10.1016/j.bbrc.2008.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 42.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Settles EW, Friesen PD. Flock house virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J Virol. 2008;82:1378–1388. doi: 10.1128/JVI.01941-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Croker B, Crozat K, Berger M, Xia Y, Sovath S, Schaffer L, Eleftherianos I, Imler JL, Beutler B. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nat Genet. 2007;39:1453–1460. doi: 10.1038/ng.2007.25. [DOI] [PubMed] [Google Scholar]
- • 45.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]; Ref. 45-46 indicate that the presence of the bacterial symbiont Wolbachia in drosophila stocks can have a dramatic effect on the susceptibility to viral infection.
- • 46.Teixeira L, Ferreira A, Asburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2009 doi: 10.1371/journal.pbio.1000002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roxstrom-Lindquist K, Terenius O, Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 2004;5:207–212. doi: 10.1038/sj.embor.7400073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. Rna. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]


