Abstract
To estimate the in vivo fitness cost of enfuvirtide resistance, we analyzed dynamic shifts in the HIV-1 quasispecies under changing selective pressure in three subjects on failing enfuvirtide-based regimens who interrupted enfuvirtide while maintaining stable background regimens. Subsequently, enfuvirtide was re-administered for four weeks as “pulse intensification”. The proportion plasma virus carrying the V38A mutation in gp41 was quantified by allele-specific real-time PCR in serial samples collected from three subjects at 1–4 week intervals. Fitness differences were calculated using a method that corrected for time-dependence of the viral replication rate. The V38A mutant made up ≥85% of the quasispecies at baseline and decayed to <5% over 12–24 weeks; plasma HIV-1 RNA levels remained stable during this time. Fitness differences for mutant versus wild type ranged from −25% to −65%, providing in vivo evidence for the reduced fitness of enfuvirtide-resistant HIV-1. The V38A mutant virus re-emerged rapidly during the enfuvirtide pulse. These results demonstrate that the HIV-1 quasispecies undergoes dynamic changes in response to withdrawal and re-initiation of fusion inhibitor therapy. The relative stability of plasma HIV-1 titers during decay of V38A suggests that factors other than viral fitness likely define viral load set-point in patients with advanced disease.
Keywords: HIV-1, enfuvirtide, treatment interruption, viral dynamics, viral fitness, allele-specific PCR
INTRODUCTION
The fusion inhibitor enfuvirtide is a potent antiretroviral drug that is most effective when used in combination with two or more agents to which the virus remains at least partially susceptible [1,2]. In the setting of partial viral suppression enfuvirtide resistance develops rapidly due to emergence of mutations in the first heptad repeat (HR-1) of the gp41 ectodomain [3,4]. The most commonly identified enfuvirtide resistance mutations include G36D or S, V38A, M or E, Q40H and N43D [5].
In vitro studies using growth competition assays have shown that enfuvirtide-resistant viruses are substantially less fit than wild type in the absence of drug [6]. Our group conducted a prospective evaluation of the virologic and immunologic consequences of enfuvirtide interruption in subjects with incomplete viral suppression on an enfuvirtide-based regimen [7]. Subjects discontinued enfuvirtide while remaining on a stable background regimen. Enfuvirtide resistance waned over time in most subjects, suggesting that enfuvirtide-resistance mutations reduce fitness of HIV-1 (as assessed in absence of drug). To determine more precisely the impact of these mutations on viral fitness in vivo, we developed a sensitive allele-specific real-time PCR assay to quantify the relative proportion of mutant HIV-1 over time. This assay allowed us to study the dynamics of enfuvirtide-resistant and wild-type virus in three subjects under changing selective pressures as subjects discontinued and subsequently resumed enfuvirtide.
METHODS
Subjects and plasma samples
Subjects were antiretroviral treatment-experienced HIV-1-infected patients enrolled in an ongoing prospective cohort study (SCOPE) [8]. This particular substudy evaluated the consequences of interrupting enfuvirtide while continuing to receive other drugs in the regimen (partial treatment interruption [PTI]). All subjects provided written informed consent, and all aspects of this study were conducted according to institutional guidelines for experiments with human subjects. Plasma samples were obtained at baseline (prior to enfuvirtide interruption) and then weekly for the first two months and every other week for the subsequent two years after enfuvirtide interruption. Several months after the enfuvirtide interruption a subset of subjects underwent pulse-intensification of their antiretroviral regimen during which enfuvirtide was re-administered for a four-week period (the “background” regimen was not modified during this time). The purpose of this pulse intensification was to determine whether a brief resumption of enfuvirtide would have beneficial effects on CD4 count stability by transiently lowering plasma HIV-1 RNA levels, or by reselecting potentially less pathogenic enfuvirtide-resistant viruses [9,10]. Plasma samples were obtained weekly during and after the pulse therapy phase. Samples were stored as 1-ml aliquots at −80° C prior to analysis. Plasma HIV-1 RNA levels were determined using a polymerase chain reaction assay (Amplicor HIV Monitor version 1.5; Roche Molecular Systems, Branchburg, NJ; lower limit of quantification 50 copies RNA/mL).
Cloning and sequencing of HIV-1 gp41
Viral RNA was extracted from plasma using the QIAamp viral RNA Kit (Qiagen, Valencia, Calif.). A 650-bp fragment of gp41 that includes the HR-1 and HR-2 coding region was amplified by a nested reverse transcriptase-coupled PCR (RT-PCR), cloned, and sequenced as described [5].
Quantification of viral populations using real-time PCR
Viral RNA was extracted from plasma samples as above and subjected to RT-PCR in order to obtain amplified cDNA products of the gp41-coding region of env. Reverse transcription was performed using a one-step process as described [11]. To quantify the proportion of mutant sequences contained within each specimen, 5 µl of the RT-PCR products were added to PCR reactions that permitted nonselective amplification of HR-1-coding sequences or selective amplification of sequences containing the V38A mutation. The limit of detection, or sensitivity of the mutant selective primer was 0.80% [11].
Calculation of relative fitness
The relative fitness advantage or disadvantage was determined based on the growth corrected method as described [12]. (Fitness differences calculated without growth correction gave qualitatively similar results, but because the growth corrected model gave the best fit, we show only those data.) We set the death rate of infected cells, δ, equal to = 0.5 ± 0.1 as this value represents approximately the mean of many independent estimates [13]. Furthermore, we assumed a coefficient of variation of 30% for the virus load measurements [14].
RESULTS
Decay of V38A mutant populations during enfuvirtide interruption
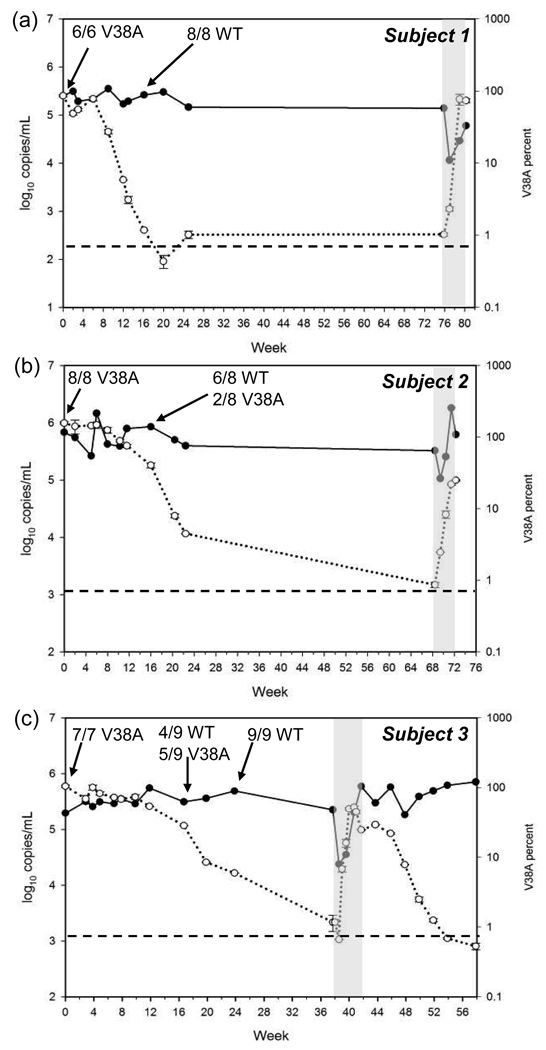
Samples from three enfuvirtide-treated subjects in whom the gp41 V38A mutation was present as the predominant population in the quasispecies (based on clonal sequence analysis) were selected for analysis by allele-specific real-time PCR (Table 1). The duration of enfuvirtide therapy in these subjects prior to the partial treatment interruption ranged from 27 to 39 weeks. The V38A mutation was present in 85% or more of the viral quasispecies at the start of the enfuvirtide interruption in all three subjects. After an initial lag of 6 to 12 weeks during which the viral population appeared to be unchanged the V38A mutant population decayed at variable rates (Fig. 1). There was little or no change in plasma HIV-1 RNA levels during this period. The time to near-complete replacement of mutant virus by wild type virus appeared to correlate with the duration of ENF treatment prior to the PTI: in subject 1, who received enfuvirtide for 27 weeks, the V38A mutants declined to less than 5% of the viral population by 12 weeks after PTI; in subject 2, who received 33 weeks of enfuvirtide, the V38A population declined to less than 5% by week 22; in subject 3, who received 39 weeks of enfuvirtide, the V38A population declined to less than 5% by week 24.
Table 1.
Clonal analysis of enfuvirtide resistance mutations in gp41 from plasma virus in subjects undergoing partial treatment interruption.
| Subject | Weeks of ENF prior to PTI |
Week after PTI |
Enfuvirtide resistance mutations at gp41 codons 36–45 |
|---|---|---|---|
| 1 | 27 | 0 | V38A, N42T (2/6)a |
| V38A, L44M (4/6) | |||
| 16 | WT (8/8) | ||
| 2 | 33 | 0 | V38A, N42T (8/8) |
| 16 | WT (5/8) | ||
| V38A (2/8) | |||
| N42T (1/8) | |||
| 22 | WT (9/9) | ||
| 3 | 39 | 0 | V38A (7/7) |
| 17 | WT (4/9) | ||
| V38A (5/9) | |||
| 24 | WT (9/9) |
ENF, enfuvirtide; PTI, partial treatment interruption.
No. of clones with indicated sequence/No. of clones sequenced.
Figure 1. Population dynamics of enfuvirtide-resistant HIV-1.
The proportion of V38A mutant virus (○) and plasma HIV-1 RNA levels (●) over time are indicated following interruption (beginning at week 0) and subsequent pulse administration (gray bar) of enfuvirtide for subjects 1 (a), 2 (b), and 3 (c). Data for the percent V38A are shown as mean ± SD of two determinations. The dashed line indicates the limit of quantification for the V38A allele-specific PCR. The results of clonal analysis of gp41 HR-1 sequences performed at the time points indicated by arrows are also shown. WT, wild-type.
Clonal analysis of HR-1 sequences from plasma HIV-1 RNA performed 16 weeks after the interruption showed persistence of the V38A mutation in none of 8 clones (0%) from subject 1, 2 of 8 clones (25%) for subject 2 and 5 of 9 clones (56%) from subject 3 (Table 1). The V38A mutation remained detectable by allele-specific PCR at low levels (approximately 1%) in the viral quasispecies of each subject for 9 to 19 months following enfuvirtide interruption, but had fallen below the limit of detection by clonal analysis.
Re-emergence of V38A during enfuvirtide pulse therapy
Each subject received a single four-week pulse of enfuvirtide at various times following the enfuvirtide interruption. Enfuvirtide was added to the regimen at weeks 76, 68 and 38 for subjects 1, 2 and 3, respectively. Pulse treatment with enfuvirtide resulted in only transient reductions in plasma HIV-1 RNA. Subjects 1 and 3 both had an initial 1.0-log10 decrease in plasma HIV-1 RNA, whereas subject 2 had a 0.5-log10 decrease. Subsequently, viral RNA increased by 0.7-log10 in subject 1 and by 1.4-log10 in subject 3. In the case of subject 2, viral RNA increases by 1.2-log10 above nadir, then decreases again to 0.4-log10 above the nadir associated with the pulse. Increases in the proportion of virus carrying the V38A mutation were detectable within 1 week of the enfuvirtide pulse, and were associated with the rebound in plasma HIV-1 RNA (Fig. 1). For example, in subject 3, the initial reduction in plasma HIV-1 RNA was accompanied by an increase in the proportion of V38A mutant from below the limit of detection to about 7%. As plasma HIV-1 RNA levels subsequently rebounded, the percent of V38A mutant virus increased in parallel, reaching approximately 52% before the end of the enfuvirtide pulse. Similar findings were observed in subject 2 (Fig. 1). Additional plasma samples available from subject 3 showed that during the second enfuvirtide interruption the V38A population decayed rapidly after a minimal lag time, declining to below the threshold of detection nearly eleven weeks after enfuvirtide was discontinued.
Determination of viral fitness in vivo
The relative fitness difference between the V38A mutant and wild-type virus was calculated for each subject (Table 2). In the absence of enfuvirtide, the V38A mutant was approximately 25% to 65% less fit than wild type. It is noteworthy that for subject 3 the V38A mutant appeared to have a greater fitness disadvantage relative to wild-type during the second enfuvirtide interruption. By contrast, in the presence of enfuvirtide the mutant viruses were substantially more fit than wild-type (107% to 498%). However, the limited number of data points available during the enfuvirtide pulse resulted in poor fits of the data to the model as indicated by the wide standard deviations. For this reason, estimates of the fitness differences during the enfuvirtide pulse phase are less reliable than those obtained during the interruption phase.
Table 2.
Fitness difference of V38A mutants relative to wild-type virus for periods on and off enfuvirtide therapy.
| Fitness Difference a | |||
|---|---|---|---|
| Subject | −ENF | + ENF | |
| 1 | −54% ± 6% | +498% ± 316% | |
| 2 | −35% ± 4% | +107% ± 20% | |
| 3 | First interruption | −25% ± 2% | +235% ± 85% |
| Second interruption | −65% ± 9% | n.d. | |
ENF, enfuvirtide; n.d., not done.
Fitness differences were determined following enfuvirtide interruption (−ENF) or during a four-week period of enfuvirtide “pulse” therapy (+ENF).
DISCUSSION
In this study, use of an allele-specific PCR assay allowed us to track the decay of HIV-1 variants carrying the V38A mutation for enfuvirtide resistance in gp41. Because we used a study design in which enfuvirtide was the only drug removed from or added to a stable background antiretroviral regimen we were able to measure directly the impact of this enfuvirtide resistance-associated mutation on viral fitness in vivo in the presence and absence of drug. The V38A mutant population remained stable for 6 to 12 weeks in absence of enfuvirtide, and then decayed rapidly to less than 5% of the virus population over an additional 6 to 12 weeks. Use of a growth corrected model for estimating fitness difference found that the V38A mutant viruses were 25% to 65% less fit than the wild-type in the absence of enfuvirtide. These results provide in vivo confirmation of earlier work from our group that demonstrated reduced fitness of the V38A mutant compared to wild-type virus in vitro [6].
Although marked differences in viral fitness led to turnover in the viral population in response to changing selection pressures, plasma HIV-1 RNA levels remained essentially unchanged during the partial treatment interruption. This observation illustrates the difference between relative fitness and replication capacity—relative fitness determines the proportion of various members of the viral quasispecies, but their absolute titer is more closely related to replication capacity. An alternative explanation is that target cell availability, rather than viral fitness and replication capacity, is the limiting factor in determining viral load [15]. If replacement of the V38A mutants by wild-type virus with a higher replication capacity results in greater depletion of available target cells, it may be difficult to discern an impact of these mutations on viral load set-point [16]. Detailed analysis on a larger number of subjects would be necessary to define more fully the relative impact of fitness and target cell availability on the level of viremia.
The V38A mutant re-emerged rapidly during the pulse intensification phase of this study and was accompanied by rapid rebound in plasma HIV-1 RNA levels. The more rapid turnover of the virus population during the enfuvirtide pulse suggests that enfuvirtide imposes stronger selective pressure on the virus population than does the partial treatment interruption, and that the advantage of the V38A mutant over wild-type in the presence of drug is greater than the disadvantage of the mutant compared to wild-type in the absence of drug. The rapid re-emergence of the V38A mutant population could also be explained by a persistent reservoir of actively replicating mutant virus [17,18]. Indirect evidence for this possibility is provided by the finding that in our study the level of V38A mutants detectable by allele-specific PCR never dropped below 1% of the population. This finding is consistent with previous studies in which persistence of drug-resistant virus as minority variants following treatment interruption was associated with virologic failure when antiretroviral therapy with the same drugs was resumed [19,20].
This study has a number of limitations. The detailed analysis performed allowed only a relatively small number of subjects to by studied. Consequently, the results presented here may not be representative of all patients with enfuvirtide failure associated with a V38A mutation. In addition, the small number of samples available during the pulse intensification phase resulted in a relatively poor fit of the data to the model used for determining fitness differences in the presence of enfuvirtide; hence, those estimates must be considered provisional. Although our analysis focused on the dynamics of the V38A mutation in the HR-1 domain of gp41, other mutations contributing to enfuvirtide resistance may have been present and could have contributed to overall viral fitness. For this reason, the presence or absence of V38A served only as a marker of viral turnover in response to enfuvirtide withdrawal or administration, and the fitness differences we observed cannot be ascribed solely to the presence of a valine or an alanine at amino acid 38. In addition, given the heterogeneity of HIV-1 env it is possible that the magnitude of the fitness differences we observed could vary depending on the specific envelope backbone in which the V38A mutation is found.
Work by others has shown that changes in virus populations accompanying the development of enfuvirtide resistance involves simultaneous or sequential emergence of viral variants that may carry similar mutations in HR-1 but may differ markedly in other regions of envelope [21]. These sub-populations may differ in replicative capacity and fitness in the absence of enfuvirtide, and could exhibit different decay rates when ENF is interrupted. Our results provide an estimate of the average fitness of the viral population.
In conclusion, the HIV-1 quasispecies undergoes dynamic changes in response to varying conditions imposed by the withdrawal and re-initiation of fusion inhibitor therapy. Differences in the kinetics of the decay and re-emergence of V38A mutant virus reflect differences in the strength of the selection pressures applied in this study. The rapid re-emergence of enfuvirtide resistance in association with virologic failure and a return to baseline in plasma HIV-1 RNA levels following the enfuvirtide “pulse” suggests that re-treatment with enfuvirtide after previous failure of this drug is unlikely to produce a durable virologic response. Further studies are needed to identify the factors that determine the lag period between treatment interruption and re-emergence of wild-type virus and to understand the relationship between treatment duration and size of the reservoir of drug-resistant virus.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants from the National Institutes of Health (K24 RR016482, R01 AI052745, R01 AI055273, R01 AI055357), the Harvard ACTG Virology Support Laboratory (U01 AI38858), the California AIDS Research Center (ID01-SF-049), the UCSF/Gladstone Institute of Virology & Immunology Center for AIDS Research (CFAR) (P30 MH59037), the Harvard Medical School CFAR (AI-060354), the General Clinical Research Center at SFGH (M01 RR00083), the Swiss National Science Foundation (PP00A-106751), "La Caixa" Fellowship grant to R.P. provided by Caixa d'Estalvis i Pensions de Barcelona, Spain, and by a research grant to S.G.D. provided by Roche and Trimeris.
Footnotes
These data were presented in part at the 1st International Workshop on HIV Entry Inhibition, December, 2005, Bethesda, MD [abstract 10], and at the 13th Conference on Retroviruses and Opportunistic Infections, 2006, February 5–8, Denver, CO [abstract M-188].
REFERENCES
- 1.Lalezari JP, Henry K, O'Hearn M, et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N Engl J Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 2.Lazzarin A, Clotet B, Cooper D, et al. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N Engl J Med. 2003;348:2186–2195. doi: 10.1056/NEJMoa035211. [DOI] [PubMed] [Google Scholar]
- 3.Sista PR, Melby T, Davison D, et al. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS. 2004;18:1787–1794. doi: 10.1097/00002030-200409030-00007. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Deeks SG, Hoh R, et al. Rapid emergence of enfuvirtide resistance in HIV-1-infected patients: Results of clonal analysis. J AIDS. 2006;43:60–64. doi: 10.1097/01.qai.0000234083.34161.55. [DOI] [PubMed] [Google Scholar]
- 5.Marcelin AG, Reynes J, Yerly S, et al. Characterization of genotypic determinants in HR-1 and HR-2 gp41 domains in individuals with persistent HIV viraemia under T-20. AIDS. 2004;18:1340–1342. doi: 10.1097/00002030-200406180-00015. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Sista P, Giguel F, Greenberg M, Kuritzkes DR. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20) J Virol. 2004;78:4628–4637. doi: 10.1128/JVI.78.9.4628-4637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deeks SG, Lu J, Hoh R, et al. Interruption of enfuvirtide in HIV-1-infected adults with incomplete viral suppression on an enfuvirtide-based regimen. J Infect Dis. 2007;195:387–391. doi: 10.1086/510531. [DOI] [PubMed] [Google Scholar]
- 8.Hunt PW, Martin JN, Sinclair E, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 9.Aquaro S, D’Arrigo R, Svicher V, et al. Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1-infected patients receiving enfuvirtide treatment. J Antimicrob Chemother. 2006;58:714–722. doi: 10.1093/jac/dkl306. [DOI] [PubMed] [Google Scholar]
- 10.Melby TE, Despirito M, Demasi RA, et al. Association between specific enfuvirtide resistance mutations and CD4 cell response during enfuvirtide-based therapy. AIDS. 2007;21:253–259. doi: 10.1097/QAD.0b013e3282f12362. [DOI] [PubMed] [Google Scholar]
- 11.Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods. 2007;146:136–146. doi: 10.1016/j.jviromet.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonhoeffer S, Barbour AD, de Boer RJ. Procedures for reliable estimation of viral fitness from time-series data. Proc Biol Sci. 2002;269:1887–1893. doi: 10.1098/rspb.2002.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonhoeffer S, Funk GA, Gunthard HF, Fischer M, Muller V. Glancing behind virus load variation in HIV-1 infection. Trends Microbiol. 2003;11:499–504. doi: 10.1016/j.tim.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Murphy DG, Fauvel M, Rene P, Vincelette J. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1/5. organize nuclisens QT with extractor and bayer quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2000;38:4034–4041. doi: 10.1128/jcm.38.11.4034-4041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean AR, Nowak MA. Competition between zidovudine-sensitive and zidovudine-resistant strains of HIV. AIDS. 1992;6:71–79. doi: 10.1097/00002030-199201000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Bonhoeffer S, Coffin JM, Nowak MA. Human Immunodeficiency virus drug therapy and virus load. J Virol. 1997;71:3275–3278. doi: 10.1128/jvi.71.4.3275-3278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derdeyn CA, Kilby M, Diego Miralles G, et al. Evaluation of distinct blood lymphocyte populations in human immunodeficiency virus type 1-infected subjects in the absence or presence of effective therapy. J Infect Dis. 1999;180:1851–1862. doi: 10.1086/315117. [DOI] [PubMed] [Google Scholar]
- 18.Lambotte O, Chaix ML, Gubler B, et al. The lymphocyte HIV reservoir in patients on long-term HAART is a memory of virus evolution. AIDS. 2004;18:1147–1158. doi: 10.1097/00002030-200405210-00008. [DOI] [PubMed] [Google Scholar]
- 19.Izopet J, Souyris C, Hance A, et al. Evolution of human immunodeficiency virus type 1 populations after resumption of therapy following treatment interruption and shift in resistance genotype. J Infect Dis. 2002;185:1506–1510. doi: 10.1086/340215. [DOI] [PubMed] [Google Scholar]
- 20.Palmer S, Boltz V, Maldarelli F, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS. 2006;20:701–710. doi: 10.1097/01.aids.0000216370.69066.7f. [DOI] [PubMed] [Google Scholar]
- 21.Labrosse B, Morand-Joubert L, Goubard A, et al. Role of the envelope genetic context in the development of enfuvirtide resistance in human immunodeficiency virus type 1-infected patients. J Virol. 2006;80:8807–8819. doi: 10.1128/JVI.02706-05. [DOI] [PMC free article] [PubMed] [Google Scholar]