Abstract
The lipid A residues of certain Gram-negative bacteria, including most strains of Salmonella and Pseudomonas, are esterified with one or two secondary S-2-hydroxyacyl chains. The S-2 hydroxylation process is O2-dependent in vivo, but the relevant enzymatic pathways have not been fully characterized because in vitro assays have not been developed. We previously reported that expression of the Salmonella lpxO gene confers upon Escherichia coli K-12 the ability to synthesize 2-hydroxymyristate modified lipid A (Gibbons, H. S., Lin, S., Cotter, R. J., and Raetz, C. R. H. J. Biol. Chem. 275, 32940–49, 2000). We now demonstrate that inactivation of lpxO, which encodes a putative Fe2+/O2/α-ketoglutarate-dependent dioxygenase, abolishes S-2-hydroxymyristate formation in S. typhimurium. Membranes of E. coli strains expressing lpxO are able to hydroxylate Kdo2-[4′-32P]-lipid A in vitro in the presence of Fe2+, O2, α-ketoglutarate, ascorbate and Triton X-100. The Fe2+ chelator 2,2′-bipyridyl inhibits the reaction. The product generated in vitro is a mono-hydroxylated Kdo2-lipid A derivative. The [4′-32P]-lipid A released by mild acid hydrolysis from the in vitro product migrates with authentic S-2-hydroxlyated lipid A isolated from 32P-labeled S. typhimurium cells. Electrospray ionization mass spectrometry and gas chromatography/mass spectrometry of the in vitro product are consistent with the 2-hydroxylation of the 3′-secondary myristoyl chain of Kdo2-lipid A. LpxO contains two predicted trans-membrane helices (one at each end of the protein), and its active site likely faces the cytoplasm. LpxO is an unusual example of an integral membrane protein that is a member of the Fe2+/O2/α-ketoglutarate-dependent dioxygenase family.
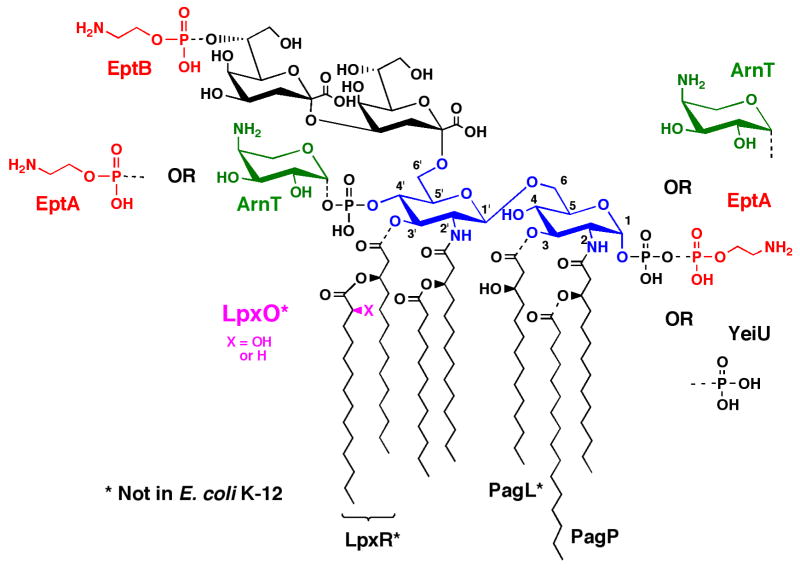
The Salmonella typhimurium genome encodes several enzymes (Fig. 1) that catalyze the covalent modification of the Kdo2-lipid A region of lipopolysaccharide (LPS)1 (1, 2). Activation of the PmrA/PmrB two component system by growth at low pH or as the result of point mutations in PmrA (3–8) induces the transcription of the enzymes EptA and ArnT, which attach phosphoethanolamine (pEtN)1 and 4-amino-4-deoxy-L-arabinose (L-Ara4N)1 units to lipid A, respectively (Fig. 1) (9–11). The active sites of EptA and ArnT are located on the outer surface of the inner membrane (12). Activation of the PhoP/PhoQ two-component system by growth of cells at low divalent cation concentrations (13–15), or in the presence of cationic antimicrobial peptides (16), induces the genes encoding the outer membrane enzymes PagP (17–19) and PagL (20, 21), which remodel the acyl chains of lipid A (Fig. 1). The addition of a pEtN unit to the outer Kdo residue by EptB (Fig. 1) is independent of PmrA/PmrB and PhoP/PhoQ, but instead is induced by 5 to 50 mM Ca2+ ions (22, 23).
Figure 1. Regulated covalent modifications of S. typhimurium and E. coli Kdo2-lipid.
A. Covalent modifications of the lipid A phosphate groups are regulated by the PmrA/PmrB two-component signaling system (1). ArnT attaches L-Ara4N mainly to the 4′-position, whereas EptA predominantly adds pEtN mainly to the 1-position. However, the selectivity of these enzymes is reversed in the absence of Kdo (73, 74), and minor species are formed in which both positions are modified either with pEtN or with L-Ara4N (1, 50). The PagP-dependent palmitoylation (1, 17–19) and the PagL catalyzed deacylation (1, 20) of the lipid A acyl chains are under the control of the PhoP/PhoQ two-component system (1). Other lipid A modifications, such as those catalyzed by another pEtN transferase EptB (26) or the 3′-O-deacylase LpxR (1, 23), are induced by high concentrations of Ca2+. The formation of 2-hydroxymyristate on lipid A by LpxO (1, 27), observed when cells are grown in the presence of O2, is not dramatically regulated by PhoP/PhoQ under our conditions (5). The properties of the indicated enzymes responsible for these modifications are reviewed elsewhere (1). The YeiU gene, which encodes a novel phosphotransferase, was recently renamed LpxT to reflect is enzymatic function (77).
The PmrA/PmrB and PhoP/PhoQ two component systems are both activated following endocytosis of live S. typhimurium cells by RAW 264.7 macrophage tumor cells, resulting in multiple partial covalent modifications of lipid A (5). Addition of the L-Ara4N and palmitate moieties to lipid A confers increased resistance to polymyxins and β-defensins, respectively (4, 15, 24). The remodeling of the acyl chains (Fig. 1) also reduces the potency of lipid A as an agonist against TLR-4 (25).
The enzymes that add the L-Ara4N, pEtN and palmitate groups to lipid A are present both in Escherichia coli K-12 and S. typhimurium (1). However, PagL (20), LpxR (26) and LpxO (27) (Fig. 1) are restricted to Salmonella. LpxR cleaves the 3′-acyloxyacyl moiety of lipid A in the presence of Ca2+ ions (26). LpxO is involved in generating the S-2-hydroxy group present on the 3′-secondary myristoyl chain of Salmonella lipid A (27), provided that the cells are grown in the presence of O2 (Fig. 1). Although the existence of S-2-hydroxymyristate in S. typhimurium lipid A has been known for many years (28), its enzymatic synthesis has not been fully elucidated (27). The S-2-hydroxymyristate moiety may increase hydrogen bonding between adjacent lipid A units, enhancing the outer membrane’s ability to resist penetration by organic ions, such as ethidium, under some growth conditions (29, 30).
In previous work we identified the Salmonella lpxO gene, the occurrence of which correlates with the presence of S-2-hydroxyacylated lipid A in diverse Gram-negative bacteria (27). Expression of Salmonella lpxO in E. coli K-12 resulted in robust O2-dependent formation of E. coli lipid A containing 2-hydroxymyristate (27), implicating LpxO as the enzyme responsible for lipid A 2-hydroxylation. The LpxO protein shares similarity with the superfamily of Fe2+/O2/α-ketoglutarate-dependent dioxygenases (27), which participate in important processes such as collagen crosslinking, transcription factor inactivation, herbicide degradation, and taurine biosynthesis (31, 32). Many Fe2+/O2/α-ketoglutarate-dependent dioxygenases catalyze hydroxylation reactions, but some catalyze ring expansions, dehydrations, and halogenations (33–35). The active sites of these enzymes contain a single ferrous ion, usually coordinated by a His-X-Asp/Glu-Xn-His facial triad motif in which n denotes at least 40 consecutive amino acid residues (36). The existence of a similar sequence motif in LpxO (27) suggested a possible enzymatic mechanism for lipid A 2-hydroxylation (37). However, an in vitro assay for LpxO was not developed (27), and therefore the substrates for the 2-hydroxylation process remained obscure.
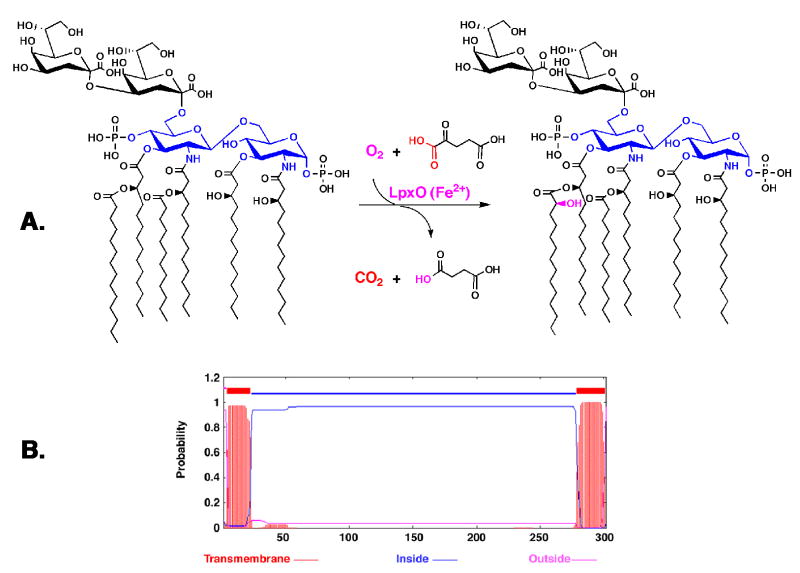
We now demonstrate that membranes of E. coli cells expressing Salmonella lpxO catalyze the hydroxylation of Kdo2-lipid A (38) in accordance with the proposed scheme shown in Fig. 2A, whereas membranes from the vector control strain do not. LpxO appears to be localized to the inner cytoplasmic membrane. LpxO activity is stimulated in vitro by the addition of Fe2+ ions and is absolutely dependent upon the presence of α-ketoglutarate and a non-ionic detergent. LpxO possesses two putative trans-membrane domains (one at its N-terminus and the other at its C-terminus) (39). Its central catalytic domain (Fig. 2B) is predicted to face the cytoplasm, consistent with the fact that LpxO activity in living cells is independent of the lipid A transporter MsbA (12, 40).
Figure 2. Proposed reaction catalyzed by LpxO.
A) The LpxO reaction is proposed to proceed by a mechanism similar to that used by other Fe2+/O2/α-ketoglutarate dependent dioxygenases, most likely catalyzing the direct 2-hydroxylation of the 3′-myristate chain of hexa-acylated Kdo2-lipid A (38), as demonstrated in this study. B) Predicted topology and trans-membrane segments of LpxO based on the TMHMM algorithm (39).
MATERIALS AND METHODS
Reagents and materials
32Pi and [γ-32P]-ATP were purchased from Perkin Elmer Life Sciences, Waltham, MA. Silica gel 60 glass-backed TLC plates (0.25 mm) were obtained from E. Merck (Darmstadt, Germany). Pyridine, methanol, 88% formic acid and KH2PO4 were from Mallinckrodt, Hazelwood, MO. Chloroform, KCl, and NaCl were from EM Science, Gibbstown, NJ. Dithiothreitol (DTT), ascorbic acid, α-ketoglutarate, Fe(NH4)2(SO4)2, catalase and bovine serum albumin were from Sigma (St. Louis, MO). Triton X-100 was purchased as the SurfactAmp 10% solution in water from Pierce/Endogen (Rockford, IL). E. coli phospholipids were obtained from Avanti Polar Lipids, Alabaster, AL. DL-3-hydroxymyristic acid and DL-2-hydroxymyristic acid were from Sigma (St. Louis, MO), and Tri-Sil reagent was from Pierce (Rockford, IL).
Bacterial strains and cultures
Unless otherwise stated, bacteria were grown in Luria-Bertani (LB) broth (41). Antibiotics were added at concentrations of 100 μg/ml (ampicillin), and 30 μg/ml (chloramphenicol and kanamycin).
Construction of an LpxO overexpression vector and isolation of membranes from overexpressing strains
The lpxO gene was excised from pHSG1(27) using NdeI and XhoI and cloned into pET21a+ (Novagen) using T4 DNA ligase (Boehringer Mannheim). The resulting plasmid (pHSG2) was transformed into E. coli XL1-Blue (Stratagene). The insert size was confirmed by restriction enzyme digest analysis. pHSG2 was subsequently transformed into E. coli BLR(DE3)/pLysS (Novagen).
To express LpxO, stationary phase cultures in LB broth, containing ampicillin and chloramphenicol, were diluted 1:100 into 1 l of fresh LB broth containing ampicillin and chloramphenicol, and grown at 37° C until the A600 reached 0.5. Isopropyl thio-β-D-galactoside (IPTG) (1mM) was then added, and the growth was continued for 3 h. The cells were harvested by centrifugation (5000 × g for 20 min). The cell pellets were washed, re-suspended in 50 mM HEPES, pH 7.5, and broken in a French pressure cell. Cell debris was removed by centrifugation at 4000 × g for 20 min. Membranes were isolated by centrifuging the clarified lysates at 100,000 × g for 1 h. The membrane pellet was re-suspended in 50 mM HEPES, pH 7.5, and centrifuged at 100,000 × g for 1 h. The washed membranes were re-suspended in 50 mM HEPES, pH 7.5, to a concentration of 1–5 mg/ml of membrane protein, as judged by the bicinchoninic acid assay with bovine serum albumin as the standard (42).
Isolation of lipid A from S. typhimurium Cultures
Cultures of S. typhimurium 14028s wild type) and HG002 (5) were grown with shaking at 250 rpm in 200 ml LB medium at 7°C to A600 = 1.0. Cells were harvested by centrifugation at 5000 ×g for 30 min. Pellets ere washed once with 30 ml phosphate-buffered saline (pH 7.4) (43), and the cells were entrifuged again at 5000 ×g for 30 min. The final cell pellet was re-suspended in 20 ml hosphate-buffered saline; chloroform (25 ml) and methanol (50 ml) were then added to enerate a single-phase Bligh/Dyer mixture (44). After 1h at room temperature, the LPS-containing precipitate was collected by centrifugation (2500 ×g for 20 min). The pellet as then washed twice with 50 mL of a single-phase Bligh/Dyer mixture (44) and collected by centrifugation. The washed pellet was dispersed in 25 ml 50 mM sodium acetate (pH 4.5) using a Branson probe sonicator. The suspension was heated to 100 °C for 30 min in a boiling water bath. After cooling to room temperature, chloroform (28 ml) and methanol (28 ml) were added to make a two-phase Bligh/Dyer mixture (44). The lower organic phase as collected after separation of the phases by centrifugation (2500 ×g for 20 min). The aqueous phase was re-extracted with fresh pre-equilibrated lower-phase, and again the organic layer was collected after centrifugation. The organic phases were pooled and dried using a rotary evaporator. The final lipid A preparations were stored at −80 °C.
Labeling of S. typhimurium cells with 32Pi and isolation of the lipid A fraction were arried out as described previously (27).
Enzymatic synthesis and purification of LpxO substrates
Preparation of radiolabeled do2-[4′-32P]-lipid A was performed according to published procedures (45). Unlabeled do2-lipid A was purified from WBB06 according to the method of Raetz et al. (38) or urchased from Avanti Polar Lipids.
In vitro assay of LpxO activity
An in vitro system for LpxO, using Kdo2-lipid A as the utative acceptor substrate, was developed based on assays previously reported for other Fe2+/O2/α-ketoglutarate-dependent hydroxylases (46, 47). The reaction conditions, unless therwise indicated, included 50 mM HEPES, pH 7.5, 1 mM α-ketoglutarate, 2 mM scorbate, 10 μM Fe(NH4)2(SO4)2, 0.2% Triton X-100, 4 mM DTT, 0.5 mg/ml E. coli hospholipid and 4 μM Kdo2-[4′-32P]-lipid A (20 000 cpm/reaction or 100,000 cpm/nmol). Assays were carried out at 30° C in a final volume of 50 μl. Reactions were initiated by adding E. coli membranes (0.01–0.1 mg/ml final concentration). In some cases, catalase 0.1 mg/ml) was also included to scavenge H2O2
To monitor the hydroxylation of lipid A, the reaction was quenched by removing a 10 μl portion of the assay mixture and adding it to 170 μl of a hydrolysis buffer, consisting of 12.5 mM sodium acetate (pH 4.5) and 1% SDS. This mixture was heated at 100°C for 30 in to cleave off the Kdo residues (48). Following hydrolysis, 400 μl of CHCl3/MeOH 1:1, v/v) were added to make a two-phase Bligh/Dyer mixture. After mixing, the phases were separated by brief centrifugation (13,000 rpm) in a tabletop micro-centrifuge. The pper phase was discarded, and the lower phase containing the released lipid A was acuum-dried, re-dissolved in 10 μl CHCl3/MeOH (4:1, v/v), and spotted at the origin of a 20 cm × 20 cm Silica Gel 60 TLC plate. Hydroxylated lipid A was resolved from unmodified lipid A by chromatography in the solvent CHCl3/pyridine/88% formic acid/water (50:50:16:5, v/v). The hydroxylated lipid A product migrates more slowly than unmodified lipid A, being resolved just enough to permit quantification (27). Lipid A species separated by TLC were quantified using a PhosphorImager, equipped with ImageQuant software (Molecular Dynamics).
Purification of hydroxylated Kdo2-lipid A generated in vitro
The LpxO hydroxylation reaction was scaled up to a 4.5 ml reaction volume under conditions otherwise identical to the assay system described above, except that the radioactive tracer was omitted and the concentration of the Kdo2-lipid A was raised up to 100 μM. Reactions were carried out in the presence of 0.1 mg/ml membrane protein, using membranes from IPTG-induced BLR(DE3)/pLysS cells containing either pET21a+ or pHSG2. After 2 h at 30 °C, the reaction mixture was converted into a single-phase Bligh/Dyer system by addition of 5.6 ml CHCl3 and 11.3 ml methanol. The Kdo2-lipid A acceptor substrate is soluble in this single-phase solvent mixture (38), whereas the endogenous LPS present in the membranes of the host strain is not. The Kdo2-lipid A was retrieved by adding appropriate volumes of CHCl3 and aqueous HCl to generate a two-phase Bligh/Dyer system, which consists of CHCl3/MeOH/0.1 M HCl (2:2:1.8, v/v) (44). After thorough mixing, the phases were separated by low-speed centrifugation for 10 min. The lower phase, which contains the Kdo2-lipid A and its hydroxylated product, was removed, washed once with fresh pre-equilibrated acidic upper phase, and then dried under N2. The Kdo2-lipid A species were then separated from glycerophospholipids by anion exchange chromatography on DEAE cellulose (22), and analyzed by electrospray ionization-mass spectrometry (ESI/MS) in the negative mode.
ESI/MS analysis of Kdo2-lipid A and lipid A samples
Mass spectra were acquired on a QSTAR-XL quadrupole time-of-flight tandem mass spectrometer (ABI/MDS-Sciex, Foster City, CA), equipped with an ESI source. Spectra were acquired in the negative-ion mode and typically were the accumulation of 60 scans over the range of 200–2500 atomic mass units (38). For MS analysis, the extracted lipids were dissolved in 500 μL of chloroform/methanol/water (2:3:1, v/v), containing piperidine (1%, v/v), and infused into the ion source at 5–10 μL/min. The negative-ion ESI was carried out at −4200V. Data acquisition and analysis were performed using the AnalystQS software.
GC/MS analysis of hydroxylated fatty acids
The DEAE-purified Kdo2-lipid A species derived from the large scale in vitro reactions described above were hydrolyzed in acidic methanol, N-acetylated, and then converted to trimethylsilyl ethers. DL-3-hydroxymyristic acid and DL-2-hydroxymyristic acid standards were processed and analyzed in parallel with the samples. Typically, 0.5–1.0 mg of sample was thoroughly dried in a Reacti-vial equipped with a Teflon-lined screw cap. Samples were hydrolyzed by adding 300 μl of 1 M HCl in methanol and heated at 80 °C for 15 h. The reaction mixtures were then cooled, and the solvents were removed under a stream of nitrogen. Next, 200 μl of anhydrous methanol, 40 μl of pyridine, and 40 μl of acetic anhydride were added to the vial. The reaction mixtures were mixed and incubated overnight at room temperature. The solvents were evaporated under a stream of nitrogen. Finally, silylation of free OH groups to generate the trimethylsilyl (TMS) derivatives was achieved by adding 200 μl of Tri-Sil reagent to the dried samples, mixing, and incubating at room temperature for an hour. The samples were dried under a gentle stream of nitrogen, redissolved in 100 μl of hexane and transferred to new vials for gas chromatography/mass spectrometry (GC/MS) analysis.
GC/MS was performed using a Finnigan Trace MS instrument coupled with a Trace GC 2000 gas chromatography system. The column was a 30 m RTX-5MS (0.25 μm internal diameter and 0.25 μm phase thickness) from Restek (Bellefonte, PA). The temperature program of the GC was as follows: the column oven temperature was initially held at 100 °C for 3 min, increased to 150 °C at a rate of 20 °C/min, then increased to 200 °C at a rate of 1 °C/min, further increased to 335 °C at a rate of 30 °C/min, and finally held at 335 °C for 2 min. The total run time was 62 min. The injector was operated in the split mode (1:20 split), and the temperature of the injection port was kept at 200 °C. Helium was the carrier gas with a constant flow rate of 1 mL/min. The instrument was operated in the electron impact (EI) mode with the electron energy set at 70 eV.
RESULTS
A mutant of S. typhimurium lacking 2-hydroxymyristate-modified lipid A
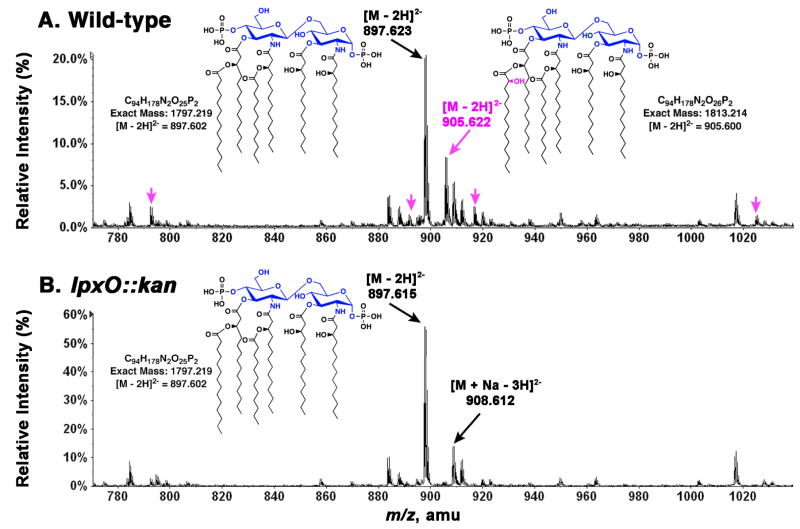
S. typhimurium mutant HG002 (lpxO::kan) was previously shown to lack 2-hydroxymyristate modified lipid A species, as judged by TLC analysis of 32Pi-labeled cells (5). This conclusion was confirmed by high-resolution ESI/MS of crude lipid A species (Fig. 3) obtained from wild-type or HG002 cells grown on LB broth. As shown in Fig. 3A, the predominant [M - 2H]2− ions in the spectrum of wild-type are consistent with enteric hexa-acylated lipid A species, bearing either myristate (m/z 897.623) or 2-hydroxymyristate (m/z 905.622) as the secondary acyl chain at position 3′. Minor ions can be attributed to penta-or hepta-acylated lipid A species, to variations in acyl chain lengths, or to sodium adducts (Fig. 3A). The prominent peak at m/z 905.622 (Fig. 3A) is absent in the spectrum of the mutant lipid A (Fig. 3B), consistent with a defect in the 2-hydroxylation of lipid A. Several minor molecular species, predicted to contain 2-hydroxymyristate based on their exact masses, are likewise missing in mutant HG002 (Fig. 3A versus 3B, magenta arrows).
Figure 3. The lpxO mutant HG002 lacks lipid A species containing 2-hydroxymyristate.
ESI/MS spectra of crude lipid A species isolated from the wild-type S. typhimurium 14028s (A) or from the lpxO::kan mutant HG002 (B). The minor peaks shown in magneta in panel A are proposed to arise from the following covalent modifications or adducts of the parent compound with [M–2H]2− at m/z 905.6: [M+C16:0–2H]2− at m/z 1024.7; [M+Na–3H]2− at m/z 916.6; [M–2CH2–2H]2− at m/z 891.6; and [M–3OHC14–2H]2− at m/z 792.5.
Deletion of lpxO did not appreciably compromise outer membrane integrity or growth of cells either on LB broth or on low-Mg2+ N-minimal medium (data not shown). Sensitivity to erythromycin, rifampicin, streptomycin, or bacitracin was not increased, as judged by disc diffusion assays (data not shown). The ability of HG002 to penetrate and multiply inside of mouse RAW 264.7 macrophage tumor cells was unaffected (5). The virulence of HG002 was comparable to that of wild-type S. typhimurium, when tested in a mouse infection model by either intravenous or oral administration (E. Romilianus and D.J. Maskell, personal communication).
Membrane localization of overexpressed LpxO in E. coli
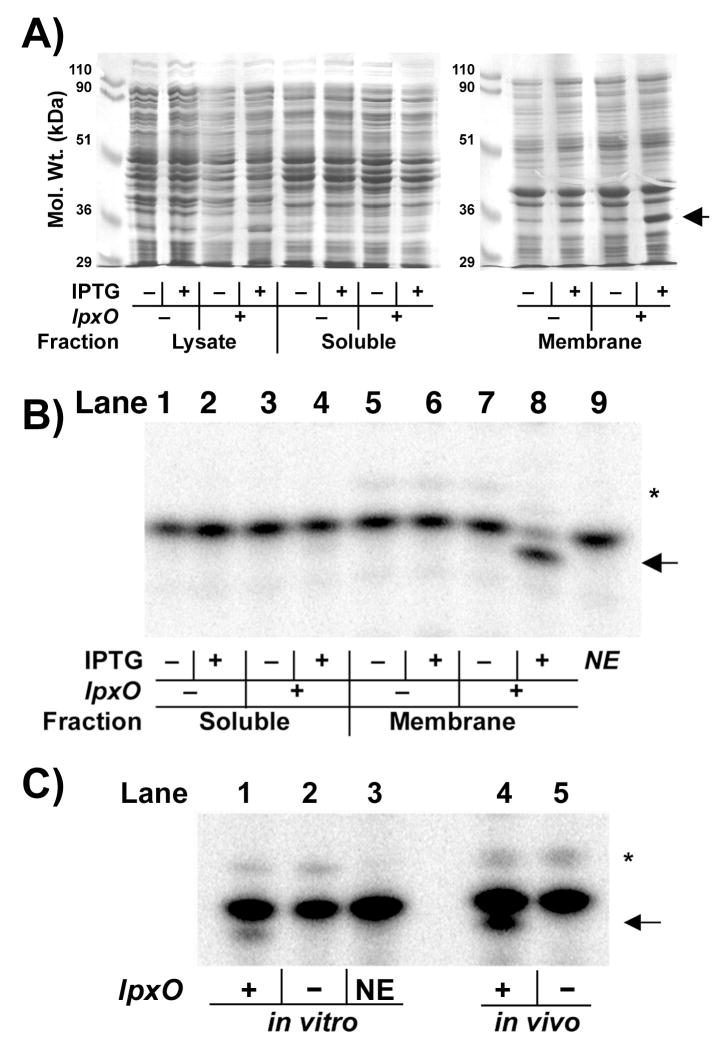
The lpxO gene was over-expressed behind the T7-lac promoter on the hybrid plasmid pHSG2 (Table 1) in E. coli BLR(DE3)/pLysS. Crude extracts of lpxO-induced cultures displayed a faint protein band on SDS gels near 35 kDa, consistent with the predicted molecular weight of LpxO (Fig. 4A). This band was absent in the cytosol but was enriched in the membrane fraction (Fig. 4A), confirming the predicted subcellular localization of LpxO (Fig. 2B) (39), which is unusual among the Fe2+/α-ketogluatarate dependent hydroxylases.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain/Plasmid | Description | Source |
|---|---|---|
| Escherichia coli | ||
| XL1-BlueMR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| BLR(DE3)pLysS | T7-lac expression strain, recA | Novagen |
| WBB06 | ΔrfaC rfaF::tet | (76) |
|
| ||
| Salmonella typhimurium | ||
| 14028s | virulent wild-type | S.I. Miller |
| HG002 | lpxO::kan derivative of 14028s | (5) |
|
| ||
| Plasmids | ||
| pHSG1 | pBluescript II SK containing the lpxO gene | (27) |
| pET21a+ | T7-lac expression vector | Novagen |
| pHSG2 | pET21a+ containing lpxO gene in NdeI and XhoI sites | This work |
Figure 4. Over-expression and catalytic activity of membrane-associated LpxO.
A) Coomassie-blue stained SDS-gel of sub-cellular fractions from LpxO-over-expressing E. coli. The LpxO protein (arrow) migrates as a 35-kDa protein and is localized in the membrane fraction. B) Incubation of E. coli Kdo2-[4′-32P]-lipid A in the LpxO assay system, described in the Materials and Methods, with 0.1 mg/ml cytosol (lanes 1–4) or 0.1 mg/ml membranes (lanes 5–8) of E. coli BLR(DE3)/pLysS harboring pET21a+ (lanes 1, 2, 5, 6) or pHSG2 (lanes 3, 4, 7, 8) for 60 min. Lipid A species were separated by TLC in CHCl3/pyridine/formic acid/H2O (50:50:16:5, v/v) after mild acid hydrolysis of the reaction product at pH 4.5 to remove the Kdo disaccharide (75). The hydroxylated lipid A product is highlighted by the arrow. NE indicates (lane 9) the no-enzyme control. C) Chromatographic comparison of lipid A 1,4′-bis-phosphate species generated in vitro by LpxO-over-expressing membranes (lane 1) at 0.01 mg/ml for 5 min to matched vector or no-enzyme controls (lanes 2 and 3), and to the lipid A species obtained from 32Pi-labeled wild-type (lane 4) or lpxO::kan mutant S. typhimurium (lane 5). The asterisk indicates the hepta-acylated product formed by the outer membrane enzyme PagP (17).
An in vitro assay for LpxO
Given its sequence similarity to the Fe2+/α-ketoglutarate-dependent dioxygenases and the O2 requirement for the formation of 2-hydroxymyristate and other 2-hydroxyacyl chains in vivo (27, 49), it seemed plausible that LpxO might catalyze the in vitro hydroxylation of hexa-acylated E. coli Kdo2-lipid A (38) in the presence of appropriate cofactors and O2 (Fig. 2A). We therefore prepared an aerobic assay system to probe for LpxO activity, consisting of 4 μM Kdo2-[4′-32P]-lipid A, 1 mM α-ketoglutarate, 2 mM ascorbate, 10 μM Fe(NH4)2(SO4)2, 0.2% Triton X-100, 4 mM DTT, and 0.5 mg/ml E. coli phospholipids in 50 mM Hepes, pH 7.5. The reaction was initiated by adding either soluble protein or membranes (typically 0.01 to 0.1 mg/ml, as indicated) from the vector control or the LpxO over-expressing strain (with or without IPTG induction). Incubation of the complete reaction mixture at 30° C resulted in LpxO-dependent formation of a new, more slowly migrating 32P-labeled lipid A species (Fig. 4B), as judged by TLC, after removal of the Kdo disaccharide by hydrolysis at pH 4.5. The Rf of the modified lipid A species is consistent with the behavior of 2-hydroxymyristate- containing lipid A isolated by hydrolysis at pH 4.5 from S. typhimurium LPS (Fig. 4C) (27, 50 ).
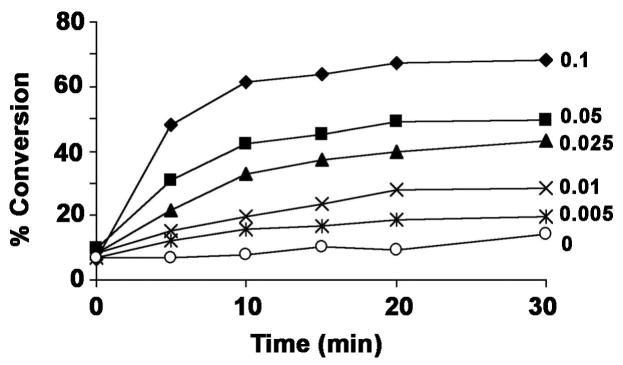
The formation of the slowly migrating [4′-32P]-lipid A product was time and protein concentration dependent (Fig. 5), although linearity of product formation with time was gradually lost after 5 min (Figs. 5 and 6A). The specific activity of membranes from LpxO-over-expressing cells, assayed at 0.01 mg/ml for 5 min, was 6.9 nmol/min/mg with Kdo2-[4 ′-32P]-lipid A as the substrate. No products were generated when LpxO was incubated with Kdo2-lipid IVA (1), lipid IVA (1) or hexa-acylated lipid A (lacking the Kdo disaccharide) under otherwise identical assay conditions (data not shown).
Figure 5. Dependence of LpxO product formation on time and protein concentration.
Increasing amounts of LpxO membrane protein were added to the assay cocktail, and the formation of hydroxylated lipid A was monitored with time. Membrane protein concentrations (mg/ml) in the final assay mixture are indicated. Diamonds, 0.1 mg/ml; squares, 0.05 mg/ml; triangles, 0.025 mg/ml; X, 0.01 mg/ml; stars, 0.005 mg/ml; open circles, no enzyme control. The small amount of apparent product formation at time 0 and in the no enzyme control is caused by slight streaking of the substrate band during TLC, which migrates just above the hydroxylated product band (Fig. 4B).
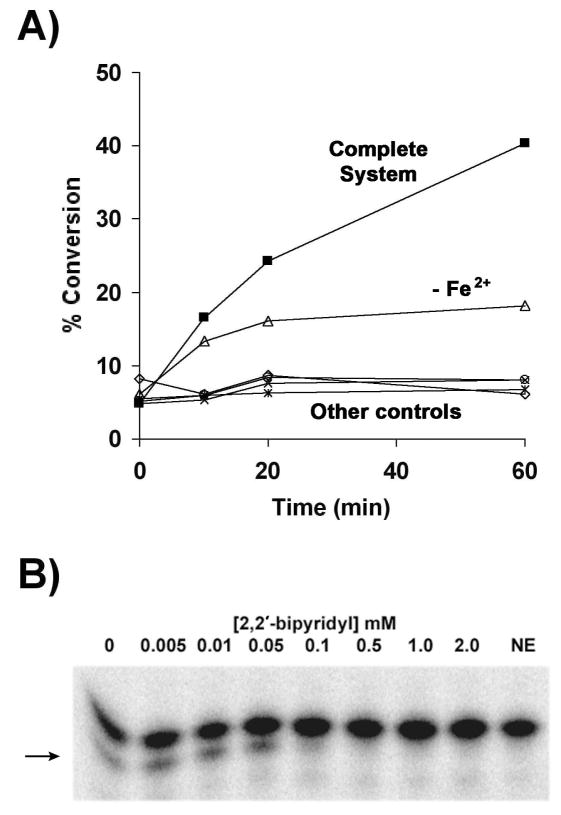
Figure 6. Dependence of LpxO activity on soluble cofactors.
A) A standard LpxO assay mixture containing 0.01 mg/ml membrane protein was prepared in which either α-ketoglutarate, ascorbate or iron was omitted. Full activity was evident only when all the cofactors were present (closed squares). Omission of α-ketoglutarate (open diamonds) or ascorbate (open circles) eliminated lipid A hydroxylation completely, as in the vector and no enzyme controls (crosses and asterisks). Omission of added Fe2+ resulted in lower activity (open triangles). B) Inhibition of LpxO activity by increasing concentrations of 2,2′-bipyridyl, an Fe2+ chelator. The standard reaction mixture containing 0.01 mg/ml protein (5 min incubation) was supplemented with the indicated concentrations of 2,2′-bipyridyl. The arrow indicates the position of the LpxO product.
Dependence of the LpxO reaction on cofactors
When either α-ketoglutarate or ascorbate was omitted from the reaction mixture (Fig. 6A, open diamonds and open circles, respectively), no LpxO product was formed, as in the no-enzyme and vector controls (Fig. 6A, crosses and asterisks). Omission of iron from the assay cocktail reduced the extent of lipid A 2-hydroxylation relative to the complete reaction mixture, but it did not entirely abolish enzymatic activity (Fig. 6A, open triangles). The membranes used to initiate the reaction probably contained some iron. When the reaction mixture was supplemented with catalase to scavenge H2O2, partial lipid A hydroxylation was observed in the absence of ascorbate, indicating that ascorbate is not absolutely necessary for LpxO activity (data not shown).
If LpxO requires Fe2+ for activity, then 2,2′-bipyridyl, an Fe2+ chelator, should inhibit lipid A hydroxylation. When 2,2′-bipyridyl was included in the reaction mixture, dose-dependent inhibition of LpxO product formation was indeed observed (Fig. 6B). Taken together, the results demonstrate a strict requirement for α-ketoglutarate and Fe2+ in the LpxO catalyzed hydroxylation of lipid A.
Detergent-requirement and stabilization of LpxO by phospholipids
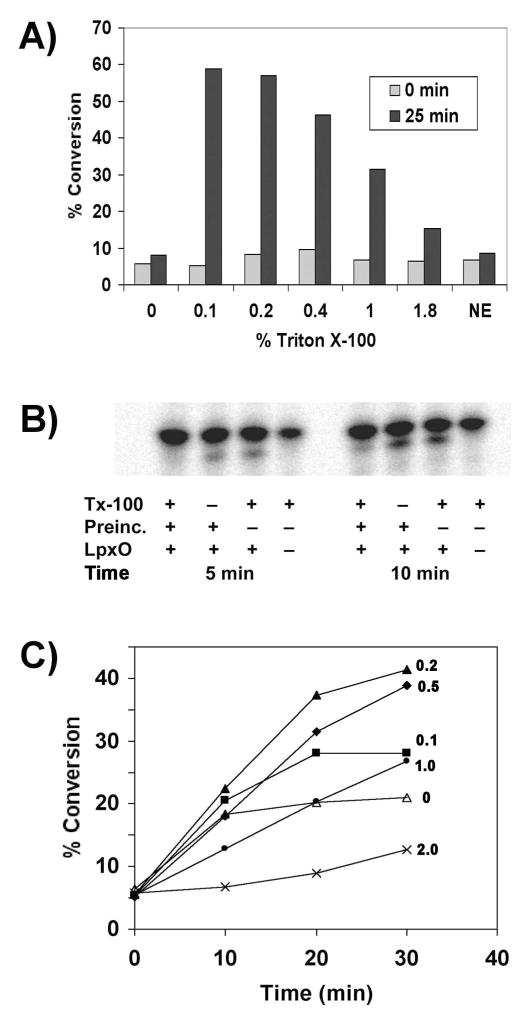
Because LpxO is an integral membrane enzyme with a hexa-acylated lipid substrate, the reaction should display an absolute requirement for detergent. Indeed, no hydroxylation was observed when the detergent was omitted, whereas 0.1–0.2 % Triton X-100 provided optimal stimulation of enzyme activity (Fig. 7A). Product formation was reduced at higher detergent concentrations (Fig. 7A), either because of surface dilution of the substrate (51) or inactivation of the enzyme.
Figure 7. Detergent and phospholipid dependence of LpxO activity.
A) LpxO-catalyzed conversion of Kdo2-lipid A to its hydroxylated derivative was monitored after 25 min in the presence of increasing amounts of Triton X-100 (% v/v) at 0.1 mg/ml membrane protein. NE, no-enzyme control. B) Inactivation of LpxO by pre-incubation for 30 min at 0.02 mg/ml membrane protein in assay buffer containing Triton X-100 prior to initiation of the reaction by addition of the full set of cofactors and substrates to give a final membrane protein concentration of 0.01 mg/ml. C) Addition of E. coli phospholipids to the LpxO assay cocktail partially stabilizes LpxO activity, when assayed at 0.01 mg/ml membrane protein. Time courses of product formation (without pre-incubation) were followed for assay mixtures supplemented with 0 (open triangles), 0.1 (closed squares), 0.2 (closed triangles), 0.5 (closed diamonds), 1.0 (closed circles), or 2 mg/ml (x x) of exogenous E. coli phospholipids (as indicated by the numbers on the graph). The rate and extent of product formation with time were deemed optimal with 0.2 or 0.5 mg/ml phospholipids.
Pre-incubation of LpxO-containing membranes with Triton X-100 and assay buffer, prior to initiation of the reaction by addition of the remaining reagents, resulted in reduced LpxO activity (Fig. 7B). Enzymes like LpxO sometimes inactivate themselves by hydroxylating tyrosine residues near their active sites (52). Alternatively, solubilization of LpxO at low protein concentrations and removal of tightly bound lipids might also inactivate the enzyme. Supplementation of the reaction mixture with additional E. coli phospholipids partially stabilized LpxO activity and improved linearity of product formation with time, but higher concentrations of phospholipids were inhibitory (Fig. 7C).
ESI/MS of the product generated by LpxO-catalyzed hydroxylation of Kdo2-lipid A
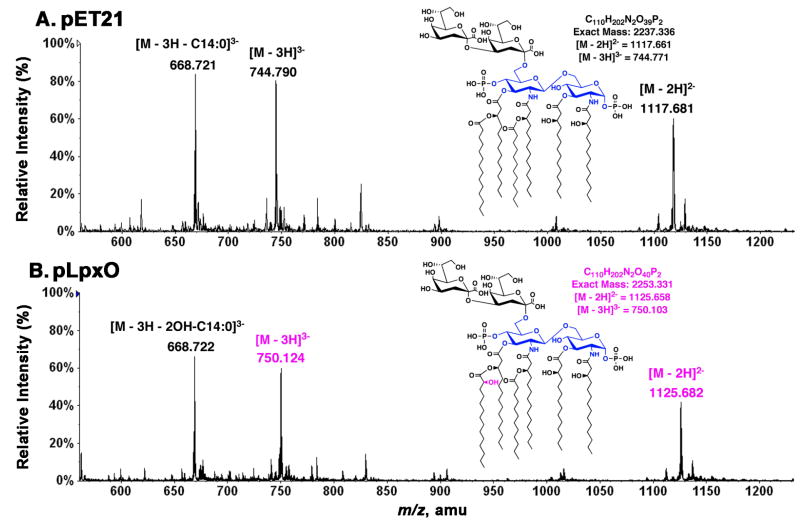
To confirm that the 3′-secondary acyl chain of Kdo2-lipid A is selectively hydroxylated in our in vitro LpxO assay system, a 4.5 ml reaction mixture, containing 0.1 mg/ml membrane protein and 100 μM Kdo2-lipid A (38), was incubated for 2 h at 30 °C. The Kdo2-lipid A species (unresolved substrate and hydroxylated product) were re-purified from the crude reaction mixture by Bligh/Dyer extraction followed by anion-exchange chromatography on DEAE cellulose (22). The Kdo2-lipid A species were then analyzed directly without pH 4.5 hydrolysis by negative ion ESI/MS (Fig. 8). The vector control sample (Fig. 8A) yielded peaks at m/z 1117.681 and m/z 744.790, which are interpreted as [M-2H]2− and [M-3H]3− of unmodified Kdo2-lipid A, respectively (38). The sample incubated with the LpxO-over- expressing membranes as the enzyme source (Fig. 8B) yielded peaks at m/z 1125.682 and m/z 750.124, which are interpreted as the [M-2H]2− and [M-3H]3− ions of mono- hydroxylated Kdo2-lipid A, respectively. The prominent triply charged ions near m/z 668.72, present in both spectra, arise from neutral loss of the 3′-secondary acyl chains during ESI/MS, as previously observed for Kdo2-lipid A (38). The extent of neutral loss from the triply charged ions is much higher than that from the doubly charged ions, which is likely attributed to the fact that the triply charged ions are subjected to more energetic collisional activation than the doubly charged ions in the ion source region. The identical mass of this species in the spectra of both the substrate and the product provides unequivocal evidence that the LpxO-catalyzed hydroxylation of Kdo2-lipid A is restricted to the 3′-secondary myristate chain, as previously observed in living cells (27).
Figure 8. ESI/MS analysis of the LpxO reaction product.
Negative-ion mode ESI/MS spectra were acquired for re-purified Kdo2-lipid A samples that had been incubated in vitro for 2 h under assay standard conditions with 0.1 mg/ml membranes from either the vector control strain harboring pET21a+ (A) or from the LpxO-over-expressing strain harboring pHSG2 (B). The [M-2H]2− ions at m/z 1117.681 and at m/z 1125.682 correspond to the substrate (Kdo2-lipid A) and its hydroxylated product, respectively. The [M-3H]3− ions of these two species (at m/z 744.790 and m/z 750.124 respectively) are also very prominent, as noted previously (38). The triply charged ions near m/z 688.72, seen in both samples, arise by neutral loss of the 3′-secondary acyl chain from the triply charged Kdo2-lipid A ions. The extensive neutral loss from the triply charged ions is likely due to the fact that the triply charged ions undergo more energetic collisional activation in the ion source region during ESI/MS than do the doubly charged ions (38). This unusual property of Kdo2-lipid A (and its hydroxylated derivative) confirms that the LpxO-dependent hydroxylation of Kdo2-lipid A occurs exclusively on the 3′-secondary acyl chain in vitro, as it does in vivo.
GC/MS analysis of the hydroxylated fatty acid generated in vitro
To prove that 2-hydroxymyristate is indeed generated in our in vitro system, the hydroxylated Kdo2-lipid A product formed by the LpxO-expressing membranes was subjected to GC/MS analysis, as described in the Materials and Methods, and compared to unmodified Kdo2-lipid A re-isolated from the vector control reaction mixture, as well as to the standards (3-hydroxymyristic acid and 2-hydroxymyristic acid). The vector control material (Fig. 9A) yielded only the TMS derivative of 3-hydroxymyristoylmethylester, as judged by its retention time, whereas the material from the LpxO expressing membranes contained an additional shoulder with the retention time (Fig. 9B) expected for the TMS derivative of 2-hydroxymyristoylmethylester. The EI/MS spectra of the major and minor peaks from Fig. 9B are shown in Fig. 9C and 9D, respectively. The spectra of these isomeric species are clearly different and correspond to the EI/MS spectra of the standards for a TMS derivative o f 3-hydroxymyristoylmethylester and of a TMS derivative of 2-hydroxymyristoylmethylester, respectively (see Supporting Information Fig. S1). The key fragmentations are indicated in the inserted structures. The fact that a 2-hydroxymyristoyl group is generated by LpxO on Kdo2-lipid A provides unequivocal evidence for the proposed location of the hydroxylation reaction (Fig. 2), based on the known structure of Kdo2-lipid A. The only remaining ambiguity is whether the in vitro system generates the physiological S-hydroxymyristate moiety or a mixture of stereoisomers.
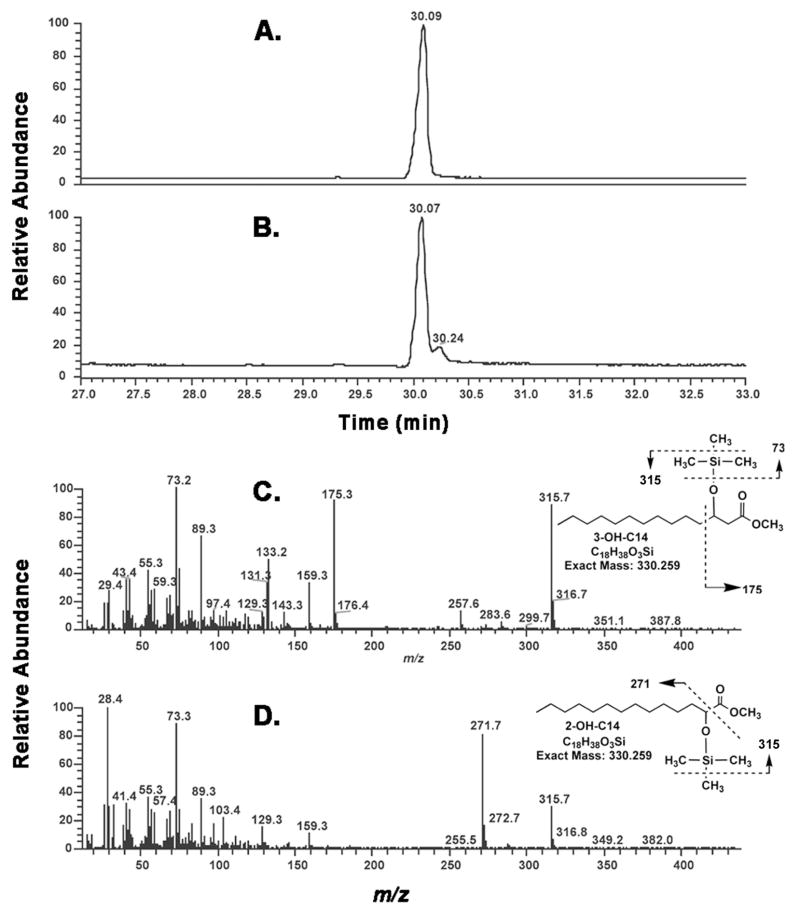
Figure 9. GC/MS analysis of the hydroxylated fatty acids in the vitro product generated by LpxO.
Kdo2-lipid A was incubated with vector control or LpxO membranes, as described in the Materials and Methods. The Kdo2-lipid A or the hydroxylated product formed by LpxO were re-isolated as a mixture from the reaction system, hydrolyzed, and converted to TMS derivatives to generate the fatty acid methylesters. Panel A. Total ion chromatogram (TIC) of the GC/MS analysis of the hydroxy fatty acids from the vector control reaction, showing the presence of a single peak consistent with a TMS-3-hydroxymyristoylmethylester. Panel B. TIC of the GC/MS analysis of the hydroxy fatty acids from the LpxO reaction system, showing the presence a shoulder peak with the expected retention time for a TMS-2-hydroxymyristoylmethylester. The peak area ratio between the TMS-3-hydroxymyristoylmethylester and the TMS-2-hydroxymyristoylmethylester is about 5 to 1. Panel C. EI/MS spectrum of the major leading peak in panel B (retention time: 30.07 min), showing identity with a TMS-3-hydroxymyristoylmethylester standard (see Supporting Information Fig. S1). Panel D. EI/MS spectrum of the minor lagging peak in panel B (retention time: 30.24 min), showing identity with a TMS-2-hydroxymyristoylmethylester standard (see Supporting Information Fig. S1). The proposed origins of the major fragments are indicated.
DISCUSSION
Although 2-hydroxyacyl chains have been recognized as constituents of some LPSs for over thirty years (28, 53–58), the biosynthesis and function of these moieties remain poorly characterized. A possible mechanism for its biosynthesis was suggested by the discovery of the Salmonella lpxO gene (27), which encodes a novel member of the Fe2+/O2/α-ketoglutarate-dependent dioxygenase family. Expression of lpxO in E. coli K-12, which normally lacks this gene, confers upon E. coli the ability to synthesize 2-hydroxymyristate modified lipid A in the presence of O2 (27). Conversely, S. typhimurium mutants lacking lpxO (5) lose the ability to generate lipid A species containing 2-hydroxymyristate (Fig. 3B).
In the present study we have devised the first in vitro enzymatic assay system for detecting and quantifying LpxO activity, predicated on the assumption that LpxO is an Fe2+/O2/α-ketoglutarate-dependent dioxygenase (Fig. 2A). Hexa-acylated Kdo2-[4′-32P]lipid A from E. coli (23, 38) was used as the acceptor substrate in the presence of appropriate cofactors (Fig. 2A) and membranes from an E. coli strain that over-expresses Salmonella LpxO. As shown in Figs. 5 and 6, LpxO activity is proportional to time and protein concentration, and it is dependent upon α-ketoglutarate and ascorbate (Fig. 6A). It is stimulated by added Fe2+ (Fig. 6A) and is inhibited by the chelator 2,2′-bis-pyridyl (Fig. 6B). The inclusion of catalase in the assay system (59, 60) partially eliminates the ascorbate requirement (data not shown), demonstrating that ascorbate is not absolutely necessary. Furthermore, E. coli cells do not synthesize ascorbate but nevertheless carry out lipid A 2-hydroxylation when lpxO is over-expressed from a hybrid plasmid (27). Some other endogenous reductant must be substituting for ascorbate in vivo.
LpxO activity is found exclusively in the particulate fraction (Fig. 4A), consistent with the hydropathy analysis (Fig. 2B). Its active site presumably is localized on the inner surface of the inner membrane. This topography would give LpxO access to its water-soluble co-substrates (Fig. 2A), particularly to Fe2+, which is readily converted to Fe3+ in an aqueous aerobic environment. Further evidence for the topography of LpxO comes from experiments in which LpxO is expressed in an E. coli strain harboring a temperature-sensitive point mutation in the ABC transporter MsbA (12). These mutants fail to transport newly synthesized core-lipid A across the inner membrane after 30 minutes at 44 °C (12, 40). However, under these conditions lipid A is still synthesized and 2-hydroxylated normally in this mutant (12). This finding demonstrates that 2-hydroxylation occurs before MsbA-driven transport of core-lipid A to the periplasmic surface of the inner membrane (1, 40). The other covalent modifications of lipid A, shown in Fig. 1, are catalyzed by extra-cellular enzymes, which are located either on the periplasmic surface of the inner membrane or in the outer membrane (1). Consequently, these enzymes are all MsbA dependent in living cells (1).
How LpxO orthologues discriminate between the secondary acyl chains of lipid A, or for that matter between lipid A and glycerophospholipids, is uncertain. One important observation is that LpxO, like LpxL (61), is dependent upon the presence of the Kdo disaccharide, thereby targeting LpxO to LPS. In S. typhimurium and S. minnesota lipid A, the secondary 2-hydroxymyristate moiety appears to be attached exclusively to the 3′-R-3-hydroxymyristate chain (Fig. 1) (14, 27, 62). P. aeruginosa contains two LpxO orthologues (63), and in fact the two secondary acyl chains of its lipid A are both modified with 2-OH groups (64, 65). Each of these LpxO orthologues is probably responsible for hydroxylating only one of the two secondary acyl chains of P. aeruginosa lipid A, which are located at positions 2 and 2′ (64). Purification of LpxO to homogeneity and structural studies will be needed to address the details of its substrate selectivity. The availability of pure enzyme would also facilitate the unequivocal demonstration of stoichiometric formation of succinate and CO2 in conjunction with lipid A 2-hydroxylation (Fig. 2A).
The phosphatidylethanolamine species of Burkholderia cepacia (66–68) and the ornithine amide lipids Rhizobium tropici (69) contain putative 2-hydroxyacyl chains. These organisms possess genes that are distantly related to lpxO (69). The single lpxO homologue present in R. tropici (known as olsC) is strictly required for the 2-hydroxylation of the ornithine lipids, as judged by characterization of an olsC mutant (69). LpxO contains the canonical HXDX40–60H motif, which is characteristic of most Fe2+/O2/α-ketoglutarate-dependent hydroxylases. In primary sequence OlsC (69) more closely resembles the mammalian aspartate β-hydroxylase (70–72), but in vitro assays for OlsC have not yet been reported.
The function of lipid A 2-hydroxylation remains uncertain. Based on its chromatographic properties, a large fraction of the lipid A species isolated from S. typhimurium grown inside of macrophage tumor cells is 2-hydroxylated (5). However, Salmonella lpxO mutants are fully virulent in RAW 264.7 macrophage tumor cells (5) and in a mouse infection model (E. Romilianus and D. Maskell, personal communication). There are no gross defects in outer membrane permeability, as judged by disc diffusion assays with erythromycin or rifampicin on LB broth plates (data not shown). However, plate-based assays may not be sensitive enough to reveal subtle roles for lpxO in regulating outer membrane permeability. Recently, Nikaido and coworkers reported that an lpxO mutant bound more of a hydrophobic dye than did the isogenic wild-type strain (29, 30), implying a role for LpxO in the regulation of outer membrane permeability to some compounds. Testing the membrane permeability of strains containing the lpxO::kan mutation in combination with mutations of other genes involved in lipid A modification (Fig. 1), as recently reported for ethidium uptake (30), might reveal additional phenotypes.
The relatively wide distribution of LpxO orthologues among diverse Gram-negative bacteria suggests that they must play an important biological role. A PSI-BLAST search of the current non-redundant protein database reveals close orthologues (> 50% amino acid sequence identity) in strains of Burkholderia, Pseudomonas, Serratia, Klebsiella, Xanthomonas, Ralstonia, Xylella, Acinetobacter, Chromobacterium, Bordetella, Azotobacter, Bradyrhizobium and many others. A mutant of R. tropici with a transposon insertion in the olsC gene lacks 2-hydroxylated ornithine amide lipids and is deficient in colonization of root nodules (69). Like LpxO, with which it shares 35% sequence identity and 51% similarity over 168 amino acids, OlsC is predicted to be an inner membrane protein (69).
We are currently attempting to solubilize and purify S. typhimurium LpxO. We have generated a C-terminal hexa-histidine fusion construct, the in vitro activity of which is indistinguishable from that of wild-type LpxO (unpublished results). Assuming that the enzyme can be stabilized in the presence of detergents, purification of LpxO and related hydroxylases to homogeneity should be possible, permitting direct assessment of its metal content and catalytic mechanism.
Supplementary Material
Fig. S1. GC/MS analysis of 3-hydroxymyristate and 2-hydroxymyristate standards as the TMS derivatives of their methyl esters. Sample preparation and GC/MS conditions are described in the manuscript.
Acknowledgments
This research was supported by National Institutes of Health Grant GM-51310 to C. R. H. Raetz. The mass spectrometry facility in the Department of Biochemistry of the Duke University Medical Center and Dr. Ziqiang Guan are supported by the LIPID MAPS Large Scale Collaborative Grant number GM-069338 from NIH.
List of Abbreviations
- L-Ara4N
4-amino-4-deoxy-L-arabinose
- BSA
bovine serum albumin
- EI/MS
electron impact/mass spectrometry
- ESI/MS
electrospray ionization/mass spectrometry
- GC/MS
gas chromatography/mass spectrometry
- HEPES
N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- IPTG
isopropyl thio-β-D-galactoside
- Kdo
3-deoxy-D-manno-octulosonic acid
- LPS
lipopolysaccharide
- pEtN
phosphoethanolamine
- TIC
total ion chromatogram
- TLC
thin layer chromatography
- TMS
trimethylsilyl
Footnotes
Supporting Information Available
This material is available free of charge via the internet at: http://pubs.acs.org
References
- 1.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 3.Groisman EA, Kayser J, Soncini FC. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons HS, Kalb SR, Cotter RJ, Raetz CRH. Role of Mg++ and pH in the modification of Salmonella lipid A following endocytosis by macrophage tumor cells. Mol Microbiol. 2005;55:425–440. doi: 10.1111/j.1365-2958.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 6.Roland KL, Martin LE, Esther CR, Spitznagel JK. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helander IM, Kilpelainen I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol. 1994;11:481–487. doi: 10.1111/j.1365-2958.1994.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 8.Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CRH. Accumulation of a polyisoprene-linked amino sugar in polymyxin resistant mutants in Salmonella typhimurium and Escherichia coli. Structural characterization and possible transfer to lipid A in the periplasm. J Biol Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- 9.Trent MS, Raetz CRH. Cloning of EptA, the lipid A phosphoethanolamine transferase associted with polymyxin resistance. J Endotoxin Res. 2002;8:158. [Google Scholar]
- 10.Lee H, Hsu FF, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. An inner membrane enzyme in Salmonella typhimurium and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A. Induction in polymyxin resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 12.Doerrler WT, Gibbons HS, Raetz CRH. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 13.Belden WJ, Miller SI. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 16.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CRH. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. Embo J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. Embo J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trent MS, Pabich W, Raetz CRH, Miller SI. A PhoP/PhoQ- induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 21.Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, de Haan A, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanipes MI, Lin S, Cotter RJ, Raetz CRH. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J Biol Chem. 2001;276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 24.Breazeale SD, Ribeiro AA, McClerren AL, Raetz CRH. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J Biol Chem. 2005;280:14154–14167. doi: 10.1074/jbc.M414265200. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki K, Ernst RK, Miller SI. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J Biol Chem. 2004;279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbons HS, Lin S, Cotter RJ, Raetz CRH. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe(II)/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275:32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- 28.Bryn K, Rietschel ET. L-2-hydroxytetradecanoic acid as a constituent of Salmonella lipopolysaccharides (lipid A) Eur J Biochem. 1978;86:311–315. doi: 10.1111/j.1432-1033.1978.tb12312.x. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ- mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prescott AG, Lloyd MD. The iron(II) and 2-oxoacid-dependent dioxygenases and their role in metabolism. Nat Prod Rep. 2000;17:367–383. doi: 10.1039/a902197c. [DOI] [PubMed] [Google Scholar]
- 32.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 33.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 34.Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc Natl Acad Sci U S A. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Nature’s inventory of halogenation catalysts: oxidative strategies predominate. Chem Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 36.Hegg EL, Que L. The 2-His-1-carboxylate facial triad - an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur J Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 37.Raetz CRH. Regulated covalent modifications of lipid A. J Endotoxin Res. 2001;7:73–78. [PubMed] [Google Scholar]
- 38.Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. (Kdo)2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting trans-membrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 40.Doerrler WT, Reedy MC, Raetz CRH. An Escherichia coli mutant defective in lipid export. J Biol Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 41.Miller JR. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 42.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 43.Dulbecco R, Vogt M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J Exp Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bligh EG, Dyer JJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 45.Tran AX, Karbarz MJ, Wang X, Raetz CRH, McGrath SC, Cotter RJ, Trent MS. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myllyharju J, Kivirikko KI. Characterization of the iron- and 2- oxoglutarate-binding sites of human prolyl 4-hydroxylase. Embo J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gronke RS, VanDusen WJ, Garsky VM, Jacobs JW, Sardana MK, Stern AM, Friedman PA. Aspartyl beta-hydroxylase: in vitro hydroxylation of a synthetic peptide based on the structure of the first growth factor-like domain of human factor IX. Proc Natl Acad Sci U S A. 1989;86:3609–3613. doi: 10.1073/pnas.86.10.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z, Lin S, Cotter RJ, Raetz CRH. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy- L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 49.Kawahara K, Uchida K, Aida K. Direct hydroxylation in the biosynthesis of hydroxy fatty acid in lipid A of Pseudomonas ovalis. Biochim Biophys Acta. 1979;572:1–8. [PubMed] [Google Scholar]
- 50.Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CRH. Lipid A modifications in polymyxin resistant Salmonella typhimurium. PmrA dependent 4-amino-4-deoxy-L-arabinose and phosphoethanolamine incorporation. J Biol Chem. 2001;276:43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- 51.Carman GM, Deems RA, Dennis EA. Lipid signaling enzymes and surface dilution kinetics. J Biol Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 52.Ryle MJ, Liu A, Muthukumaran RB, Ho RY, Koehntop KD, McCracken J, Que L, Jr, Hausinger RP. O2- and alpha-ketoglutarate-dependent tyrosyl radical formation in TauD, an alpha-keto acid-dependent non-heme iron dioxygenase. Biochemistry. 2003;42:1854–1862. doi: 10.1021/bi026832m. [DOI] [PubMed] [Google Scholar]
- 53.Humphreys GO, Hancock IC, Meadow PM. Synthesis of the hydroxyacids in lipid A of Pseudomonas aeruginosa. J Gen Microbiol. 1972;71:221–230. doi: 10.1099/00221287-71-2-221. [DOI] [PubMed] [Google Scholar]
- 54.Hancock JC, Humphreys GO, Meadow PM. Characterization of the hydroxy acids of Pseudomonas aeruginosa 8602. Biochim Biophys Acta. 1970;202:389–391. doi: 10.1016/0005-2760(70)90204-3. [DOI] [PubMed] [Google Scholar]
- 55.Welch DF. Applications of cellular fatty acid analysis. Clin Microbiol Rev. 1991;4:422–438. doi: 10.1128/cmr.4.4.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawai Y, Moribayashi A. Characteristic lipids of Bordetella pertussis: simple fatty acid composition, hydroxy fatty acids, and an ornithine-containing lipid. J Bacteriol. 1982;151:996–1005. doi: 10.1128/jb.151.2.996-1005.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rietschel ET. Absolute configuration of 3-hydroxy fatty acids present in lipopolysaccharides from various bacterial groups. Eur J Biochem. 1976;64:423–8. doi: 10.1111/j.1432-1033.1976.tb10318.x. [DOI] [PubMed] [Google Scholar]
- 58.Jantzen E, Knudsen E, Winsnes R. Fatty acid analysis for differentiation or Bordetella and Brucella species. Acta Pathol Microbiol Immunol Scand [B] 1982;90:353–359. doi: 10.1111/j.1699-0463.1982.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 59.De Jong L, Kemp A. Stoicheiometry and kinetics of the prolyl 4- hydroxylase partial reaction. Biochim Biophys Acta. 1984;787:105–111. doi: 10.1016/0167-4838(84)90113-4. [DOI] [PubMed] [Google Scholar]
- 60.Myllyla R, Kaska DD, Kivirikko KI. The catalytic mechanism of the hydroxylation reaction of peptidyl proline and lysine does not require protein disulphide-isomerase activity. Biochem J. 1989;263:609–611. doi: 10.1042/bj2630609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clementz T, Bednarski JJ, Raetz CRH. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A. HtrB catalyzed incorporation of laurate. J Biol Chem. 1996;271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 62.Zahringer U, Lindner B, Rietschel ET. Chemical structure of lipid A: recent advances in structural analysis of biologically active molecules. In: Brade H, Opal SM, Vogel SN, Morrison DC, editors. Endotoxin in Health and Disease. Marcel Dekker, Inc; New York: 1999. pp. 93–114. [Google Scholar]
- 63.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 64.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 65.Kulshin VA, Zahringer U, Lindner B, Jager KE, Dmitriev BA, Rietschel ET. Structural characterization of the lipid A component of Pseudomonas aeruginosa wild-type and rough mutant lipopolysaccharides. Eur J Biochem. 1991;198:697–704. doi: 10.1111/j.1432-1033.1991.tb16069.x. [DOI] [PubMed] [Google Scholar]
- 66.Phung LV, Tran TB, Hotta H, Yabuuchi E, Yano I. Cellular lipid and fatty acid compositions of Burkholderia pseudomallei strains isolated from human and environment in Viet Nam. Microbiol Immunol. 1995;39:105–116. doi: 10.1111/j.1348-0421.1995.tb02176.x. [DOI] [PubMed] [Google Scholar]
- 67.Taylor CJ, Anderson AJ, Wilkinson SG. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology. 1998;144(Pt 7):1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- 68.Cox AD, Wilkinson SG. Polar lipids and fatty acids of Pseudomonas cepacia. Biochim Biophys Acta. 1989;1001:60–67. doi: 10.1016/0005-2760(89)90307-x. [DOI] [PubMed] [Google Scholar]
- 69.Rojas-Jimenez K, Sohlenkamp C, Geiger O, Martinez-Romero E, Werner D, Vinuesa P. A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol Plant Microbe Interact. 2005;18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- 70.Jia S, VanDusen WJ, Diehl RE, Kohl NE, Dixon RA, Elliston KO, Stern AM, Friedman PA. cDNA cloning and expression of bovine aspartyl (asparaginyl) beta-hydroxylase. J Biol Chem. 1992;267:14322–14327. [PubMed] [Google Scholar]
- 71.Lancaster DE, McDonough MA, Schofield CJ. Factor inhibiting hypoxia-inducible factor (FIH) and other asparaginyl hydroxylases. Biochem Soc Trans. 2004;32:943–945. doi: 10.1042/BST0320943. [DOI] [PubMed] [Google Scholar]
- 72.McGinnis K, Ku GM, VanDusen WJ, Fu J, Garsky V, Stern AM, Friedman PA. Site-directed mutagenesis of residues in a conserved region of bovine aspartyl (asparaginyl) beta-hydroxylase: evidence that histidine 675 has a role in binding Fe2+ Biochemistry. 1996;35:3957–3962. doi: 10.1021/bi951520n. [DOI] [PubMed] [Google Scholar]
- 73.Strain SM, Armitage IM, Anderson L, Takayama K, Qureshi N, Raetz CRH. Location of polar substituents and fatty acyl chains on lipid A precursors from a 3-deoxy-D-manno-octulosonic acid-deficient mutant of Salmonella typhimurium: Studies by 1H, 13C and 31P nuclear magnetic resonance. J Biol Chem. 1985;260:16089–16098. [PubMed] [Google Scholar]
- 74.Zhou Z, Ribeiro AA, Raetz CRH. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-L-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J Biol Chem. 2000;275:13542–13551. doi: 10.1074/jbc.275.18.13542. [DOI] [PubMed] [Google Scholar]
- 75.Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the B. pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 76.Brabetz W, Muller-Loennies S, Holst O, Brade H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur J Biochem. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 77.Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2007.06044.x. 2008 in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. GC/MS analysis of 3-hydroxymyristate and 2-hydroxymyristate standards as the TMS derivatives of their methyl esters. Sample preparation and GC/MS conditions are described in the manuscript.