Abstract
Recently, a potentially powerful strategy based on the of phage-display libraries has been presented to target tumors via homing peptides attached to nanoparticles. The Cys-Arg-Glu-Lys-Ala (CREKA) peptide sequence has been identified as a tumor-homing peptide that binds to clotted plasmas proteins present in tumor vessels and interstitium. The aim of this work consists of mapping the conformational profile of CREKA to identify the bioactive conformation. For this purpose, a conformational search procedure based on modified Simulated Annealing combined with Molecular Dynamics was applied to three systems that mimic the experimentally used conditions: (i) the free peptide; (ii) the peptide attached to a nanoparticle; and (iii) the peptide inserted in a phage display protein. In addition, the free peptide was simulated in an ionized aqueous solution environment, which mimics the ionic strength of the physiological medium. Accessible minima of all simulated systems reveal a multiple interaction pattern involving the ionized side chains of Arg, Glu and Lys, which induces a β-turn motif in the backbone observed in all simulated CREKA systems.
Introduction
Over the last few years Ruoslahti and coworkers1–3 have identified a series of tumor-homing peptides by using in vivo screening of peptide libraries. These tumor-homing peptides were obtained from phage display libraries targeting highly-expressed molecular receptors in tumor blood vessels. Nanoparticles coated by the homing peptides were shown to efficiently focus at these sites and the potential of these nanoparticles for imaging and as drug-carriers has already been indicated. Among the homing peptides discovered by in vivo phage screening is a linear peptide that contains only 5 amino acids with the sequence CREKA (Cys-Arg-Glu-Lys-Ala). Synthesized CREKA peptide labeled with an attached fluorescent dye was detectable in human tumors from minutes to hours after intravenous injection, while it was essentially undetectable in normal tissues.3 In vivo experiments revealed that CREKA bound to clotted plasma proteins, establishing its being a clot-binding peptide. The CREKA peptide was further linked to amino dextran-coated iron oxide (SPIO) nanoparticles.3 Strikingly, experiments with CREKA-SPIO nanoparticles indicated that they not only bind to blood and plasma clots but can also effectively induce further localized tumor clotting, thus amplifying the nanoparticle homing. It was further found that the chemical nature of the nanoparticle is not important for this activity since both CREKA-SPIO nanoparticles and CREKA-coated liposomes were found to cause clotting in tumor vessels. A similar activity, i.e. binding to plasma clot, was observed when CREKA was inserted into phage display proteins.4 The clotting caused by CREKA in tumor vessels is expected to be useful in improving tumor detection by microscopic- and whole body-imaging techniques as well to tumor treatment by physical blockage of vessels through local embolism and drug-carrying nanoparticles.
At the same time, in spite of the potential interest in CREKA in cancer diagnostics and therapeutics, the application of this and other homing peptide sequences might be endangered by short half life times; after injection endogenous proteolytic enzymes could rapidly digest the peptides. Thus, protection from proteases is an important step in the development of potential applications of tumor-homing peptides. Several strategies have been proposed to protect proteogenic peptide sequences from proteases; among these, targeted replacements with synthetic amino acids have been the most successful.5–12 Selective incorporation of synthetic amino acids induces a significant resistance against proteases not only at the mutated position but also at neighboring amino acids. Non-proteogenic amino acids are compounds with many potential applications in Nanobiology, and have been found to be very useful for the re-engineering of physical protein modules and the generation of nanodevices.13–15.
Within the framework of a broad project devoted to improve the affinity of CREKA to its targets as well as to impart resistance against proteases, we have started by performing a computational analysis on such peptide sequence. Thus, the first step prior to the design of synthetic analogs which incorporate non-proteogenic amino acids, consists of the identification of the conformational profile and, if possible, the bioactive conformation of the peptide. Although no information about the conformational profile of this peptide is available, that is, the CREKA sequence does not occur in the Protein Data Base, experiments using the free peptide, the CREKA-SPIO nanoparticles and phage display proteins containing expressed CREKA suggest that the bioactive conformation for this sequence does not depend on the molecular environment.1–4 Identification of the low-energy regions of the conformational profiles of the natural CREKA sequence is important since any candidate synthetic analogue should not present significant differences with respect to the native. Theoretical methods based on computer simulations of atomic models have proven to be powerful techniques to sample the conformational space of biomolecules.16–18
In this work we use computer simulation methods based on Molecular Dynamics (MD) to characterize the conformational profile of the CREKA sequence and identify the corresponding bioactive conformation. For this purpose we used a multiple conformational search strategy considering the three experimentally tested environments: free peptide, peptide attached to a nanoparticle, and a peptide inserted in a phage display protein. The influence of the ionic strength on the conformational preferences of CREKA has also been examined. We note that computer simulation methods are usually applied considering a dilute solution of pure water while physiological media consists of ionized aqueous solutions. Accordingly, in this work we decided to examine how the addition of NaCl molecules to the aqueous environment affects the three-dimensional organization of the peptide.
Simulated Systems and Bioactive Conformation
The conformational profiles of four different molecular systems based on the CREKA peptide sequence have been determined using atomistic computational methods. The four systems under consideration are the following:
System I
The CREKA linear peptide was capped at the N- and C-terminus with acetyl (Ac) and N-methyl amide (NHMe) groups respectively, and was immersed in a dilute aqueous solution.
System II
CREKA was blocked at the ends with Ac and NHMe and was immersed in a solution that mimics the ionic strength of the physiological medium. The ionized solution was obtained by adding sodium and chloride ions to the environment. Comparison between the conformational profiles obtained for systems I and II should permit testing the influence of the simulation conditions on the conformational preferences of the peptide.
System III
CREKA was attached to a surface through the sulfhydryl group of the Cys residue. This surface was formed by 100 spherical particles, which were distributed in a 10×10 square (47.5 Å2), with van der Waals parameters [R= 2.35 Å and ε= 0.90 kcal/mol] and without electric charge. The whole system was immersed in a dilute aqueous solution. System III mimics the situation in which CREKA is bound to the surface of a SPIO nanoparticle. It should be noted that, in this simple model, electrostatic charges were omitted for the particles used to represent the surface. This is because many other chemical species that should induce a significant screening in the electrostatic interaction between the surface and the peptide have been also neglected in this model, e.g. neighboring peptides attached to the particle (about 8000 peptides were attached to each particle), ionic salts. The conformational freedom of the peptide is expected to be lower in III than in I and II because of the effect of having the end tethered to the surface.
System IV
This molecular system mimics the CREKA peptide when it is inserted in a phage display protein. Copies of the expressed peptides are usually presented on the flexible regions of the phage display proteins, for example in loops. In order to mimic such flexible regions, two Gly were added to each termini of CREKA, making the peptide sequence of system IV GGCREKAGG. The two Gly located at both ends of the chain were attached to spherical particles similar to those used in III to construct the surface. Such particles represent the secondary structure motifs that tether the flexible regions to the protein. The separation between the two added particles was restrained to a distance D through a force constant, k= 5 kcal·mol−1·Å−2, and the whole system was immersed in a dilute aqueous solution. The conformational profiles of IV were characterized considering D= 5.0, 7.0, 10.0, 15.0 and 20.0 Å. It is expected that such wide range of values will allow sampling a large number of potential positions and configurations in which the CREKA peptide is inserted in the flexible regions of phage display proteins.
Although the conformational profiles of I, II, III and IV may be significantly different, the bioactive conformation of the CREKA peptide sequence should appear within the set of lower-energy structures of each system. As stated in the Introduction, the experimental results suggest that the bioactive conformation is unique for this sequence independently of the molecular environment, i.e. the bioactive conformations when the peptide is free, attached to a SPIO nanoparticle or inserted in a phage display protein are practically identical. At the same time, the bioactive conformation does not necessarily correspond to the lowest minimum energy conformation, even though it must be contained within the set of accessible energy minima, i.e. the energy of the bioactive conformation is relatively close to that of the global minimum. Accordingly, a thorough exploration of the conformational space of the four CREKA-based systems is necessary to identify such a bioactive conformation.
Methods
Conformational Search
The structural features of the four CREKA-based systems studied in this work were explored using a modified simulated annealing combined with MD (SA-MD) as a sampling technique.19,20 In the SA-MD strategy, which relies on temperature evolution in time, the starting temperature is gradually reduced during the simulation. Thus, the system is given the opportunity to surmount energy barriers in a search for conformations with energies lower than the local-minimum typically found by energy minimization. However, in practice, when used with a regular force-field, SA-MD does not always lead the system to the most stable region at the end of the simulation. Recent studies demonstrated that very low energies are obtained by minimizing the energy of structures generated at the initial and intermediate states of a SA-MD process.21,22 In the present study we applied the latter strategy to locate the lower-energy minimum structures of the CREKA-based systems. A large number of structures (500) were selected for energy minimization from each SA-MD cycle, which is justified since the temperature was decreased very slowly along a relatively large trajectory (see below). As mentioned above, the bioactive conformation does not necessarily correspond to the lowest energy minimum, even though it should be a low-energy structure. Accordingly, the sampling technique applied to the four systems under study must be robust enough to obtain low-energy structures that may be quasi-degenerate with the global minimum but situated in different valleys of the peptide landscape.
Molecular Dynamics and Computational Details
Each simulated system was placed in the center of a cubic simulation box (a= 47.5Å) filled with at least 3405 explicit water molecules, which were represented using the TIP3 model.23 One negatively charged chloride and two positively charged sodium atoms were added to the simulation box to reach electric neutrality (net charges were considered for Arg, Lys and Glu residues at neutral pH). For the conformational search of free CREKA under physiological conditions (system II), four additional NaCl molecules were added to the simulation box.
Before the production cycles with the modified SA-MD, the content and the density of the simulation box were equilibrated. For this purpose, each system was initially minimized to relax the conformational and structural tension using the conjugate gradient method. Next, different consecutive rounds of short MD runs were performed. Thus, 0.5 ns of NVT-MD at 500 K were used to homogeneously distribute the solvent and ions in the box. Next, 0.5 ns of NVT-MD at 298 K (thermal equilibration) and 0.5 ns of NPT-MD at 298 K (density relaxation) were run. The last snapshot of the NPT-MD was used as the starting point for the conformational search process. This initial structure was quickly heated to 900 K at a rate of 50 K/ps to force the molecule to jump to a different region of the conformational space. Along 10 ns, the 900 K-structure was slowly cooled to 500 K at a rate of 1 K per 25 ps. A total of 500 structures were selected and subsequently minimized during the first cycle of modified SA-MD. The resulting minimum energy conformations were stored in a rank-ordered library of low energy structures. The lowest energy minimum generated in a modified SA-MD cycle was used as starting conformation of the next cycle.
The 100 spherical particles used to construct the surface in system III were kept fixed at the initial positions in all MD simulations and energy minimizations.
The energy was calculated using the AMBER force-field,24,25 with the required parameters taken from the AMBER libraries. Atom pair distance cutoffs were applied at 14.0 Å to compute the van der Waals and electrostatic interactions. Both temperature and pressure were controlled by the weak coupling method, the Berendsen thermo-barostat,26 using a time constant for heat bath coupling and a pressure relaxation time of 1 ps. Bond lengths were constrained using the SHAKE algorithm27 with a the numerical integration step of 2 fs.
Conformation Classification and Clustering Analysis
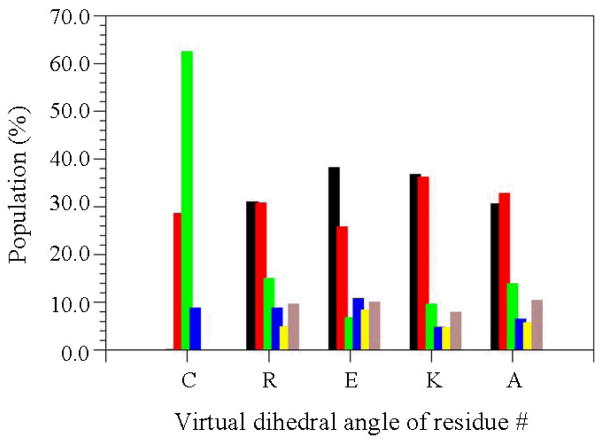
In order to construct a list of unique minimum energy conformations, the 500 minimized structures provided by each cycle of modified SA-MD were compared not only among themselves but also with unique minima generated in previous cycles. The list was organized by rank ordering all the unique minimum energy conformations found following an increasing order or energy. Previously listed conformations were discarded. After testing different criteria, the one selected to identify unique minimum energy conformations was based on the values of virtual dihedral angles, which were defined using main-chain atoms, and the presence of interaction patterns, i.e. salt bridges, hydrogen bonds and dipole-dipole interactions.
Five virtual dihedral angles were defined considering the α-carbon atoms of the CREKA peptide and one methyl hydrogen atom of the Ac and NHMe capping groups. The hydrogen atoms of Ac and NHMe were chosen instead of the methyl carbon atoms, to retain the two-particle separation imposed by the consecutive α-carbon atoms of the peptide sequence. Each of these virtual dihedral angles was associated with the conformation of one amino acid of the CREKA peptide. The existence of different interactions was accepted on the basis of the following geometric criteria: a) salt bridges: distance between the centers of the interacting groups shorter than 4.50 Å; b) hydrogen bonds: distance H···O (dH···O) shorter than 2.50 Å and angle ∠N-H···O higher than 120.0°; and (c) dipole-dipole: distance between dipoles shorter than 3.00 Å and the interaction has not been counted as hydrogen bond. Two structures were considered different when they differ in at least one of their virtual dihedral angles by more than 60° or in at least one of the interactions counted.
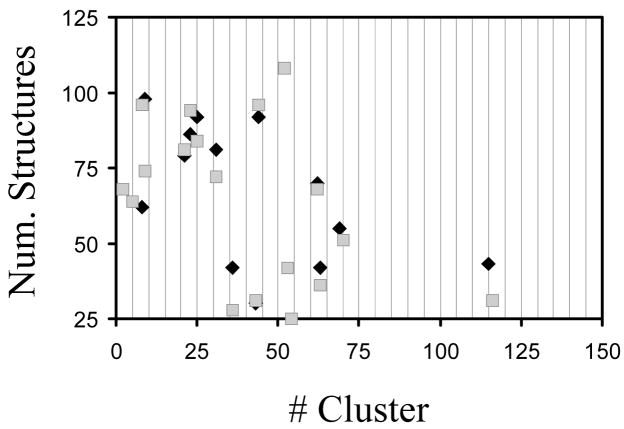
For each system, all the structures categorized as different were subsequently clustered according to a criterion based on the presence of the intramolecular interactions mentioned above: salt bridges and hydrogen bonds. For each studied system, a representation of the number of clusters versus the number of structures within a given cluster is provided in the Supporting Information.
Results
Free CREKA in Unionized and Ionized Aqueous Solution (Systems I and II)
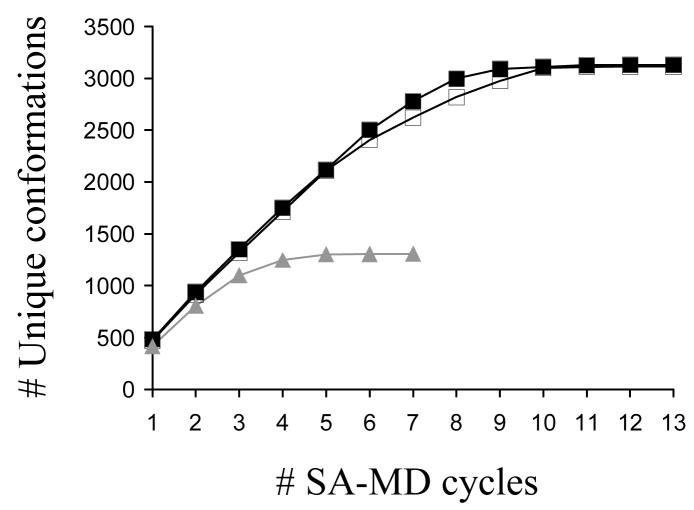
Thirteen production cycles of modified SA-MD were run for each of the two systems, with 500 selected structures minimized from each cycle. This procedure led to 3130 and 3114 unique minimum energy conformations for systems I and II, respectively. Figure 1, which represents the evolution of the number of unique conformations against the number of SA-MD cycles, clearly indicates that the conformational search converges for the two systems. However, interestingly, the molecular dynamics of the conformational search procedure was influenced by the chemical nature of the aqueous solution. Thus, inspection of the energies of the unique conformations obtained in each SA-MD cycle indicates that in unionized aqueous solution (system I) the energy of at least one new unique conformation is lower than those of the conformations previously generated for cycles 2, 3, 4 and 8, i.e. the global minimum appears in the 8th cycle of modified SA-MD. In contrast, in the ionized solution (system II) the lowest energy conformation of free CREKA appears after only 3 cycles, and no improvement in the energy has been detected subsequently. Analysis of the number of new unique conformations generated by each cycle reveals that it is lower than 1% after the 9th and 11th cycle for I and II, respectively.
Figure 1.
Number of unique minimum energy conformations found for systems I (black line, solid squares), II (black line, empty squares) and III (grey line, solid triangles) against the number of modified SA-MD cycles used in the conformational search process.
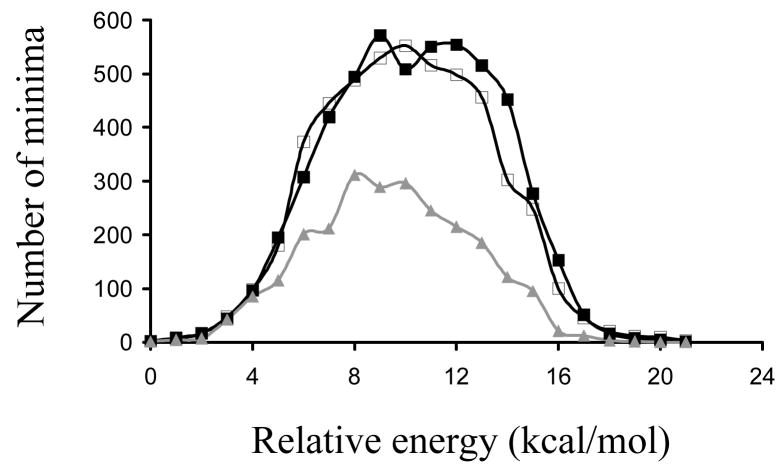
Figure 2 shows the distribution of energies, which are relative to that of the global minimum, for the unique minimum energy conformations of I and II. As can be seen, the two profiles are very similar, with the highest energy bordering ~23 kcal/mol in both cases. The histograms displayed in Figure 3, which represent the distribution of the virtual dihedral angles used to define the conformation of the five CREKA residues (see Methods), permit a comparison of the similarity between the sets of unique minima detected for the free peptide in aqueous and ionized solutions. Thus, the incorporation of additional NaCl molecules into the aqueous medium does not alter, in general terms, the conformational profile of the free CREKA peptide. A more detailed comparison between the energy minima of systems I and II is provided in the Supporting Information section (Figure S1), which shows the distribution of the main chain dihedral angles Ψ,ϕ of the five CREKA residues. It is worth noting that special attention has been paid to the comparison of the structures with a relative energy lower than 2.0 kcal/mol, i.e. 27 and 23 structures for I and II respectively, which are expected to be the more significant ones. Accordingly, the values of Ψ,ϕ for these accessible energy minima are highlighted in the Ramachandran maps of Figure S1 (see Supporting Information). As can be seen, a very notable resemblance between the results reached for the two systems is indicated again. Overall, these results allow us to conclude that the conformational profile of the peptide is not affected by the increase of the ionic concentration.
Figure 2.
Distribution of energies for the minimum energy conformations found for systems I (black line, solid squares), II (black line, empty squares) and III (grey line, solid triangles). In each case represented energies are relative to the corresponding global minimum.
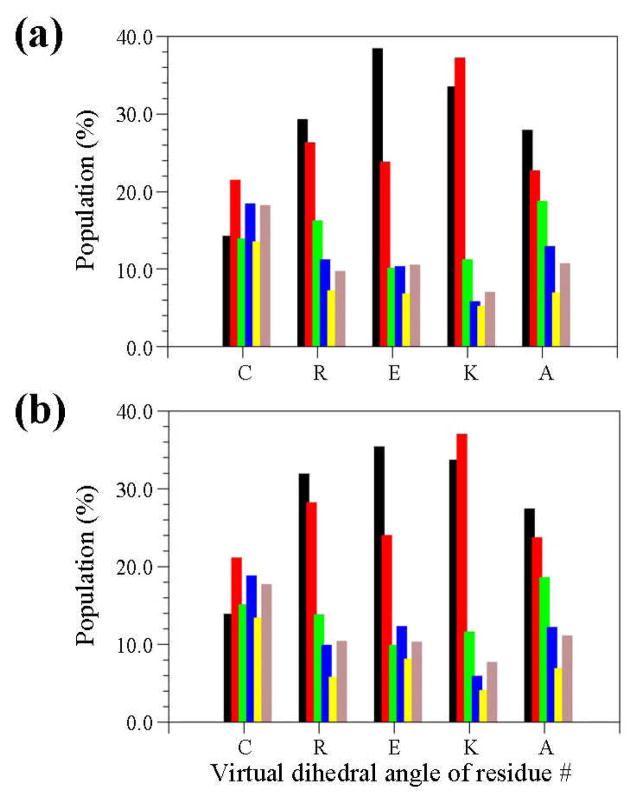
Figure 3.
Distribution of the virtual dihedral angles (see Methods) used to define the conformation of five CREKA residues in all the unique minima obtained for systems I (a) and II (b). Colour code for the bars is: black for dihedral angle values ranging from 0°-to-60°, red from 60°-to-120°, green from 120°-to-180°, blue from 180°-to-240°, yellow from 240°-to-300° and grey from 300°-to-360°.
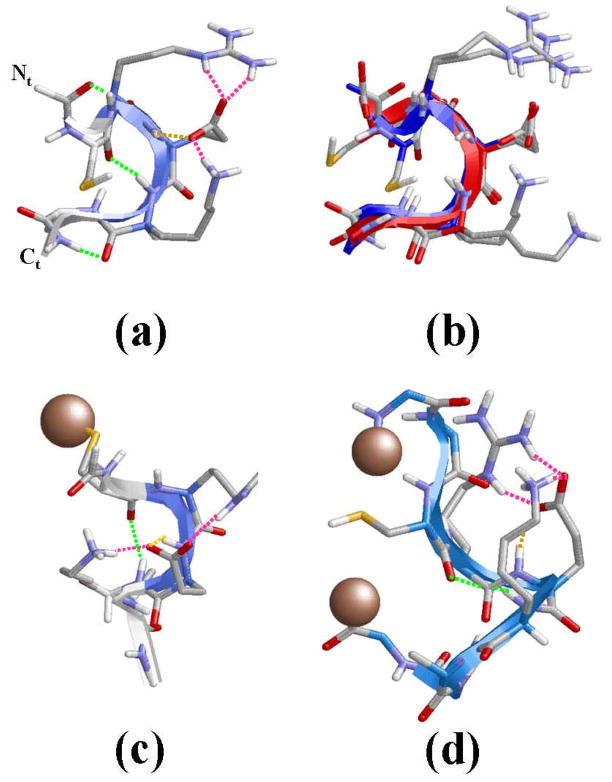
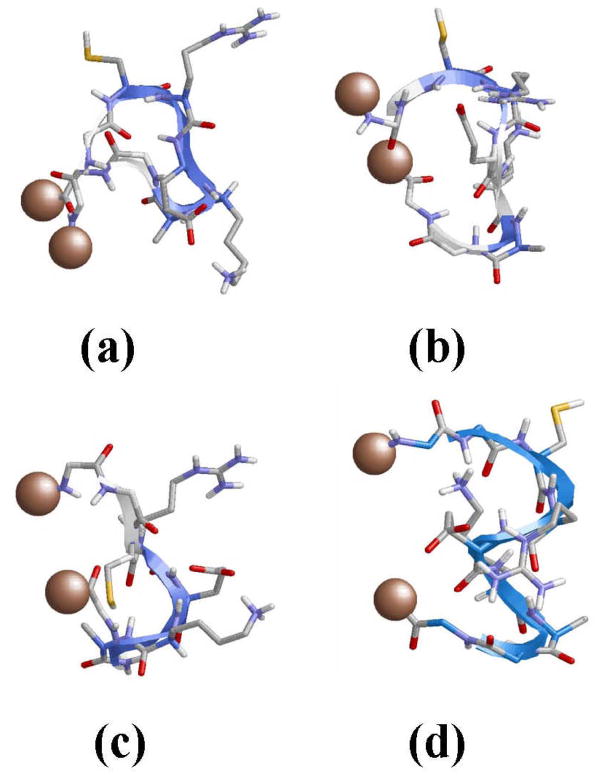
Figure 4a shows the lowest energy minimum obtained for system I. In this structure the backbone amide groups form intramolecular hydrogen bonds leading to the formation of two γ-turns and one β-turn. Specifically, γ-turns (7-membered intramolecular hydrogen bonds) are formed between the (Ac)C=O···H-N(Arg) [dH···O= 1.999 Å, ∠N-H···O= 149.2°] and (Lys)C=O···H-N(NHMe) [dH···O= 1.828 Å, ∠N-H···O= 153.8°], where Ac and NHMe are the blocking groups at the Nt- and Ct-termini, respectively. The β-turn (10-membered intramolecular hydrogen bond) involves the Cys and Lys residues: (Cys)C=O···H-N(Lys) [dH···O= 1.917 Å, ∠N-H···O= 168.0°]. The view provided in Figure 4a of the global minimum suggests that the (Cys)C=O and H-N(Ala) moieties form a 13-membered intramolecular hydrogen bond, which is the interaction typically found α-helices. However, the poor geometric parameters found in this case, [dH···O= 2.375 Å, ∠N-H···O= 112.3°], indicates that it should considered as a simple dipole-dipole interaction. Inspection of the side chains reveals a multiple interaction pattern formed by salt bridges between the negatively charged carboxylate group of Glu and the positively charged side chains of Arg and Lys. Furthermore, the carboxylate group of Glu also forms a hydrogen bond with its own backbone N-H amide group, i.e. one oxygen atom of the carboxylate forms two interactions with the guanidium group of Arg while the second oxygen participates in one salt bridge with the Lys side chain and one intra-residue hydrogen bond (Figure 4a).
Figure 4.
(a) Lowest energy minimum obtained for free CREKA in unionized aqueous solution (system I). (b) Superposition of the global minimum found for systems I (blue) and II (green). (c) Lowest energy minimum obtained for CREKA attached to a SPIO nanoparticle (system III). (d) Lowest energy minimum obtained for GGCREKAGG inserted in a phage display protein (system IV; D= 20.0 A). The surface used to mimic the nanoparticle in system III and the connections to the phage display protein in system IV have been represented using a single grey ball. Intramolecular hydrogen bonds between the backbone amide groups are displayed using green dashed lines, while side chain···side chain and side chain···backbone interactions are marked with pink (salt bridges) and orange (hydrogen bonds) dashed lines.
Figure 4b shows the superposition of the global minimum found for systems I and II. As can be seen, the two structures show a remarkable similarity in the backbone conformation, the only difference between them corresponding to the orientation of the Lys side chain. Specifically, in the global minimum of system II no salt bridge interaction between the Glu(C=O) and H-N(Lys) was detected. However, this small difference does not allow us to conclude that ionic concentration leads to structural changes in the accessible conformations.
A detailed analysis of the minima obtained for systems I and II below a relative energy threshold of 2 kcal/mol shows two common structural patterns for at least 90% of the relevant conformations: (i) a β-turn involving the Cys and Lys residues is systematically formed; and (ii) the ionized side chains of the three central residue are arranged to form a multiple interaction pattern, which is dominated predominantly by salt bridges.
Clustering analyses of systems I and II indicate that 69.5% (2165 structures) and 76.4% (2380 structures) minima present at least one intramolecular interaction (see Supporting Information). As can be seen, only 13 (I) and 19 (II) clusters contain more than 25 structures grouping 37.9% and 56.9% of the minima stabilized by intramolecular interactions, i.e. many clusters are constituted by a very small number of minimum. A more detailed analysis of the structures contained in the different clusters reveals that a total of 4243 and 4522 interactions were detected in the 2165 and 2380 minima of I and II, respectively, stabilized by intramolecular interactions, which is consistent with ~ 2 interactions per minimum. These interactions are distributed as follows:
System I. Main chain···main chain hydrogen bonds: 84.7% (3593 interactions); main chain···side chain hydrogen bonds: 6.7% (285 interactions); and side chain···side chain salt bridges: 8.6% (365 interactions).
System II. Main chain···main chain hydrogen bonds: 80.9% (3658 interactions); main chain···side chain hydrogen bonds: 5.7% (258 interactions); and side chain···side chain salt bridges: 13.4% (605 interactions).
Figure 5 compares the number of minimum energy conformations found for the more populated clusters of I and II. As can be seen, there is a quantitative agreement between the interaction patterns detected in the two systems. Thus, the more populated patterns are clearly conserved in the two systems. The overall of the results reflects a significant resemblance between the conformational profiles of I and II, which is also consistent with the small influence of the ionic concentration in the conformational profile of CREKA (see above).
Figure 5.
Number of minimum energy conformations found for the more populated clusters, i.e. those containing more than 25 structures, of I and II.
CREKA Attached to the Surface of a SPIO-Nanoparticle (System III)
After seven production cycles of modified SA-MD, which provided 1306 unique minima, the conformational search of system III fully converged. This is seen in Figure 1, which indicates that the last 3 cycles (minimization of 1500 selected structures) only provided 4 new unique conformations. The global minimum was obtained in the 2nd cycle of SA-MD reflecting the efficiency of the conformational search procedure. The number of unique minimum energy conformations is about 40% of those obtained for systems I and II indicating that, as expected, the conformational flexibility of the peptide becomes severely restricted when the sulfhydryl group of the Cys residue is attached to a surface. On the other hand, inspection of the distribution of the relative energies displayed in Figure 2 shows that the tethering of the peptide to the SPIO nanoparticle induces a significant reduction in both the height and width of the peak. The number of minima with a relative energy smaller than 2 kcal/mol is only 12, which means that the number of significant minima decreases by about 50–55% with respect to the free CREKA peptide.
The distribution of the virtual dihedral angles used to define the conformation of CREKA attached to a SPIO nanoparticle is displayed in Figure 6. Comparison with the histograms obtained for free CREKA (Figure 3) reveals very interesting features. First, the conformation of the Cys residue is completely different in the free peptide as compared to that of system III. Thus, the linking of the sulfhydryl group to the surface used to represent the SPIO nanoparticle induces significant perturbations in the backbone conformation of the Cys. Second, and most importantly, the conformational distribution of the backbone for the remaining four residues is very similar in systems I, II and III. Thus, the surface of the nanoparticle does not alter the conformational preferences of the polar amino acids, which are dominated by the strong interactions between the ionized side chains. This interesting result is fully consistent with the binding to plasma clot of both free CREKA and CREKA-SPIO nanoparticle found by Ruoslahti and coworkers.1–3 The resemblance between the conformational preferences of the free peptide and system III is also reflected in Figure S2 (see Supporting Information), which shows the distribution of the dihedral angles Ψ,ϕ for the five residues for the latter system. As can be seen, the regions of the Ramachandran map visited by the unique minimum energy conformations are very similar to those displayed in Figure S1. In addition, the tethering to the surface does not influence the positions of the more significant minima of CREKA, which are highlighted in the same Figure. Thus, with the exception of the Cys residue, the positions of the accessible minima for the other four residues is similar in Figures 4 and 8, even though the dispersion is significantly lower in the latter. The latter feature is a consequence of the reduction of the conformational flexibility induced by the linking to the nanoparticle.
Figure 6.
Distribution of the virtual dihedral angles (Methods) used to define the conformation of five CREKA residues in all the unique minima obtained for system III. Colour code for the bars is identical to that of Figure 3.
Figure 8.
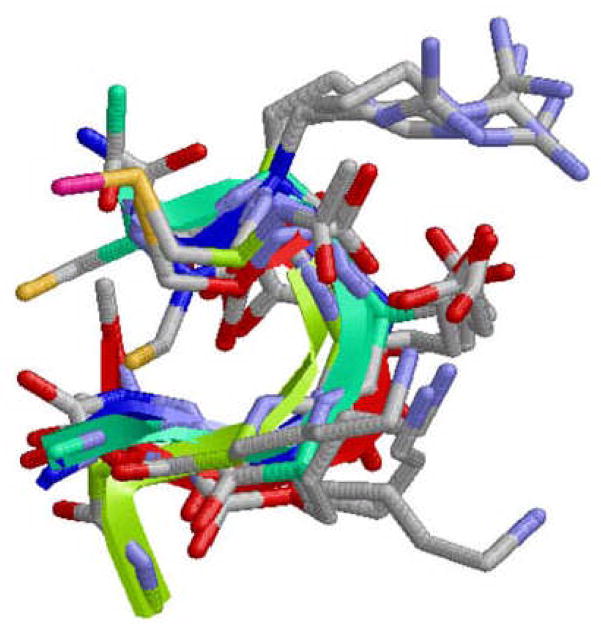
Superposition of the lowest energy minimum obtained for systems I, II, III and IV (D= 20.0 A). The CREKA peptide is the only represented, the other elements included in simulations of systems III and IV being omitted for clarity.
The global minimum found for system III is depicted in Figure 4c. As can be seen, there is a notable resemblance between the global minimum of the three systems. In spite of this, there is a critical difference between the most stable structure of CREKA attached to the nanoparticle and that obtained for the free peptide: the two γ-turns found at the ends of the latter system are not detected when the peptide is tethered to the surface. Thus, the only backbone···backbone intramolecular interaction detected in the global minimum of system III corresponds to a β-turn, which involves (Cys)C=O···H-N(Lys) [dH···O= 2.048 A, ∠N-H···O= 147.0°]. This structural motif, which is topologically identical to that obtained for I and II, is also detected in many of the minima with relative energies below 2 kcal/mol. On the other hand, the carboxylate group of the Glu is involved in a multiple interaction pattern similar to that described above for the free peptide. The only difference with respect to the latter is that the salt bridge between the oxygen atom of the carboxylate group and the guanidinium group of Arg involves two centers rather than three. Accordingly, one oxygen atom of the carboxylate forms a salt bridge with the Arg while the second oxygen participates in one salt bridge and one hydrogen bond (Figure 4c). Furthermore, multiple interaction patterns involving the ionized side chains of Arg, Glu and Lys were also found in the 12 significant minima, the main difference among them being the number of centers involved in the salt bridges.
Clustering analysis of system III indicates that 82.1% (1072 structures) of the minimum energy conformations present at least one intramolecular interaction (see Supporting Information). Interestingly, only 8 clusters contain more than 25 structures grouping 25.5% of the minima stabilized by intramolecular interactions. A more detailed analysis of the structures contained in the different clusters reveals that a total of 2198 interactions were detected in the 1072 minima of III stabilized by intramolecular interactions, i.e. ~2 interactions per minimum. These interactions are distributed as follows: (i) main chain···main chain hydrogen bonds, 83.6% (1839 interactions); (ii) main chain···side chain hydrogen bonds, 4.7% (102 interactions); and side chain···side chain salt bridges, 11.7% (257 interactions). It is worth noting that these results, i.e. both the average number of interactions per minima and the distribution of the interactions, are fully consistent with those discussed above for I and II.
CREKA Inserted in a Phage Display Protein (System IV)
After four cycles of modified SA-MD for each value of D (minimization of 2000 selected structures), we observed that the energies of the minima obtained for system IV are considerably higher when D= 5.0, 7.0, 10.0 and 15.0 Å. Thus, a significant strain is induced in the peptide if the distance between the two ending particles is too short. The reduction of the strain of the peptide when the distance between the particles that tether CREKA to the phage display protein increases is clearly reflected in Figure 7, which depicts the lowest energy minimum for each such D value. For the unique minima obtained for D= 20.0 Å, the corresponding energies show reasonable values although still higher than those of systems I, II and III. This led us to focus on the analysis of the 527 resulting unique minimum energy conformations obtained using the latter distance. However, due to the extra energy penalty observed for these structures with respect to those of the previously discussed systems, no additional modified SA-MD cycle was performed. Despite this shortcoming, the high efficiency shown by this conformational search procedure combined with the restricted conformational freedom of system IV, which is imposed by the restraint at the ends, suggests that the number of minima provided in four cycles is enough to provide qualitative information about the influence of the phage display protein on the conformational preferences of CREKA. On the other hand, the lack of experimental information about the structure of the loops of phage display proteins precludes a more realistic modeling of system IV.
Figure 7.
Lowest energy minimum obtained for GGCREKAGG attached to two particles (system IV), which represents the tethering to a phage display protein, separated by D= 5 (a), 7 (b), 10 (c) and 15 Å (d).
The distribution of the backbone dihedral angles Ψ,ϕ for the CREKA peptide obtained using D= 20.0 Å is provided in the Supporting Information section (Figure S3), omitting the analysis of the four Gly residues included in system IV. As can be seen, the Ramachandran maps are very similar to those reported for the free peptide (Figure S1) and the peptide attached to a SPIO-nanoparticle (Figure S2). This coincidence, which also extends to the accessible energy minima, allows us to conclude that the chemical environment does not influence the conformational preferences of CREKA. Thus, analysis of the unique energy minima obtained for the four systems under study provides evidence that the conformational profile of this peptide is mainly characterized by both the β-turn motif and the interactions that form the side chains of the three charged residues.
Figure 4d depicts the lowest energy minimum obtained for system IV with D= 20.0 Å. Again, this structure is dominated by the (Cys)C=O···H-N(Lys) [dH···O= 2.226 Å, ∠N-H···O= 152.1°] β-turn and the multiple interaction that involve the ionized side chains of Arg, Glu and Lys. Furthermore, the intraresidue hydrogen bond between the amide N-H and the oxygen atom of the carboxylate group of Glu is also detected in this conformation. The noticeable agreement between the lowest energy minimum of systems I, II, III and IV is graphically displayed in Figure 8, which provides the superposition of all structures.
Clustering analysis of system IV reveals that 75.0% (395 structures) minima show one or more intramolecular interactions (see Supporting Information). Analysis of these minima evidence a total of 715 interactions distributed in such 395 structures, i.e. ~2 interactions per minimum, which can be categorized as follows: (i) main chain···main chain hydrogen bonds, 89.0% (636 interactions); (ii) main chain···side chain hydrogen bonds, 2.2% (16 interactions); and side chain···side chain salt bridges, 8.8% (63 interactions). The overall of these results are in excellent agreement with those reported for I, II and III.
Stability of the β-Turn Motif in CREKA
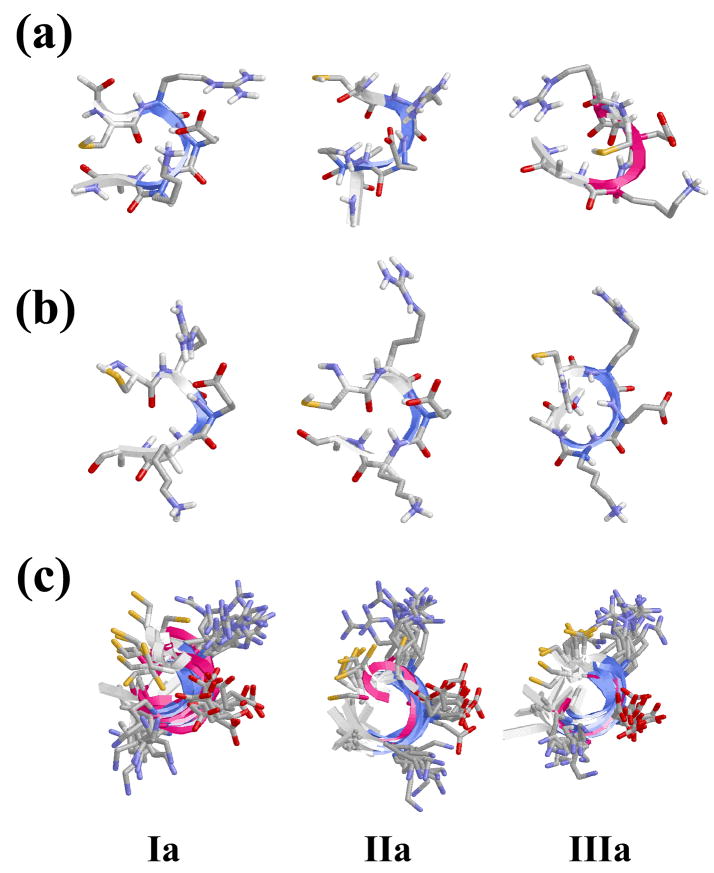
Results obtained in previous sections indicate that the β-turn motif is significantly favored for the CREKA peptide sequence. Thus, this conformation was found to be present in many of the minima found for systems I-IV evidencing its accessibility. In this section we demonstrate that the β-turn motif is not only an accessible conformation but also a stable one. For this purpose, NPT MD simulations were performed in aqueous solution at 310 K (physiological temperature) and P= 1 atm using as starting points three of the minima found for I. Specifically, we considered two different minima arranged in a β-turn conformation, hereafter denoted Ia and Ib, and one minimum with a helical-like conformation, Ic. These three minima are schematically depicted in Figure 9a. As can be seen, minimum Ia corresponds to the lowest energy minimum of system I (Figure 4a).
Figure 9.
(a) Minimum energy conformations (labelled as Ia, Ib and Ic) of free CREKA in unionized aqueous solution (system I) selected for MD studies at 310 K (physiological temperature). (b) Structure of Ia, Ib and Ic after the equilibration period. (c) Superimposition of the snapshots recorded at intervals of 1 ns during the MD simulations of Ia, Ib and Ic.
Before the production series, the thermodynamic variables of the system were equilibrated using the protocol indicated in the Methods section. Specifically, consecutive rounds of short MD runs were performed as follows: (i) 0.5 ns of NVT-MD at 500 K were used to homogeneously distribute the solvent and ions in the box; (ii) 0.5 ns of NVT-MD at 298 K (thermal equilibration); and (iii) 0.5 ns of NPT-MD at 298 K (density relaxation) were run. The conformations obtained for Ia, Ib and Ic after equilibration are displayed in Figure 9b. It is worth noting that both dynamical and thermal effects induce some re-arrangements in the three minima, even although the more important motifs of the three initial conformations are retained, especially of Ia and Ib. After equilibration, a NPT-MD in aqueous solution was run during 10 ns for each structure.
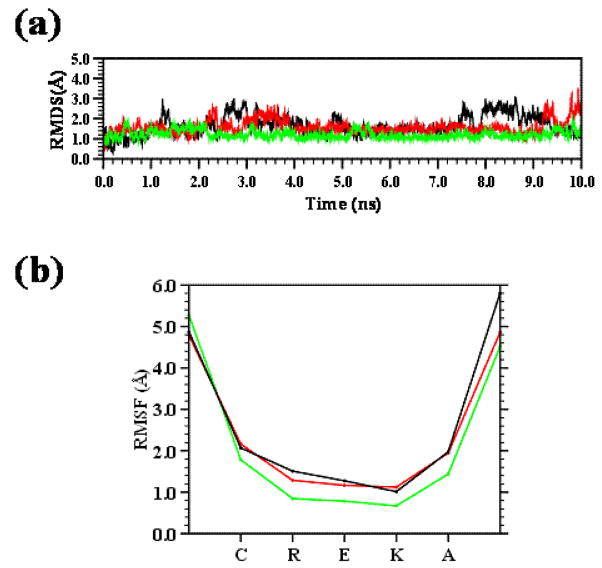
Figure 10a compares the root mean square deviation (RMSD) of the three minima, while Figure 10b displays the root mean square fluctuation (RMSF) of individual residues averaged over the whole simulation. Both RMSD and RMSF were computed with respect to the backbone atoms (-N-Cα-C-). Inspection to the RMSD, which were calculated without considering the Ac and NHMe capping groups, indicates that the three minima are very stable, the average RMSD being around 1.5 Å in all cases. Similarly, the RMSF profiles clearly show that the only distortions are located at the capping groups. This feature is also reflected in Figure 9c, which shows the superimposed snapshots that were recorded every 1 ns. These results indicate that the more main structural characteristics of these accessible minima are retained through the whole trajectories evidencing the high stability of the β-turn motif.
Figure 10.
(a) Temporal evolution of the backbone RMSD and (b) RMSF of the minimum energy conformations Ia (black), Ib (green) and Ic (red) of the CREKA peptide.
Discussion and Conclusions
Considering all the results reported in this work, it is clear that the modified SA-MD is a powerful method to explore the conformational space of peptides. Complete convergence was reached for systems I, II and III after a few simulation cycles. The limited influence exerted, in this case, by the NaCl molecules used to mimic the ionic strength of the physiological medium was confirmed by comparing the conformational profiles of systems I and II. Unique structures produced by independent searches for these two systems reveal a strong degree of similarity, as was also evidence by the clustering analyses. Accordingly, these results demonstrate that simulations in unionized aqueous solution are able to represent the behavior of CREKA in the ionized physiological medium.
The conformational space of both the free peptide and the peptide attached to a nanoparticle has been explored exhaustively. Results indicate that the conformational profile of the REKA sequence is very similar in both cases, the only difference being the Cys residue. This coincidence is particularly evident for the accessible minima, as can be deduced from the Figures 4 and 8. Thus, comparison between all the minimum energy conformations with relative energies lower than 2.0 kcal/mol reveals a high degree of concordance. Furthermore, complete clustering analyses also reflect to a significant agreement, as is demonstrated by the similarities in the average number of interactions per minimum and the distribution of hydrogen bonding interactions and salt bridges. The overall of these results allows us to conclude that the conformational profile of the peptide is independent of the chemical environment, i.e. the conformational preferences of the peptide are very similar when it is free or attached to a SPIO nanoparticle. This agreement is expected to facilitate the future design of synthetic analogs to protect the peptide from the attack of proteases since the choice of the synthetic amino acid will be based on a general conformational profile rather than on a specific conformation. It should be noted that the conformational preferences of synthetic amino acids are typically controlled by the introduction of chemical restraints, e.g. Cα↔Cα cyclization and/or the introduction of side groups. Therefore, in recent years it has been demonstrated that synthetic amino acids with restrained and well-defined conformational preferences can be designed for a given application.13,15,28
On the other hand, the pattern of multiple interactions which involves salt bridges and hydrogen bonds, defined by the ionized side chains of Arg, Glu and Lys determines the backbone conformation of the peptide. This is clearly reflected in the lowest energy minimum, which is very similar for both the free CREKA and the CREKA-SPIO nanoparticle. Thus, the β-turn motif that characterizes the global minimum and many of the accessible minima is a consequence of such interactions. Moreover, the ionized side chains of Arg, Glu and Lys are probably essential for the binding activity of CREKA. Thus, peptide sequences used to recognize tumors are frequently formed by charged amino acids.29 Therefore, the multiple interaction pattern mentioned above together with the backbone β-turn should be preserved in the engineered CREKA synthetic analogs.
Calculations on system IV have shown that it is not easy to mimic peptide sequences expressed in phage display proteins. Thus, the lack of available structural and chemical information about such proteins makes their simulation extremely difficult. Our results indicate that, when the dimensions used mimic the loop that contains the expressed peptide are too short, the produced minimum energy conformations are significantly strained. Accordingly, the results obtained for system IV should be considered with caution. In spite of these limitations, the results are fully consistent with those obtained for the free peptide and the peptide attached to a nanoparticle when the separation between the two extremes of the loop that contains CREKA is 20.0 Å. This separation provides the lowest strain, even though an unfavorable energy penalty is still involved in the resulting minima. However, the good agreement with the conformational profiles of systems I, II and III supports the conclusion reached above: the conformational preferences of CREKA are not determined by the chemical environment but by the strong interactions between the charged residues.
The coincidence between the global minimum of the four studied systems, which is reflected in Figure 8, together with the analysis of the accessible conformations suggest that the bioactive conformation of CREKA involves both a β-turn motif and strong interactions involving the side chains of Arg, Glu and Lys. Furthermore, MD simulations show that this is a very stable conformation. Thus, overall, the results indicate that the shape of the global minimum is appropriate to form intermolecular interactions with a receptor, i.e. it is like a pocket with the charged groups pointing outwards. Accordingly, the future design of CREKA synthetic analogs should take care to retain these elements to preserve the peptide binding activity.
Supplementary Material
Acknowledgments
The authors are indebted to the Barcelona Supercomputer Center (BSC) for computational resources. The authors wish to thank Prof. E. Ruoslahti for the careful reading of the paper and the excellent suggestions for future work. DZ thanks financial support from the Ramon y Cajal program of the Spanish “Ministerio de Educacion y Ciencia” (MEC). This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number N01-CO-12400. The content of this publication does not necessarily reflect the view of the policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
For systems I, II, III and IV: distribution of the main chain dihedral angles (Ψ,ϕ) for the five residues of CREKA in the set of unique minimum energy conformations provided by modified SA-MD and clustering analyses. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pasqualini R, Ruoslahti E. Organ Targeting in Vivo Using Phage Display Peptide Libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman JA, Giraudo E, Singh M, Zhang L, Inoue M, Porkka K, Hanahan D, Ruoslathi E. Progressive Vascular Changes in a Transgenic Mouse Model of Squamous Cell Carcinoma. Cancer Cell. 2003;4:383–391. doi: 10.1016/s1535-6108(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 3.Ruoslahti E. Personal communication (unpublished results) 2007. [Google Scholar]
- 4.Simberg D, Duza T, Park JH, Essier M, Pilch J, Zhang L, Derfus AM, Yang M, Hoffman RM, Bathia S, Sailor MJ, Ruoslahti E. Biomimetic Amplification of Nanoparticle Homing to Tumors. Proc Natl Acad Sci. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adessi C, Soto C. Converting a Peptide into a Drug: Strategies to improve Stability and Bioavailability. Curr Med Chem. 2002;9:963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 6.Lelais G, Seebach D. Beta(2)-Amino Acids - Syntheses, occurrence in Natural Products, and Components of Bbeta-Peptides. Biopolymers. 2004;76:206–243. doi: 10.1002/bip.20088. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi H, Kodama H, Osada S, Kato F, Jelokhani-Niaraki M, Kondo M. Effect of alpha, alpha-dialkyl Amino Acids on the Protease Resistance of Peptides. Biosci Biotech Biochem. 2003;67:2269–2272. doi: 10.1271/bbb.67.2269. [DOI] [PubMed] [Google Scholar]
- 8.Sadowsky JD, Murray JK, Tomita Y, Gellman SH. Exploration of backbone space in foldamers containing alpha- and beta-amino acid residues: Developing protease-resistant oligomers that bind tightly to the BH3-recognition cleft of Bcl-x(L) . Chem Bio Chem. 2007;8:903–916. doi: 10.1002/cbic.200600546. [DOI] [PubMed] [Google Scholar]
- 9.Talele TT, McLaughlin ML. Asymmetric syntheses of enantiomerically pure alpha,alpha-dialkylated glycines as core structural units for novel HIV-1 protease inhibitors. Biopolymers. 2003;71:344–344. [Google Scholar]
- 10.Banerjee R, Basu G, Chene P, Roy S. Aib-based Peptide Backbone as Scaffolds for Helical Peptide Mimics. J Pept Res. 2002;60:88–94. doi: 10.1034/j.1399-3011.2002.201005.x. [DOI] [PubMed] [Google Scholar]
- 11.Webb AI, Dunstone MA, Williamson NA, Price JD, de Kauwe A, Chen WS, Oakley A, Perlmutter P, McCluskey J, Aguilar MI, Rossjohn J, Purcell AW. T Cell determinants incorporating Beta-Amino Acid residues are Protease resistant and remain Immunogenic in Vivo. J Immun. 2005;175:3810–3818. doi: 10.4049/jimmunol.175.6.3810. [DOI] [PubMed] [Google Scholar]
- 12.Bannwarth L, Kessler A, Pethe S, Collinet B, Merabet N, Boggetto N, Sicsic S, Reboud-Ravaux M, Ongeri S. Molecular Tongs containing Amino Acid Mimetic Fragments: New Inhibitors of Wild-Type and Mutated HIV-1 Protease Dimerization. J Med Chem. 2006;49:4657–4664. doi: 10.1021/jm060576k. [DOI] [PubMed] [Google Scholar]
- 13.Aleman C, Zanuy D, Jimenez AI, Cativiela C, Haspel N, Zheng J, Casanovas J, Wolfson H, Nussinov R. Concepts and Schemes for the Re- Engineering of Physical Protein Modules: generating Nanodevices via targeted replacements with Constrained Amino Acids. Phys Biol. 2006;3:S54–S62. doi: 10.1088/1478-3975/3/1/S06. [DOI] [PubMed] [Google Scholar]
- 14.Tsai CJ, Zheng J, Zanuy D, Haspel N, Wolfson H, Aleman C, Nussinov R. Principles of Nanostructure Design with Protein Building Blocks. Proteins. 2007;68:1–12. doi: 10.1002/prot.21413. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Zanuy D, Haspel N, Tsai CJ, Aleman C, Nussinov R. Nanostructure Design using Protein Building Blocks enhanced by Conformationally Constrained Synthetic Residues. Biochemistry. 2007;46:1205–1218. doi: 10.1021/bi061674a. [DOI] [PubMed] [Google Scholar]
- 16.Agrafiotis DM, Gibbs AC, Zhu F, Izrailev S, Martin E. Conformational Sampling of Bioactive Molecules: A Comparative Study. J Chem Inf Model. 2007;47:1067–1086. doi: 10.1021/ci6005454. [DOI] [PubMed] [Google Scholar]
- 17.Steinbach PJ. Exploring Peptide Energy Landscapes: A test of Force Fields and Implicit Solvent Models. Proteins. 2004;57:665–677. doi: 10.1002/prot.20247. [DOI] [PubMed] [Google Scholar]
- 18.Swaminathan P, Hariharan M, Murali R, Singh CU. Molecular Structure, Conformational Analysis, and Structure-Activity studies of Dendrotoxin and its Homologues using Molecular Mechanics and Molecular Dynamics Techniques. J Med Chem. 1996;39:2141–2155. doi: 10.1021/jm950579p. [DOI] [PubMed] [Google Scholar]
- 19.Kirkpatrick S, Gelatt CD, Jr, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 20.Steinbach PJ, Brooks BR. Protein Simulation below the Glass-Transition Temperature Dependence on Cooling Protocol. Chem Phys Letters. 1994;226:447–452. [Google Scholar]
- 21.Baysal C, Meirovitch H. Efficiency of Simulated Annealing for Peptides with Increasing Geometrical Restrictions. J Comput Chem. 1999;20:1659–1670. [Google Scholar]
- 22.Simmerling C, Elber R. Hydrophobic Collapse in a Cyclic Hexapeptide -Computer-Simulations of Chdlfc and Caaaac in Water. J Am Chem Soc. 1994;116:2534–2547. [Google Scholar]
- 23.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 24.Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 25.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman P. A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 26.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 27.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints Molecular Dynamics of n-Alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 28.Zanuy D, Jimenez AI, Cativiela C, Nussinov R, Aleman C. Use of Constrained Synthetic Amino acids in β-Helix Proteins for Conformational Control. J Phys Chem B. 2007;111:3236–3242. doi: 10.1021/jp065025k. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Giraudo E, Hoffman JA, Hanahan D, Ruoslahti E. Lymphatic Zip Codes in Premalignant Lesions and Tumors. Cancer Res. 2006;66:5696–5706. doi: 10.1158/0008-5472.CAN-05-3876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.