Abstract
Aims/hypothesis
We investigated whether variation in MTNR1B, which was recently identified as a common genetic determinant of fasting glucose levels in healthy, diabetes-free individuals, is associated with measures of beta cell function and whole-body insulin sensitivity.
Methods
We studied 1,276 healthy individuals of European ancestry at 19 centres of the Relationship between Insulin Sensitivity and Cardiovascular disease (RISC) study. Whole-body insulin sensitivity was assessed by euglycaemic–hyperinsulinaemic clamp and indices of beta cell function were derived from a 75 g oral glucose tolerance test (including 30 min insulin response and glucose sensitivity). We studied rs10830963 in MTNR1B using additive genetic models, adjusting for age, sex and recruitment centre.
Results
The minor (G) allele of rs10830963 in MTNR1B (frequency 0.30 in HapMap Centre d’Etude du Polymorphisme [Utah residents with northern and western European ancestry] [CEU]; 0.29 in RISC participants) was associated with higher levels of fasting plasma glucose (standardised beta [95% CI] 0.17 [0.085, 0.25] per G allele, p = 5.8 × 10−5), consistent with recent observations. In addition, the G-allele was significantly associated with lower early insulin response (−0.19 [−0.28, −0.10], p = 1.7 × 10−5), as well as with decreased beta cell glucose sensitivity (−0.11 [−0.20, −0.027], p = 0.010). No associations were observed with clamp-assessed insulin sensitivity (p = 0.15) or different measures of body size (p > 0.7 for all).
Conclusions/interpretation
Genetic variation in MTNR1B is associated with defective early insulin response and decreased beta cell glucose sensitivity, which may contribute to the higher glucose levels of non-diabetic individuals carrying the minor G allele of rs10830963 in MTNR1B.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-009-1392-x) contains a list of the members of the RISC Consortium, which is available to authorised users.
Keywords: Genome-wide association, Glucose sensitivity, Insulin resistance, Insulin sensitivity, Melatonin receptor 1B, MTNR1B
Introduction
Compared with recent progress in the discovery of genes for type 2 diabetes, knowledge of genetic influences on fasting glucose levels in healthy individuals is limited. Common sequence variants related to the GCK promoter [1–3], G6PC2 [2, 3] and GCKR [4–6] are the most significant determinants of fasting glucose levels identified in recent large-scale genome-wide association studies (GWAS), yet without demonstrable consistent effects on the risk of type 2 diabetes [7]. Vice-versa, none of the established type 2 diabetes genes have emerged as convincing loci for fasting glucose within the normal range in recent GWAS [2, 7]. This suggests that common variants contributing to small physiological variation in fasting glucose may be different from those that increase type 2 diabetes susceptibility.
One exception is noteworthy; in a recently published exchange of top fasting glucose hits from four large GWAS consortia, variants in the gene encoding the melatonin receptor 1B (MTNR1B) were not only consistently associated with fasting glucose across all studies totalling 36,610 healthy adult participants in the meta-analysis of the MTNR1B region [7], but carriers of the risk allele of the most significant overall signal at rs10830963 (minor G allele; frequency 0.30 in HapMap Centre d’Etude du Polymorphisme [Utah residents with northern and western European ancestry] [CEU]) were also at increased risk of type 2 diabetes (odds ratio [95% CI] 1.09 [1.05, 1.12]) in a separate meta-analysis of case–control studies [7]. The novel link between MTNR1B and type 2 diabetes was confirmed in two other studies, one investigating the same variant [8], and another reporting rs1387153 (r2 with rs10830963 = 0.70) to be the most significant single nucleotide polymorphism (SNP) in a GWAS of 2,151 French participants [2]. Investigating the mechanisms through which MTNR1B contributes to variation in fasting glucose levels in healthy, non-diabetic individuals may help to understand what underlies the MTNR1B-related risk of progression to clinical diabetes. Lyssenko et al. have reported that the risk genotype was associated with impairment of early insulin response to oral and intravenous glucose, and with faster deterioration of insulin secretion over time [8].
Thus our objective was to study whether the association between MTNR1B and fasting glucose is mediated through reduced pancreatic beta cell function, insulin secretion or whole-body insulin sensitivity in individuals participating in the Relationship between Insulin Sensitivity and Cardiovascular disease (RISC) study [9].
Methods
The recruitment, methods, and inclusion and exclusion criteria of the RISC cohort have been described previously [9]. Briefly, healthy men and women of European ancestry, aged between 30 and 60 years, were recruited from 19 centres in 14 European countries. Individuals with diabetes, hypertension or dyslipidaemia were excluded [9]. Analyses presented in this study are based on 1,276 participants who met the eligibility criteria and for whom complete genotype data were available. Local ethics committee approval was obtained by each recruitment centre and written consent was obtained from all participants.
Participants underwent a 75 g OGTT with blood sampling before and at 30, 60, 90 and 120 min after the oral glucose load. On a separate day within 1 month of the OGTT, participants underwent a euglycaemic–hyperinsulinaemic clamp as previously reported [9]. To ensure consistency across study centres, the clamp procedure was standardised and each centre underwent prestudy training. Clamp data were then transferred and analysed at the RISC coordinating centre (Pisa, Italy) and quality assured against preset criteria. These were as follows: clamped glucose levels within 20% of target (fasting glucose concentration), coefficient of variation of ≤15%, avoidance of hypoglycaemia (glucose <3.5 mmol/l). Insulin sensitivity was assessed as the mean glucose infusion rate over the last 40 min of the clamp, corrected for the mean plasma insulin levels achieved during the same period. Pancreatic beta cell function was assessed using the OGTT data. The 30 min insulin response was calculated as the ratio of the insulin concentration increment to the 30 min glucose concentration (30 min insulin −0 min insulin/30 min glucose) [10]. We additionally performed sensitivity analyses using other commonly derived indices of early insulin response, including insulinogenic index (30 min insulin −0 min insulin/30 min glucose −0 min glucose) and corrected insulin response [11].
Indices of beta cell function variables were derived from mathematical analysis of plasma glucose and C-peptide, using C-peptide deconvolution, as previously described in more detail [12].
In addition, detailed anthropometric assessment was performed and fat mass determined as the difference between body weight and fat-free mass determined by bioimpedance (Tanita International Division, Yiewsley, UK).
Samples were processed and stored locally before being transferred to the central assay laboratories and analysed as previously reported [9]. Genomic DNA was extracted using a kit (Nucleon BACC2; Tepnel Life Sciences, Manchester, UK). All samples were genotyped at KBiosciences (KBiosciences, Hoddesdon, UK) [12]; the call rate was 98% and genotype frequencies were in Hardy–Weinberg equilibrium (p = 0.33). Duplicate genotyping of 5% of DNA samples was conducted with 100% success.
We normalised the distributions of OGTT and clamp outcomes using the natural logarithm; these data are presented as median and interquartile range. Linear regression analyses using an additive genetic model were performed to test for associations between rs10830963 and selected phenotypes, adjusting for age, sex and recruitment centre. Standardised measures were used in regression analyses; these were calculated by subtracting the mean of each outcome from an individual’s value and dividing this by the standard deviation, separately by sex, and using loge transformed measures. This results in a normal distribution for each measure with a mean of 0 and standard deviation of 1, and allows the strength of the genotype effect to be compared across several outcomes with different distributions and units. As previously reported [12], the cohort had 80% power at p = 0.01 to detect differences of 0.18 SD per allele for a minor allele frequency of 0.30.
Results
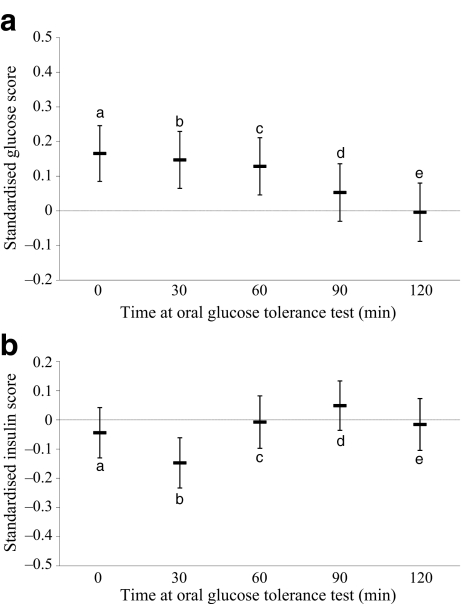
The minor (G) allele of rs10830963 was significantly associated with higher fasting glucose levels (standardised beta [95% CI] 0.17 [0.085, 0.25] per G allele, p = 5.8 × 10−5) (Fig. 1a) consistent with recent observations [7]. In addition, we observed significantly higher glucose levels at 30 and 60 min (standardised beta [95% CI] 0.15 [0.065, 0.23], p = 4.7 × 10−4 and 0.13 [0.046, 0.21], p = 2.3 × 10−3, respectively) (Fig. 1a), but not at 90 or 120 min during the OGTT (p = 0.21 and 0.93, respectively).
Fig. 1.
Effect of rs10830963 in MTNR1B on glucose (a) and insulin (b) levels during the OGTT (per allele difference and 95% CI). aap = 5.8 × 10−5; bp = 4.7 × 10−4; cp = 2.3 × 10−3; dp = 0.211; ep = 0.926. bap = 0.135; bp = 8.0 × 10−4; cp = 0.868; dp = 0.259; ep = 0.727
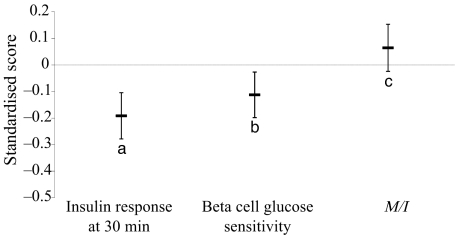
Variation at rs10830963 was not associated with whole-body insulin sensitivity measured by euglycaemic–hyperinsulinaemic clamps (standardised beta [95% CI] 0.064 [−0.024, 0.15], p = 0.15) (Fig. 2). In contrast, significant differences in indices of pancreatic beta cell function were found. Individuals carrying the minor (G) allele had significantly lower 30 min insulin response (standardised beta [95% CI] −0.19 [−0.28, −0.10], p = 1.7 × 10−5) (Fig. 2), as well as beta cell glucose sensitivity (standardised beta [95% CI] −0.11 [−0.20, −0.027], p = 0.010). These associations remained significant after additional adjustment for whole-body insulin sensitivity (p = 0.0001 and 0.014 respectively). Similar results were obtained in sensitivity analyses using the insulinogenic index (p = 6.8 × 10−5) or corrected insulin response (p = 2.7 × 10−3) as alternative measures of early insulin response. Associations with insulin (Fig. 1b) and C-peptide (data not shown) mirrored these observations, with the only time point during the OGTT at which significant differences were found being at 30 min (standardised beta [95% CI] −0.15 [−0.23, −0.061], p = 8.0 × 10−4 for insulin; −0.089 [−0.17, −0.0044], p = 0.039 for C-peptide). No significant associations were observed between variation in MTNR1B and different measures of body size (Table 1).
Fig. 2.
Effect of rs10830963 in MTNR1B on early insulin response, beta cell glucose sensitivity and whole-body insulin sensitivity (M/I) (per allele difference and 95% CI). ap = 1.7 × 10−5; bp = 0.010; cp = 0.154
Table 1.
Relationships between rs10830963 genotypes and key phenotypic traits
| Phenotype | Mean (SD)/median (IQR) | Mean (SD), median (IQR) by genotype | p value | ||
|---|---|---|---|---|---|
| CC (max n = 655) | GC (max n = 505) | GG (max n = 114) | |||
| Sex (% women) | 55.2 | 58.0 | 54.0 | 43.9 | 0.016 |
| Age (years) | 43.9 (8.3) | 44 (8.2) | 44 (8.2) | 43 (9.3) | 0.34 |
| BMI (kg/m2) | 25.7 (4.1) | 25.5 (4.0) | 25.6 (4.1) | 25.8 (4.1) | 0.81 |
| Waist (cm) | 86 (77–96) | 85 (76–95) | 86 (78–96) | 88 (79–96) | 0.93 |
| Fat mass (kg) | 21.2 (8.9) | 21.2 (8.9) | 21.0 (8.9) | 20.2 (9.2) | 0.78 |
| Glucose (mmol/l) | |||||
| OGTT 0 min (fasting) | 5.1 (4.7–5.4) | 5.0 (4.7–5.3) | 5.1 (4.8–5.5) | 5.2 (4.8–5.5) | 5.8 × 10−5 |
| OGTT 30 min | 8.0 (6.9–9.3) | 7.8 (6.7–8.9) | 8.2 (7.1–9.4) | 7.9 (7.0–9.5) | 4.7 × 10−4 |
| OGTT 60 min | 7.5 (6.0–9.3) | 7.2 (5.8–8.8) | 7.8 (6.3–9.4) | 7.7 (6.1–9.7) | 2.3 × 10−3 |
| OGTT 90 min | 6.2 (5.0–7.5) | 6.1 (5.0–7.3) | 6.4 (5.2–7.7) | 6.2 (5.1–7.6) | 0.21 |
| OGTT 120 min | 5.6 (4.7–6.7) | 5.6 (4.7–6.6) | 5.6 (4.6–6.6) | 5.5 (4.6–6.9) | 0.93 |
| Insulin (pmol/l) | |||||
| OGTT 0 min (fasting) | 31.0 (21.0–46.0) | 31.0 (21.0–44.0) | 31.0 (21.0–45.5) | 28.5 (19.0–42.0) | 0.32 |
| OGTT 30 min | 240 (168–355) | 244 (173–358) | 238 (167–347) | 199 (152–297) | 8.0 × 10−4 |
| OGTT 60 min | 269 (178–413) | 265 (172–399) | 266 (177–406) | 244 (166–385) | 0.87 |
| OGTT 90 min | 200 (127–316) | 196 (122–309) | 210 (136–330) | 197 (123–295) | 0.26 |
| OGTT 120 min | 153 (89–257) | 147 (89–249) | 151 (85–251) | 159 (73–226) | 0.73 |
| C-peptide (pmol/l) | |||||
| OGTT 0 min (fasting) | 540 (410–703) | 528 (399–690) | 562 (420–727) | 525 (391–682) | 0.88 |
| OGTT 30 min | 1,891 (1,502–2,387) | 1,910 (1,517–2,403) | 1,894 (1,511–2,391) | 1,704 (1,416–2,095) | 0.039 |
| OGTT 60 min | 2,509 (2,008–3,098) | 2,440 (1,961–3,029) | 2,558 (2,066–3,182) | 2,562 (1,927–3,022) | 0.78 |
| OGTT 90 min | 2,406 (1,863–3,036) | 2,336 (1,789–2,958) | 2,509 (1,955–3,116) | 2,512 (1,843–3,148) | 0.24 |
| OGTT 120 min | 2,154 (1,642–2,777) | 2,121 (1,598–2,743) | 2,213 (1,677–2,789) | 2,252 (1,603–2,777) | 0.56 |
| 30 min insulin response (pmol/mmol) | 26.3 (18.5–38.7) | 28.0 (19.3–41.4) | 25.9 (18.4–37.0) | 22.0 (14.9–33.8) | 1.7 × 10−5 |
| Beta cell glucose sensitivitya | 113 (79–158) | 118 (82–169) | 109 (74–150) | 102 (74–148) | 0.010 |
| Insulin sensitivityb | 128 (92–177) | 128 (90–175) | 128 (92–180) | 135 (99–175) | 0.15 |
Comparisons between genotypes (additive model) are based on linear regression analysis of loge-transformed data (where applicable) and using sex-specific standardised outcomes for OGTT and clamp measures, adjusting for age, sex and recruitment centre
Sex differences were tested using a χ2 test
aIn pmol min−1 m−2 mmol−1
bIn μmol min−1 (kg fat-free mass)−1 nmol−1
IQR, interquartile range
Discussion
Recent evidence from large-scale meta-analysis of GWAS showed that variation in MTNR1B is a common genetic determinant of fasting glucose in healthy, diabetes-free individuals. We show here that variation in MTNR1B is significantly associated with early insulin response and beta cell glucose sensitivity, while no effect on whole-body insulin sensitivity was observed.
The minor (G) risk allele of rs10830963 in MTNR1B was associated with lower beta cell glucose sensitivity and 30 min insulin response before and after accounting for whole-body insulin sensitivity levels. These findings are in keeping with a primary defect of beta cell function rather than secondary changes in response to altered insulin sensitivity, and support the observations of other studies, which have reported decreased early insulin response and decreased disposition index in G allele carriers of the same variant [8, 13]. Interestingly, we found significant associations of rs10830963 with insulin and C-peptide at 30 min during the OGTT, but not at any other time point, again highlighting that the main effect appears to be on early-phase insulin response. The emerging evidence strongly suggests that the melatonin system directly modulates the insulin secretory response to glucose. It has been shown that MTNR1B is expressed in human islets, and specifically in pancreatic beta and alpha cells [8, 14, 15]. Furthermore, MTNR1B gene expression was increased in isolated islets from older (>45 years of age) G allele carriers of rs10830963, and exposure of clonal beta cells to melatonin decreased the acute insulin secretory response to glucose [8]. It has also been postulated that melatonin might influence insulin secretion through a paracrine effect of glucagon [14]. We found that variation in MTNR1B was not associated with fasting glucagon levels (data not shown), but we did not measure the glucagon response during the OGTT.
Melatonin plays a role in regulation of the circadian clock, and melatonin and insulin both show marked circadian variability [16, 17]. Data from human and rodent studies suggest that disturbances of circadian rhythmicity may affect metabolic control and the risk of diabetes [18, 19]. Moreover, overexpression of melatonin receptors has been observed in islets from patients with type 2 diabetes compared with non-diabetic controls [20]. Taken together, these findings suggest that an effect of MTNR1B on the insulin secretory response to glucose may underlie the reported associations with fasting glucose and the risk of type 2 diabetes, adding to the body of evidence linking circadian rhythm and metabolic control and disease.
A key observation of our study is that there was no significant association between the MTNR1B variant and whole-body insulin sensitivity, despite the fact that this is one of the largest collections of healthy people of European ancestry to be phenotyped using the gold standard euglycaemic–hyperinsulinaemic clamp technique. In addition, we have replicated with the RISC cohort the observations that a common FTO variant and the Pro12Ala PPARG variant influence whole-body insulin sensitivity in man [12]. This would suggest that if variation in MTNR1B does affect insulin sensitivity, then it is likely to be functionally weak and of questionable clinical significance. In support of this, Staiger et al. recently reported that none of five tagging SNPs covering all common genetic variation of the MTNR1B locus showed an association with clamp-derived insulin sensitivity in a selected group of 513 individuals at increased risk of type 2 diabetes [13].
We also found no significant associations between MTNR1B and different measures of body size, suggesting that the effects seen on beta cell function are not influenced by an alteration in adiposity.
As we recently reported, individual type 2 diabetes risk alleles in TCF7L2, HHEX, IDE and CDKAL1 combine in an additive manner to impact upon pancreatic beta cell function [21]. Beta cell glucose sensitivity was decreased by 39% in individuals with five or more risk alleles compared with individuals who had no risk alleles. Inclusion of the MTNR1B risk variant in the analysis led to a 47% difference (p = 1.5 × 10−7) between the zero-allele group those with more than six alleles. A similar change was noted for the 30 min insulin response. We had previously found a 43% decrease between the zero-allele group and the group with more than five alleles. Inclusion of the MTNR1B variant increased that value to 49% between the zero-allele group and those with more than six alleles.
We conclude that MTNR1B is associated with defective early insulin response and decreased beta cell glucose sensitivity, both of which may contribute to the higher glucose levels and increased diabetes risk of individuals carrying the minor G allele of rs10830963. In contrast, no association with whole-body insulin sensitivity was observed in this large collection of healthy people of European ancestry.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 8 kb)
Acknowledgements
The RISC Study is supported by European Union grant QLG1-CT-2001-01252 and AstraZeneca. L. Pascoe is the recipient of a joint BBSRC and Unilever UK case PhD studentship. I. Barroso acknowledges funding from The Wellcome Trust grant 077016/Z/05/Z.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- GWAS
Genome-wide association studies
- RISC
Relationship between Insulin Sensitivity and Cardiovascular disease
- SNP
Single nucleotide polymorphism
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-009-1392-x) contains a list of the members of the RISC Consortium, which is available to authorised users.
References
- 1.Weedon MN, Clark VJ, Qian Y et al (2006) A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am J Hum Genet 79:991–1001 [DOI] [PMC free article] [PubMed]
- 2.Bouatia-Naji N, Rocheleau G, Van LL et al (2008) A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320:1085–1088 [DOI] [PubMed]
- 3.Chen WM, Erdos MR, Jackson AU et al (2008) Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest 118:2620–2628 [DOI] [PMC free article] [PubMed]
- 4.Saxena R, Voight BF, Lyssenko V et al (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336 [DOI] [PubMed]
- 5.Vaxillaire M, Cavalcanti-Proenca C, Dechaume A et al (2008) The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes 57:2253–2257 [DOI] [PMC free article] [PubMed]
- 6.Orho-Melander M, Melander O, Guiducci C et al (2008) Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 57:3112–3121 [DOI] [PMC free article] [PubMed]
- 7.Prokopenko I, Langenberg C, Florez JC et al (2009) Variants in MTNR1B influence fasting glucose levels. Nature genetics 41:77–81 [DOI] [PMC free article] [PubMed]
- 8.Lyssenko V, Nagorny CL, Erdos MR et al (2009) Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nature genetics 41:82–88 [DOI] [PMC free article] [PubMed]
- 9.Hills SA, Balkau B, Coppack SW et al (2004) The EGIR-RISC STUDY (The European group for the study of insulin resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives. Diabetologia 47:566–570 [DOI] [PubMed]
- 10.Wareham NJ, Phillips DI, Byrne CD, Hales CN (1995) The 30 minute insulin incremental response in an oral glucose tolerance test as a measure of insulin secretion. Diabet Med 12:931 [DOI] [PubMed]
- 11.Hanson RL, Pratley RE, Bogardus C et al (2000) Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol 151:190–198 [DOI] [PubMed]
- 12.Pascoe L, Tura A, Patel SK et al (2007) Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes 56:3101–3104 [DOI] [PubMed]
- 13.Staiger H, Machicao F, Schafer SA et al (2008) Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS ONE 3:e3962 [DOI] [PMC free article] [PubMed]
- 14.Ramracheya RD, Muller DS, Squires PE et al (2008) Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res 44:273–279 [DOI] [PubMed]
- 15.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C et al (2009) A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nature genetics 41:89–94 [DOI] [PubMed]
- 16.Boden G, Ruiz J, Urbain JL, Chen X (1996) Evidence for a circadian rhythm of insulin secretion. Am J Physiol 271:E246–E252 [DOI] [PubMed]
- 17.Stumpf I, Muhlbauer E, Peschke E (2008) Involvement of the cGMP pathway in mediating the insulin-inhibitory effect of melatonin in pancreatic beta-cells. J Pineal Res 45:318–327 [DOI] [PubMed]
- 18.Turek FW, Joshu C, Kohsaka A et al (2005) Obesity and metabolic syndrome in circadian clock mutant mice. Science 308:1043–1045 [DOI] [PMC free article] [PubMed]
- 19.Spiegel K, Leproult R, Van Cauter E (1999) Impact of sleep debt on metabolic and endocrine function. Lancet 354:1435–1439 [DOI] [PubMed]
- 20.Peschke E, Stumpf I, Bazwinsky I, Litvak L, Dralle H, Muhlbauer E (2007) Melatonin and type 2 diabetes—a possible link? J Pineal Res 42:350–358 [DOI] [PubMed]
- 21.Pascoe L, Frayling TM, Weedon MN et al (2008) Beta cell glucose sensitivity is decreased by 39% in non-diabetic individuals carrying multiple diabetes-risk alleles compared with those with no risk alleles. Diabetologia 51:1989–1992 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(PDF 8 kb)