Abstract
Aims
There are a large number of common genetic variants that have been robustly associated with low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, or triglyceride concentrations. The majority of these have been identified or confirmed in recent genome-wide association studies, but few studies have assessed the combined effect of known lipid variants. We hypothesized that these variants would influence both the need for interventions and myocardial infarction (MI) outcomes. We aimed to estimate combined effects of proven SNPs on LDL, HDL, and triglyceride concentrations and MI history in a representative older population.
Methods and results
In the InCHIANTI Study of Aging (age ≥65 years), we calculated individual dyslipidaemia risk allele counts for increased LDL (range 4–14, n = 594), reduced HDL (5–16, n = 635), and increased triglycerides (7–16, n = 611). Lipid levels were compared with ATPIII National Cholesterol Education Panel (NCEP) intervention guidelines. Individual variants and the APOE haplotype explained <2.1% of the variance in their respective lipid concentrations, with the exception of the CETP SNP rs1800775 and HDL levels (4.76%). Combined risk allele counts outperformed the largest single-SNP effects for LDL (explaining 7.1% of variance) and triglycerides (4.8%), but not HDL (3.4%). Risk alleles were divided as near as possible into quartiles. The 31% of respondents with 10 or more LDL increasing alleles were more likely to have LDL levels above the intervention threshold (OR 3.00, 95% CI 1.67–5.39, P = 2.5 × 10−4), compared with the 21% with 7 or less risk alleles. Similarly, the 35% with 13 or more triglyceride risk alleles were more likely to exceed NCEP intervention thresholds (OR 2.98, 95% CI 1.43–6.22, P = 0.004) compared with the 24% with 10 or less alleles. The number of individuals reporting an MI event was small (n = 67), but an event was more common in the 36% of respondents who had the highest combined risk allele score for all three lipids (OR 3.68, 95% CI 1.21–11.2, P = 0.021) compared with the lowest risk 22%.
Conclusion
In a representative older population, the cumulative effects of proven LDL- and triglyceride-altering genetic variants increased the odds of crossing the lipid-level threshold for therapeutic intervention by approximately three-fold.
Keywords: Lipid, SNP, Genome-wide association, HDL, LDL, Triglyceride, Myocardial infarction
Introduction
Raised total and low-density lipoprotein (LDL) cholesterol levels are established risk factors for coronary artery disease (CAD), and interventions to lower these, most commonly statin treatment, are part of standard medical practice.1 However, there is increasing evidence that low levels of high-density lipoprotein (HDL) cholesterol are an independent risk factor for cardiovascular disease2–5 and stroke.6 In addition, there is some evidence that raised triglyceride levels7,8 independently increase cardiovascular and diabetes risk.9,10
The National Institutes of Health Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults11 recommended that lifestyle and treatment interventions should be considered in individuals with LDL levels ≥130 mg/dL who have two or more additional risk factors (e.g. smoking, hypertension, age, family history of CAD) and ≥160 mg/dL for individuals with less than two additional risk factors. High-density lipoprotein levels <40 mg/dL were considered an additional risk factor for CAD, whereas levels >60 mg/dL are protective. Women have a higher recommended HDL threshold than men in the diagnosis of metabolic syndrome (≥50 mg/dL compared with ≥40 mg/dL, respectively), due to the influence of oestrogen on HDL levels. Treatment of triglyceride levels should be initiated when >150 mg/dL, first using lifestyle intervention, but if levels exceed 200 mg/dL, drug treatment should also be considered.
Over the last 18 months, genome-wide association scans (GWAS) have robustly identified or confirmed 21 independent variants affecting LDL, HDL, and triglyceride levels.12,13 Most of these variants are associated with small individual changes in their respective lipid levels and therefore studies have also assessed the impact of combining information from many variants. There is no evidence for interaction between individual variants, and a simple approach of counting risk alleles has been adopted. For example, Kathiresan14 recently showed that an allele count combining nine variants for LDL and HDL levels was associated with myocardial infarction (MI) in a sample with a mean age of 57 years.
In our study, we aimed to assess the combined effect of the variants identified/confirmed by GWAS to be associated with HDL, LDL, and triglyceride levels in a representative older population. We examined associations with recommended ‘need for intervention’ levels, and history of MI in the population-based InCHIANTI Study of Aging. As secondary outcomes, we also measured associations with stroke and peripheral arterial disease, hypothesizing that lipid-altering SNPs may be also associated with these vascular outcomes.
Methods
Study sample
The InCHIANTI Study of Aging recruited a total of 1453 individuals in a population representative sample from two towns in Tuscany (Italy). Of these, 1155 respondents were aged ≥65 years and were eligible for inclusion in our study. The study design and protocol have been described elsewhere.15 Subjects had an initial assessment at home between September 1998 and March 2000, which was followed by two separate clinic visits 2–3 weeks later. Follow-up assessments were made in 2001–03 and 2004–06. DNA was extracted from blood samples from the initial clinic visit, and all individuals in the study were asked to give blood. The participants were all of white European origin. The Italian National Institute of Research and Care of Aging Ethical Committee approved the study.
Serum measures
Blood samples were collected in the morning after participants had been fasting for at least 8 h and serum was separated. Commercial enzyme tests (Roche Diagnostics, GmbH, Manheim, Germany) were used to determine HDL and triglyceride levels, whereas LDL was calculated from the Friedewald formula. Mean levels are given in Table 1.
Table 1.
Summary characteristics of the InCHIANTI study sample (aged ≥65 years)
| Characteristic | All | Males | Females |
|---|---|---|---|
| Number genotyped | 635 | 274 | 361 |
| Male | 305 (43.4) | — | — |
| Mean age (years) | 75.0 (7.4) | 74.0 (6.9) | 75.7 (7.7) |
| BMI (kg/m2) | 27.4 (4.1) | 27.1 (3.4) | 27.6 (4.5) |
| Smokers | 103 (14.7) | 67 (22.0) | 36 (9.1) |
| Alcohol (g/day) | 13.8 (19.3) | 23.1 (24.0) | 6.6 (9.9) |
| Lipid levels (mg/dL) | |||
| Mean HDL | 55.2 (15.1) | 50.2 (12.5) | 58.9 (15.7) |
| Low HDL levels ≤40 | 97 (13.8) | 61 (20.7) | 36 (9.1) |
| High HDL levels ≥60 | 244 (34.8) | 70 (23.0) | 174 (43.7) |
| Mean LDL | 134.5 (33.3) | 130.2 (33.1) | 137.8 (33.2) |
| Borderline high LDL levels ≥130 | 390 (55.6) | 159 (52.3) | 231 (58.0) |
| High LDL levels ≥160 | 156 (22.2) | 57 (18.8) | 99 (24.9) |
| Mean triglyceride | 128.6 (71.9) | 133.7 (76.2) | 124.7 (68.3) |
| Borderline high triglyceride levels ≥150 | 177 (25.2) | 89 (29.3) | 88 (22.1) |
| High triglyceride levels ≥200 | 78 (11.1) | 46 (15.1) | 32 (8.0) |
| Myocardial infarction history | |||
| Cases at baseline and within 6 year follow-up | 67 (10.6) | 35 (12.8) | 32 (8.9) |
| Stroke history | |||
| Cases at baseline and within 6 year follow-up | 57 (9.2) | 33 (12.3) | 24 (6.8) |
| Peripheral artery disease history | |||
| Cases at baseline and within 6 year follow-up | 127 (21.6) | 81 (31.2) | 46 (13.9) |
| Hypertensives | 135 (21.3) | 43 (15.7) | 92 (25.5) |
| Diabetics | 64 (10.1) | 30 (10.9) | 34 (9.4) |
| Medication affecting lipid levelsa | 68 (10.7) | 23 (8.4) | 45 (12.5) |
All values are mean (SD) or n (%)
aIncludes those prescribed with diuretic and beta-blocker medication.
Myocardial infarction, stroke, and peripheral arterial disease outcomes
An MI or stroke event was recorded using evidence from self-report questionnaires with the question ‘Have you ever had an event?’ at the baseline assessment and also ‘Has your doctor reported an event since last interview?’ at the two follow-up interviews. Information was combined from both questions, to record individuals who had had an MI or stroke at baseline or during the 6 year follow-up period.
Peripheral artery disease (PAD) was defined using the ankle brachial index (ABI), defined as the ratio of systolic blood pressure in the ankle to systolic blood pressure in the arm. A measurement was taken from each leg, with the minimum value used in the analysis. Those with an ABI measure greater than 1.40 were removed, as this is indicative of non-compressible, calcified arteries,16 and inclusion of these individuals could have led to a misclassification of arterial extremity disease. An ABI measure less than 0.90 in either leg was used to define the presence of PAD, as this has been reported to be highly sensitive and specific for defining angiographically documented PAD.17
SNPs selected and genotyping
We selected 21 variants, of which 7 were associated with LDL (APOB, APOE haplotype, HMGCR, PCSK9, LDLR, NCAN/CILP2 and CELSR2-PSRC1-SORT1), 9 with HDL (ABCA1, the APOA1-C3-A4-A5 gene cluster, CETP, GALNT2, LIPG, LPL, MVK-MMAB and 2 in LIPC), and 11 with triglycerides (GCKR, APOB, 2 in the APOA1-C3-A4-A5 gene cluster, LPL, TRIB1, ANGPTL3, GALNT2, NCAN/CILP, LIPC and MLXIPL). Six of the variants were associated with more than one trait (APOB, APOA1-C3-A4-A5, GALNT2, LPL, NCAN-CLIP2-PBX4 and LIPC). We genotyped all of the 21 variants using 22 SNPs, as the APOE variant is defined by a two-SNP haplotype. Of the initial sample set of 1155 individuals aged ≥65, 635 were successfully genotyped for all variants and had lipid levels measured.
Genotyping was performed using version 1 and version 3 Illumina Infinium HumanHap550 genotyping chips, at the Laboratory of Neurogenetics of the US National Institute on Aging. This product assays >500k unique SNPs mostly derived from stages I and II of the International Haplotype Map Project (www.hapmap.org). The genotyping was performed according to the manufacturer’s instructions. After processing, the chips were scanned on Illumina BeadStation scanners. All of the data were analysed in BeadStudio (version 3; Illumina), with automated genotype calls made using the standard cluster files provided by Illumina. We used only SNPs that were called in >98% of samples and had minor allele frequencies in our sample of >1%. SNPs deviating appreciably from the expected population distribution (Hardy–Weinberg Equilibrium P < 1 × 10−4) were also excluded from the analyses. There were 496 032 SNPs that passed these QC criteria. Seven SNPs were not directly genotyped on the Illumina chip, but data were available for proxy SNPs in linkage disequilibrium with the original signal with r2 > 0.8 (Table 2). Six SNPs (rs3135506, rs1800588, rs328, rs662799, rs16996148, and rs4775041) were reported in the literature, but were not included or captured on the chip and were genotyped separately, using endpoint TaqMan PCR with assays designed by Applied Biosystems and genotypes called using Klustercaller software (KBioscience). In addition, the APOE (E2/E3/E4) haplotype, which is well known to influence lipid levels and is a CAD risk factor,18 but is poorly captured on the chip, was genotyped in-house.
Table 2.
The effect of validated polymorphisms included in the simple risk allele count on levels of high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglyceride in the InChianti sample, including proxy markers
| Validated SNP | Nearest gene(s) | Allele included in risk count | Hapmap allele frequency | Genotyped proxy | r2a | Per allele effect β (95% CI) | P-valueb | Lipid level variance explained (%) |
|---|---|---|---|---|---|---|---|---|
| LDL-raising variants | ||||||||
| rs693 | APOB | A | 0.492 | 3.25 (0.23, 6.27) | 0.035 | 0.40 | ||
| APOE | ϵ4 | NA | 5.66 (−1.06, 12.4) | 0.098 | 0.51 | |||
| rs646776 | CELSR2-PSRC1-SORT1 | T | 0.712 | 6.70 (2.89, 10.5) | 0.001 | 1.35 | ||
| rs12654264 | HMGCR | T | 0.417 | rs6896136 | 0.87 | 4.57 (1.61, 7.54) | 0.003 | 0.87 |
| rs6511720 | LDLR | G | 0.095 | 8.86 (4.37, 13.3) | 1.1 × 10−4 | 1.29 | ||
| rs11206510 | PCSK9 | T | 0.850 | 4.18 (0.34, 8.02) | 0.033 | 0.54 | ||
| rs16996148 | NCAN/CILP2 | G | 0.939 | 8.25 (1.82, 14.7) | 0.012 | |||
| HDL-lowering variants | ||||||||
| rs4149268 | ABCA1 | C | 0.267 | 0.015 (−0.01, 0.04) | 0.252 | 0.12 | ||
| rs3135506 | APOA1-C3-A4-A5 | G | 0.06 | −0.065 (−0.12, −0.01) | 0.017 | 0.52 | ||
| rs1800775 | CETP | C | 0.425 | −0.078 (−0.10, −0.05) | 1.4 × 10−10 | 4.76 | ||
| rs2144300 | GALNT2 | C | 0.417 | rs10779835 | 0.97 | −0.029 (−0.05, −0.005) | 0.018 | 0.75 |
| rs1800588 | LIPC | C | 0.742 | −0.049 (−0.08, −0.02) | 0.001 | 1.13 | ||
| rs2156552 | LIPG | A | 0.80 | rs4939883 | 0.95 | −0.014 (−0.05, 0.02) | 0.421 | 0.07 |
| rs328 | LPL | C | 0.875 | −0.059 (−0.09, −0.03) | 0.001 | 1.38 | ||
| rs2238104 | MVK-MMAB | C | 0.558 | −0.013 (−0.04, 0.01) | 0.282 | 0.11 | ||
| rs4775041 | LIPC | G | 0.737 | −0.036 (−0.06, −0.01) | 0.012 | |||
| Triglyceride-raising variants | ||||||||
| rs1748195 | ANGPTL3 | C | 0.675 | rs1167998 | 1.00 | 0.022 (−0.02, 0.07) | 0.302 | 0.09 |
| rs662799 | APOA1-C3-A4-A5 | G | 0.017 | 0.118 (0.05, 0.019) | 0.015 | 1.06 | ||
| rs3135506 | APOA1-C3-A4-A5 | G | 0.06 | 0.205 (0.11, 0.30) | 2.2 × 10−5 | 2.02 | ||
| rs693 | APOB | A | 0.492 | 0.041 (−0.0001, 0.08) | 0.009 | 0.40 | ||
| rs2144300 | GALNT2 | C | 0.417 | rs10779835 | 0.97 | 0.018 (−0.02, 0.06) | 0.166 | 0.08 |
| rs780094 | GCKR | T | 0.383 | 0.057 (0.02, 0.10) | 0.016 | 0.75 | ||
| rs328 | LPL | C | 0.875 | 0.104 (0.05, 0.16) | 0.013 | 1.24 | ||
| rs17145738 | MLXIPL | C | 0.881 | rs2240466 | 1.00 | 0.111 (0.03, 0.19) | 0.036 | 0.79 |
| rs17321515 | TRIB1 | A | 0.602 | rs6982636 | 1.00 | 0.052 (0.01, 0.09) | 0.041 | 0.65 |
| rs4775041 | LIPC | C | 0.263 | 0.006 (−0.04, 0.06) | 0.806 | |||
| rs16996148 | NCAN/CILP2 | G | 0.939 | 0.145 (0.06, 0.23) | 0.001 | |||
ar2 Values between validated SNP and genotyped proxy obtained from Hapmap CEU population.
bSignificance level for association between lipid level-altering SNP and associated serum lipid level.
All SNPs passed quality control criteria, which included Hardy–Weinberg Equilibrium P > 1 × 10−4 and a duplicate error rate of <1%. All studied variants represented independent signals, i.e. were not in linkage disequilibrium with each other.
Statistical analysis
Data from respondents aged 65 years and over were included in this analysis. Variables which deviated from normality were log-transformed before analysis. The effect of each validated SNP on the level of the appropriate lipid variable was determined (Table 2). Interactions between each pair of SNPs were checked for each serum lipid level in linear regression models. There were no statistical interactions between SNPs (at P < 0.01), and therefore simple counts of the number of alleles lowering HDL levels and raising both LDL and triglyceride levels were computed by adding up the numbers of dyslipidaemia risk alleles each individual carried. Groups including less than 10 respondents, at the tails of the distribution, were combined.
Lipid levels for each risk allele score were compared with recommended guidelines from The National Cholesterol Education Panel.11 Levels outside the normal range suggest that an individual will need a form of intervention, lifestyle, or otherwise. The thresholds are also quoted in the definition of the metabolic syndrome and recognized as risk factors for coronary heart disease. Specifically, these cut-points are defined as follows: HDL ≤40 mg/dL, LDL ≥130 mg/dL, and triglycerides ≥150 mg/dL.
To determine the impact of the combined effect of SNPs on lipid levels, we performed two analyses: (i) risk-altering alleles were treated as a continuous variable; (ii) we divided allele counts as near as possible into quartiles. In the latter analysis, we determined the odds ratios for crossing the intervention threshold. To determine whether the genetic risk counts were independent of conventional risk factors for raised lipid levels, we then examined associations adjusting for age, body mass index (BMI), cigarette smoking, and alcohol consumption (grams per day). Data were analysed with STATA SE 9.2 using linear regression with nominal two-sided significance taken as P < 0.05.
Our genotyped sample (n = 635) had a mean age of 75 years (Table 1): 22.2% had LDL levels ≥60 mg/dL and 11.1% had triglyceride levels ≥200 mg/dL. In total, 73 respondents (10.4%) reported having had an MI at baseline or during the 6 years of follow-up.
We examined associations of 7 variants with LDL, 9 with HDL, and 11 with triglycerides in an additive model (Table 2). We next computed simple risk allele counts (see Methods): these ranged from 4 to 14 (n = 594) for LDL-raising alleles, 5 to 16 (n = 635) for HDL-lowering alleles, and 7 to 16 (n = 611) for triglyceride-raising alleles, respectively (Figures 1–3).
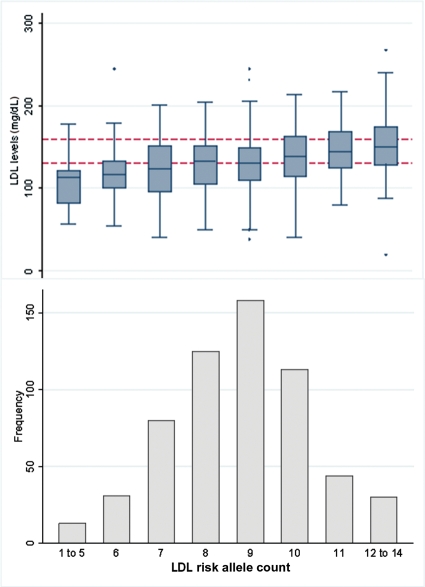
Figure 1.
Frequency of respondents by low-density lipoprotein (LDL) cholesterol allele count, with box-plots of the distribution of serum LDL cholesterol levels. Dotted lines indicate intervention levels of >130 mg/dL for borderline high levels and >160 mg/dL for high levels. The boxes indicate median and middle quartiles, whiskers include 1.5 times the inter-quartile range.
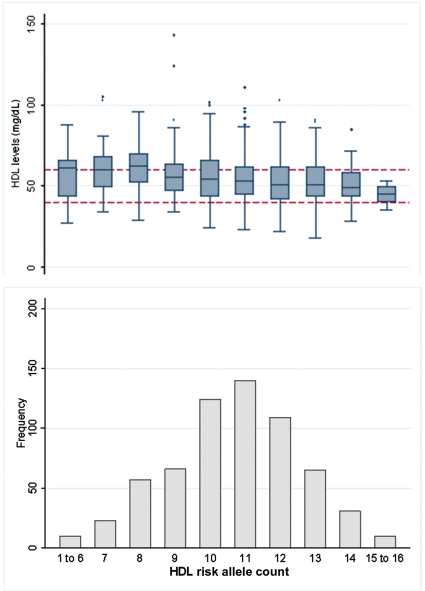
Figure 2.
Frequency of individuals by high-density lipoprotein (HDL) allele count, with box-plots of the distribution of HDL serum levels. Dotted lines indicate recommended levels, with <40 mg/dL being a risk factor for coronary artery disease and >60 mg/dL being protective.
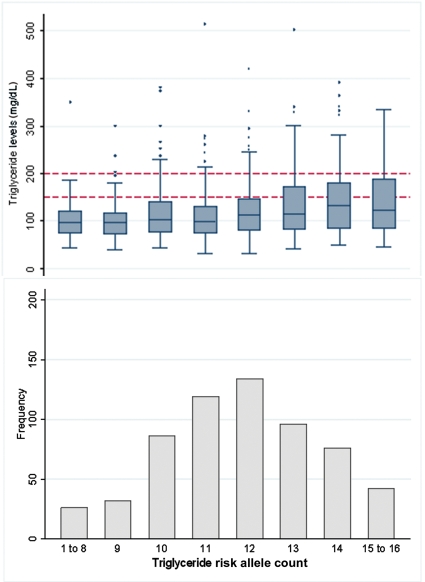
Figure 3.
Frequency of respondents by triglyceride allele count, with box-plots of the distribution of logged serum triglyceride levels. Dotted lines indicate recommended intervention levels of >150 and 200 mg/dL.
Results
Low-density lipoprotein levels
The LDL allele count was associated with an increase of 5.59 mg/dL in serum LDL levels per additional risk allele (P = 4.1 × 10−11) (Figure 1 and Table 3). This estimate was similar after adjustment for smoking status, alcohol consumption, and BMI. The allele count explained 7.1% (95% CI 2.5–9.4) of the variance in serum LDL levels, which was more than that explained by BMI (0.95%, 95% CI 0.1–2.5).
Table 3.
Linear regression models for continuous lipid levels against increasing number of lipid-altering risk alleles, including adjustments for conventional risk factors
| Lipid risk allele count | Lipid concentration (mg/dL) | Unadjusted |
Adjusteda |
||||
|---|---|---|---|---|---|---|---|
| Per allele effect size β (95% CI) | P-value | Variability explained (%) | Per allele effect size β (95% CI) | P-value | Variability explained (%) | ||
| LDL raising | LDL levels | 5.59 (3.96, 7.22) | 4.1 × 10−11 | 7.14 | 4.85 (2.80, 6.89) | 4.4 × 10−6 | 8.51 |
| HDL lowering | Logged HDL levels | −0.026 (−0.04, −0.02) | 3.2 × 10−6 | 3.38 | −0.021 (−0.03, 0.01) | 0.001 | 20.1 |
| Triglyceride raising | Logged triglyceride levels | 0.056 (0.04, 0.08) | 4.0 × 10−8 | 4.84 | 0.047 (0.02, 0.07) | 1.5 × 10−4 | 11.4 |
aAge (years), gender, smoking status, alcohol consumption (g/day), and body mass index (kg/m2) adjusted.
We examined the effect of allele counts on the likelihood of crossing the intervention threshold in adjusted logistic regression models (Table 4). Individuals with an LDL-raising allele count of 10 or more (comprising 31%, or approximately the top quartile, of the sample) formed the highest genetic risk group, and those with 7 or less alleles formed the lowest risk group (21% of sample or bottom quartile). The highest allele count group were nearly three times more likely to qualify for intervention (OR 3.37, 95% CI 1.64–6.94, P = 0.001, in fully adjusted model) compared with the lowest group.
Table 4.
Logistic regression odds ratios for lipid levels crossing the therapeutic intervention threshold, comparing highest risk alleles count group against the lowest, with adjustment for conventional risk factors
| Lipid measure, Intervention thresholds | Lipid risk allele count |
Lipid risk alleles as quartiles |
|||||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | Highest vs. lowest quartile (age- and sex-adjusted OR) |
Highest vs. lowest quartile (fully adjusteda) |
P-value for trend across quartiles | |||
| OR (95% CI) | P-value | OR (95% CI) | P-value | ||||
| LDL cholesterol (mg/dL) | |||||||
| Borderline high: >130 | 1.32 (1.19, 1.48) | 5.7 × 10−7 | 3.38 (2.09, 5.47) | 7.2 × 10−7 | 3.28 (1.81, 5.93) | 8.9 × 10−5 | <0.001 |
| High: >160 | 1.34 (1.18, 1.53) | 7.1 × 10−6 | 3.00 (1.67, 5.39) | 2.5 × 10−4 | 3.37 (1.64, 6.94) | 0.001 | <0.001 |
| HDL cholesterol (mg/dL) | |||||||
| Low levels: <40 | 1.14 (1.01, 1.29) | 0.04 | 2.13 (1.10, 4.12) | 0.025 | 1.41 (0.62, 3.24) | 0.416 | 0.022 |
| High levels: >60 | 0.83 (0.76, 0.91) | 7.9 × 10−5 | 0.49 (0.31, 0.76) | 0.002 | 0.55 (0.31, 0.99) | 0.045 | <0.001 |
| Triglycerides (mg/dL) | |||||||
| Borderline high: >150 | 1.36 (1.22, 1.52) | 3.8 × 10−8 | 3.50 (2.06, 5.93) | 3.3 × 10−6 | 2.77 (1.48, 5.19) | 0.001 | <0.001 |
| High: >200 | 1.35 (1.16, 1.57) | 8.2 × 10−5 | 2.98 (1.43, 6.22) | 0.004 | 2.58 (1.09, 6.09) | 0.030 | <0.001 |
aAge (years), gender, smoking status, alcohol consumption (g/day), and body mass index (kg/m2) adjusted.
High-density lipoprotein levels
The HDL related allele count was associated with (logged) HDL levels (β: −0.026, 95% CI −0.04 to −0.02, P = 3.2 × 10−6) (Table 3), which relates to a 2.6% decrease per allele in average HDL levels (2.0 mg/dL). The allele count explained 3.4% (95% CI 1.2–6.4) of the variance in HDL levels. This was less than the variance explained by the CETP rs1800775 SNP alone (4.76%).
For HDL, a score of 12 or more HDL-lowering alleles (33% of individuals) formed the highest risk group, whereas those with 9 or less alleles formed the lowest risk group (25% of sample) (Table 4). The HDL allele groups were not associated with HDL levels below the treatment threshold (HDL <40 mg/dL, OR 1.41, 95% CI 0.62–3.24, P = 0.416), but there was a trend towards those with the highest HDL genetic risk scores being less likely to be above the protective levels of HDL (≥60 mg/dL) (OR 0.55, 95% CI 0.31–0.99, P = 0.045) (Table 4 and Figure 2), in fully adjusted models.
We also analysed the data separately in men and women using the cut-points of 40 mg/dL for men and 50 mg/dL for women according to the guidelines for the definition of metabolic syndrome. There was a strong association between the HDL-C allele count and serum levels in men, with each additional HDL-lowering allele was associated with a 5.2 mg/dL reduction in HDL levels (P = 4.4 × 10−4) (see Supplementary material online, Figure S1). In women, each allele was associated with only a 1.05 mg/dL reduction (P = 0.386) (see Supplementary material online, Figure S1). Males in the highest HDL-C genetic risk group (27% of the sample) had an adjusted odds ratio of 2.83 (95% CI 1.44–6.18, P = 0.054) for having HDL-C levels ≤40 mg/dL. Females in the highest risk group (35% of the sample) had an adjusted odds ratio of 1.35 (95% CI 0.57–3.18, P = 0.499) for having serum levels ≤50 mg/dL.
Triglyceride levels
Serum triglyceride levels were strongly associated with the triglyceride-raising allele count, each additional allele raising average triglyceride levels by 5.6%, which relates to a 1.45 mg/dL increase in mean levels (P = 4.0 × 10−8) (Figure 3 and Table 3). The count of combined triglyceride-raising alleles explained 4.8% of the variance, again comparable to the 4.6% explained by BMI.
A triglyceride allele count of 13 or more formed the highest genetic risk group (35% of sample), whereas a count of 10 or less formed the lowest risk group (24% of the sample) (Table 4). The highest count group had an odds ratio of 2.77 (95% CI 1.48–5.19, P = 0.001) for having serum triglyceride levels above the recommended limit of 150 mg/dL, compared with those in the lowest group, in fully adjusted models.
Associations with myocardial infarction
We had only limited power to detect associations with disease outcomes because the number of affected individuals was relatively small. However, we examined associations between allele risk counts and 67 patients with MI (Table 5). Combining the allele scores for all three lipid measures, in a total allele count score that included all 21 risk variants, resulted in an association between combined allele count and MI with an OR of 1.16 (95% CI 1.04–1.30, P = 0.007): all but the LDL risk variant score gave nominally significant P-values (<0.05). Individuals with the highest combined number of risk alleles (count ≥25, 36% of sample) had an odds ratio of 3.68 (95% CI 1.21–11.2, P = 0.021) for a history of MI compared with those with the lowest number of risk alleles (count ≤21, 22% of sample). We found no associations between allele counts and the occurrence of stoke or PAD.
Table 5.
Odds ratios for vascular disease outcomes, comparing highest risk alleles count group against the lowest
| Dyslipidaemia risk allele score | Risk allele groups |
MI |
Stroke |
Peripheral arterial disease |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Highest vs. lowest, allele group |
Risk allele count |
Highest vs. lowest allele group |
Risk allele count |
Highest vs. lowest allele group |
Risk allele count |
|||||||||
| Highest | Lowest | |||||||||||||
| Cut-point (%) | Cut-point (%) | OR (95% CI) | P-valuea | OR (95% CI) | P-value | OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | |
| LDL-increasing variants | ≥10 (31) | ≤7 (21) | 1.64 (0.72, 3.73) | 0.234 | 1.06 (0.90, 1.25) | 0.514 | 1.76 (0.78, 3.99) | 0.172 | 1.06 (0.93,1.22) | 0.375 | 1.45 (0.89, 3.49) | 0.265 | 1.14 (0.96, 1.36) | 0.138 |
| HDL reducing variants | ≥12 (33) | ≤9 (25) | 1.74 (0.89, 3.38) | 0.104 | 1.20 (1.04, 1.38) | 0.010 | 1.40 (0.64, 3.04) | 0.397 | 1.14 (0.97,1.33) | 0.457 | 1.18 (0.67, 2.07) | 0.568 | 1.14 (0.97,1.33) | 0.102 |
| Triglyceride-increasing variants | ≥13 (35) | ≤10 (24) | 2.44 (1.07, 5.56) | 0.033 | 1.19 (1.03,1.37) | 0.020 | 1.71 (0.76, 3.85) | 0.193 | 1.02 (0.91,1.14) | 0.767 | 1.15 (0.65, 2.03) | 0.627 | 1.14 (0.98, 1.33) | 0.092 |
| HDL + LDL | ≥21 (35) | ≤17 (21) | 1.83 (0.79, 4.27) | 0.160 | 1.15 (1.03,1.30) | 0.018 | 2.39 (0.89, 6.38) | 0.082 | 1.16 (1.01,1.32) | 0.030 | 1.02 (0.52, 1.98) | 0.959 | 1.05 (0.95,1.15) | 0.329 |
| HDL + LDL + Triglycerides (21 variantsb) | ≥25 (36) | ≤21 (22) | 3.68 (1.21, 11.2) | 0.021 | 1.16 (1.04,1.30) | 0.007 | 1.60 (0.58, 4.43) | 0.367 | 1.11 (0.98,1.26) | 0.099 | 1.36 (0.67, 2.74) | 0.389 | 1.08 (0.99,1.17) | 0.102 |
aAge (years)- and gender-adjusted.
brs4775041 risk allele is counted as G allele, which lowers HDL (β = −0.036) and not the C allele, which raises triglycerides (β = 0.006).
Discussion
Low HDL and raised LDL and triglyceride levels have emerged as important independent risk factors for cardiovascular disease, especially in older people. Recent genome-wide scans have robustly replicated associations between lipid levels and many genetic loci. We have demonstrated that the cumulative effect of the 21 common genetic variants are clinically relevant: those in the approximate top quartile of LDL and triglyceride genetic risk have clearly raised odds of being outside recommended lipid levels compared with the bottom quartile. In addition, we have shown an association between the overall lipid risk allele count and MI outcomes, similar to findings in younger samples. Although the SNP counts explain <6% of the unadjusted variance in their respective lipid levels, this is similar or greater than the variance explained by the most powerful conventional risk factor, BMI, and therefore it should not be surprising that it is clinically relevant. By combining genetic risk variants, we have demonstrated that we can explain clinically relevant changes in lipid levels in older people, raising the possibility that genetic variants could be used for prognostic purposes, particularly as more variants are discovered. Further work is required, but it is possible that genetic variants may be a better prognostic indicator than measuring lipid levels directly, as they are invariant over time and thus represent lifetime exposure.
The association of the 21 lipid variants with their respective lipid levels has been very robustly demonstrated in younger samples.19 Many of these SNPs have only recently been discovered, and to the best of our knowledge, there are no other analyses covering as complete a set of variants, especially from older populations. As noted, Kathiresan14 recently showed that an allele count combining nine variants for LDL and HDL levels was associated with lipid levels and MI in a cardiovascular cohort of 5414 subjects with a mean age of 57 years. A combined LDL and HDL genotype score was associated with incident cardiovascular disease (n = 238 cases), in models adjusted for covariates (P < 0.001). Similarly, the LDL-increasing variants identified from GWAS were all more common in a sample of approximately 2000 people with CAD compared with controls. However, in that study, other lipid-altering genetic variants were not more common in CAD cases, with the exception of an SNP near TRIB1, which is associated with triglyceride levels.13 A recent meta-analysis18 of the LDL-altering APOE haplotype including 121 studies (37 850 cases and 82 727 controls) found an association with coronary disease: OR 1.06 (95% CI 0.99–1.13) in e4 carriers and OR 0.80 (95% CI 0.70–0.90) in e2 carriers, compared with e3/e3 group. In addition, a meta-analysis of variants in the CETP gene confirmed effects on HDL levels and an association with MI.20 In our analysis of associations with MI outcomes, one shortcoming is clearly the limited sample size available (n = 67 cases). As a result, the confidence intervals on the associations for the individual lipids are wide, with the LDL estimates not reaching formal significance. Clearly, a much larger replication study in older people is justified.
Using allele counts to compute the combined effects of variants can be criticized as being somewhat crude. Tiret et al.21 pointed out that such un-weighted counts make two major assumptions: first, that polymorphisms act independently and additively on the risk of disease, and secondly that their effects are approximately interchangeable. We have included only independent markers in our allele risk scores and found no evidence of statistical interactions. However, accurate estimates of the weights to be used in older populations for the effects of each SNP on their respective lipid level and on outcomes are not available. Using the effect sizes from our own sample would introduce an element of circular logic, magnifying any random fluctuations in the InCHIANTI study. We have therefore followed Kathiresan et al.14 in using a simple allele count. We would expect that the strengths of associations will increase, as more accurate ways of combining the studied SNP effects become available. In the case of the HDL count, the CETP rs1800775 SNP alone did explain more of the variance than the HDL allele score in our data set, illustrating that the allele count is an approximate measure with shortcomings, as well as the strength of simplicity.
Conclusions
In a representative older population, the cumulative effects of proven LDL- and triglyceride-altering genetic variants had a substantial effect on serum lipid levels, increasing the odds by approximately three-fold of an individual crossing the therapeutic intervention threshold of lipid levels, based on ATP-NCEP guidelines.
Supplementary material
Supplementary Material is available at European Heart Journal Online.
Funding
This work is supported in part by National Institutes of Health/NIA grant R01 AG24233, and by the Intramural Research Program, National Institute on Aging, NIH. The InCHIANTI study was supported as a ‘targeted project’ (ICS 110.1\RS97.71) by the Italian Ministry of Health, by the US National Institute on Aging (Contracts N01-AG-916413, N01-AG-821336 and Contracts 263 MD 9164 13 and 263 MD 821336). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by the Peninsula College of Medicine and Dentistry.
Conflict of interest: none declared.
References
- 1.Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes R. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 2.Barter P, Gotto AM, LaRosa JC, Maroni J, Szarek M, Grundy SM, Kastelein JJ, Bittner V, Fruchart JC. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357:1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- 3.Brewer HB., Jr Increasing HDL cholesterol Levels. N Engl J Med. 2004;350:1491–1494. doi: 10.1056/NEJMp048023. [DOI] [PubMed] [Google Scholar]
- 4.Corti MC, Guralnik JM, Salive ME, Harris T, Field TS, Wallace RB, Berkman LF, Seeman TE, Glynn RJ, Hennekens CH, et al. HDL cholesterol predicts coronary heart disease mortality in older persons. JAMA. 1995;274:539–544. [PubMed] [Google Scholar]
- 5.Packard CJ, Ford I, Robertson M, Shepherd J, Blauw GJ, Murphy MB, Bollen EL, Buckley BM, Cobbe SM, Gaw A, Hyland M, Jukema JW, Kamper AM, Macfarlane PW, Perry IJ, Stott DJ, Sweeney BJ, Twomey C, Westendorp RG. Plasma lipoproteins and apolipoproteins as predictors of cardiovascular risk and treatment benefit in the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER) Circulation. 2005;112:3058–3065. doi: 10.1161/CIRCULATIONAHA.104.526848. [DOI] [PubMed] [Google Scholar]
- 6.Sanossian N, Saver JL, Navab M, Ovbiagele B. High-density lipoprotein cholesterol: an emerging target for stroke treatment. Stroke. 2007;38:1104–1109. doi: 10.1161/01.STR.0000258347.19449.0f. [DOI] [PubMed] [Google Scholar]
- 7.Bansal S, Buring JE, Rifai N, Mora S, Sacks FM, Ridker PM. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA. 2007;298:309–316. doi: 10.1001/jama.298.3.309. [DOI] [PubMed] [Google Scholar]
- 8.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 9.Hokanson JE, Austin MA. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J Cardiovasc Risk. 1996;3:213–219. [PubMed] [Google Scholar]
- 10.Yarnell JW, Patterson CC, Sweetnam PM, Thomas HF, Bainton D, Elwood PC, Bolton CH, Miller NE. Do total and high density lipoprotein cholesterol and triglycerides act independently in the prediction of ischemic heart disease? Ten-year follow-up of Caerphilly and Speedwell Cohorts. Arterioscler Thromb Vasc Biol. 2001;21:1340–1345. doi: 10.1161/hq0801.093505. [DOI] [PubMed] [Google Scholar]
- 11.National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Final Report. Circulation. 2002;106:3143–3421. [PubMed] [Google Scholar]
- 12.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 15.Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, Guralnik JM. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 16.Resnick HE, Lindsay RS, McDermott MM, Devereux RB, Jones KL, Fabsitz RR, Howard BV. Relationship of high and low ankle brachial index to all-cause and cardiovascular disease mortality: the Strong Heart Study. Circulation. 2004;109:733–739. doi: 10.1161/01.CIR.0000112642.63927.54. [DOI] [PubMed] [Google Scholar]
- 17.Lijmer JG, Hunink MG, van den Dungen JJ, Loonstra J, Smit AJ. ROC analysis of noninvasive tests for peripheral arterial disease. Ultrasound Med Biol. 1996;22:391–398. doi: 10.1016/0301-5629(96)00036-1. [DOI] [PubMed] [Google Scholar]
- 18.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, Danesh J. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 19.Kathiresan S, Musunuru K, Orho-Melander M. Defining the spectrum of alleles that contribute to blood lipid concentrations in humans. Curr Opin Lipidol. 2008;19:122–127. doi: 10.1097/MOL.0b013e3282f70296. [DOI] [PubMed] [Google Scholar]
- 20.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 21.Tiret L, Tregouet DA, Cambien F. Cholesterol gene polymorphisms and cardiovascular events. N Engl J Med. 2008;359:92–93. [author reply 93] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.