Abstract
Malignant melanoma represents one of the most aggressive malignancies but outcome is highly variable with early tumor lesions having an excellent prognosis following resection. We review here the data on identification of genes involved in the progression of melanoma as a result of expression array studies, genomic profiling, and genetic models. We focus on the role of tumor suppressors involved in cell cycle function, DNA repair, and genome maintenance. Highlighted are the roles of loss of p16 in promoting neoplasia in cooperation with deregulated MAPK signaling, and the role of loss of the RASSF1A protein in promoting chromosomal instability. The interactions between point mutation in growth signaling molecules and epigenetic changes in genes involved in DNA repair and cell division are discussed.
Key Words: Melanoma, genetic progression, tumor suppressors, cell cycle, alkylating agents, mitotic spindle, mitotic spindle poisons.
I. HETEROGENEITY IN MELANOMA: CLINICAL PRESENTATION AND OUTCOMES
Melanoma arises from malignant transformation of melanocytes and has an aggressive course once the tumor has spread beyond the superficial skin. However, like all tumor types there is considerable heterogeneity in outcome and molecular pathogenesis. Clinically distinct patterns of melanoma include acral lentiginous (AL) presenting in the distal extremities, and superficial spreading (SS), lentigo maligna, and nodular types. Almost all histologic and clinical patterns of melanoma are increased in patients with a history of heavy sun exposure, particularly discrete serious sunburn episodes but other risk factors are poorly understood. Mucosal and soft tissue presentations of melanoma while rare appear to have a distinct pathogenesis. There are also a variety of different histologic appearances in melanoma including the typical epithelioid forms, as well as desmoplastic (spindle cell) and anaplastic variants. Approximately 5-10% of melanomas have a strong familial link, and the molecular defects in most of these cases involve cell cycle regulators, particularly cyclin-dependent kinases (CDKs) and the CDK inhibitor p16 (CDKN2A) in their molecular pathogenesis [1-5].
II. THERAPEUTIC OPTIONS FOR MELANOMA
Outcome in melanoma is highly dependent on the depth of invasion seen in the primary lesion. The predominant therapeutic modality in melanoma remains surgical resection with adequate margins. However, patients with residual disease after resection, with lymph node metastases, or with distant metastatic spread usually receive adjuvant therapy [6]. Radiation therapy is employed for symptomatic relief of brain and visceral metastases that cannot be resected. Advanced melanoma is highly resistant to most forms of chemotherapy and response rates to DTIC (dacarbazine) or other alkylating agents such as temozolomide (TMZ) [7], carmustine and lomustine in combination with spindle poisons such as vincristine are seen in approximately 10 to 20% [8-11]. Biologic treatments are also employed for metastatic melanoma, with trials using interferon-alpha and interleukin-2 generally showing responses in 10 to 20% of patients [12-14]. However, these treatment strategies have not yet been optimized due to a lack of biomarker predictors of response to either approach.
III. MOLECULAR MARKERS OF DYSREGULATED GROWTH FACTOR SIGNALING AND THE USE OF TYROSINE KINASE INHIBITORS
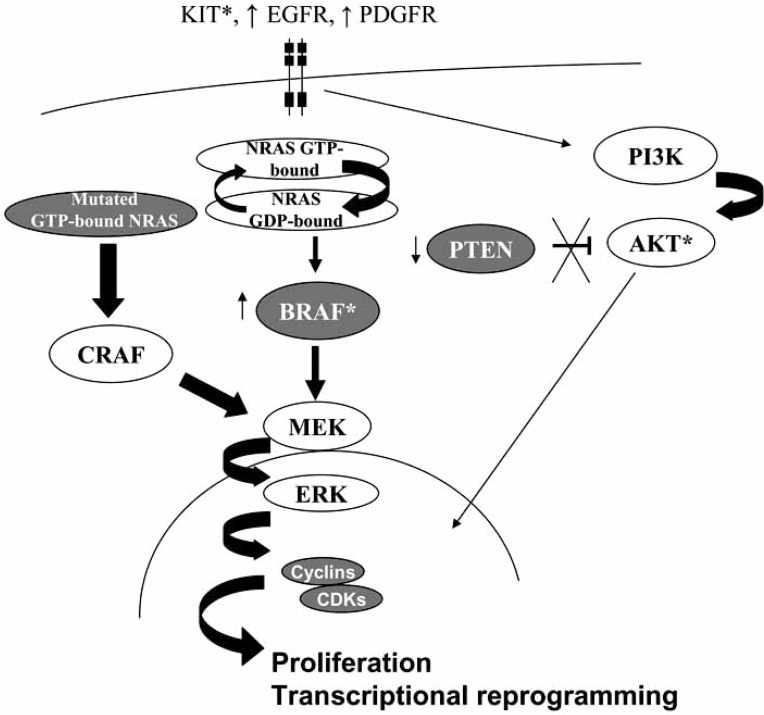
Many of the best characterized somatic mutation and epigenetic changes in melanoma involve the receptor tyrosine kinase (RTK) signaling pathways, which are particularly implicated in sun-exposed (cutaneous) cases [15]. These genetic alterations include BRAF, NRAS, KIT and (rarely) AKT1/3 point mutation, EGFR and PDGRA genomic amplification, and epigenetic silencing or mutation of the tumor suppressors PTEN and FGFR2 (Fig. 1).
Fig. (1).
Receptor tyrosine kinase (RTK) pathway dysregulation in melanoma pathogenesis. Genetic alterations seen in different subsets of melanoma include point mutation (*) of the RTK KIT, genomic amplification (↑) of EGFR and PDGRFA, point mutation of NRAS and BRAF, complete loss of PTEN expression (↓), and rarely point mutation of AKT1 and AKT3 (*).
BRAF is a serine/threonine kinase that signals downstream of RTKs and ras proteins. 30-70% of melanomas show BRAF point mutations which alter the autoregulatory activation of the kinase, of which the V600E mutation is by far the most common. BRAF mutations are most common in the nodular and SS types, and rare in AL (5-10% of cases) and non-cutaneous melanomas [16-18] (Table 1). BRAF mutation correlates with distinct histopathologic features, such as intraepidermal melanoma nest formation and a larger rounder border of the tumor with surrounding skin, suggesting surrogate markers can be used to select cases for molecular testing [19]. BRAF mutations also arise more commonly in patients with younger age at presentation and lymph node metastasis (rather than satellite tumors or visceral metastasis) [19]. However, benign nevi show similar or higher rates of the V600E BRAF mutation [20, 21], and cell line and transgenic mouse models of melanoma do not clearly demonstrate the transforming power of this mutation. Germline BRAF mutations do not occur in melanoma [22].
Table 1.
Incidence of Growth Factor Mutations in Different Melanoma Subtypes
| Site of Melanocytic Lesions | Incidence of RTK Mutations/Amplification | Incident of Genetic/Epigenetic Cell Cycle Alterations | References |
|---|---|---|---|
| Chronic sun-damaged sites | BRAF: 10% NRAS: 20% KIT mut/amp: 28% |
CDKN2A/p16 loss: 88% | [17, 26, 70] |
| Nodular sun-exposed sites | BRAF: 36.3% NRAS: 54.5% PTEN loss of expression: 7% |
CDKN2A/p16 loss: 78.9% p27KIP1 LOH: 38.9% |
[18, 37, 40, 67, 69] |
| Acral melanoma | PDGFRA amp: 18% KIT mut/amp: 36% FGFR2 mutation: 4% |
CDK4 amp: 23% CCND1 (cyclin D1) amp: 23% PDGFRA amp: 18% |
[26, 34, 37] |
| Mucosal melanoma | KIT mut/amp: 53.3% FGFR2 mutation: 6% |
CDKN2A/p16 LOH: 33% | [16, 26, 37, 68] |
| Uveal melanoma | PTEN LOH: 39.5% PTEN loss of expression: 16% |
CDKN2A/p16 methylation: 32% | [39, 53] |
| Congenital nevi | NRAS: 81.3% | [25] | |
| Sporadic nevi | BRAF: 20% | [20] | |
| Inherited melanoma syndromes | CDKN2A/p16 mutation: 70% CDK4 amp: (infrequent) |
[1-4, 58] |
Abbreviations: mut: point mutation; amp: genomic amplification; LOH: loss of heterozygosity (genomic deletion).
NRAS is a GTPase protein which functions to integrate signals from multiple RTKs. 10-20% of melanoma have point mutations in codon 12, 13 or 61 of NRAS, which are almost always mutually exclusive with BRAF mutation [15, 17, 18, 23]. This is possibly because NRAS-mutated melanoma may bypass BRAF and signal through CRAF [24]. NRAS mutations are rarely found in benign acquired nevi (although seen in congenital nevi) [25], arise later in melanoma development, and can produce melanoma in certain animal models and are thus are more clearly implicated in oncogenesis than BRAF mutations.
KIT is a RTK that is essential for normal neural crest/melanocyte differentiation. Activating KIT mutations are typically seen in mucosal and AL melanoma subtypes (5-20% of cases) but not in cases arising from chronic sun damage. Point mutations frequently occur in KIT exons 11, 13 and 17 [26]. The L576P mutation is most common (comprising 50% of mutations). KIT gene amplification also occurs [21, 27]. KIT mutations are mutually exclusive with BRAF and NRAS, and may identify a subset of melanoma that preferentially respond to the KIT inhibitors such as imatinib (Gleevec) [28, 29] or sorafenib [30, 31]. There are also regulatory changes in KIT expression during melanoma progression. For example, the highest levels of KIT expression are seen at the leading edge of tissue invasion which may indicate a role for dynamic RTK activation in metastasis [26].
EGFR is a RTK implicated in normal epithelial and melanocyte maturation. It is often overexpressed by gene amplification (usually whole gain of chromosome 7) in metastatic melanomas [32], but the prognostic impact of detection of gene amplification remains unresolved. Rare cases of melanoma (often of desmoplastic type) may show activating EGFR mutations. The RTKs platelet-derived growth factor receptor (PDGR)-alpha and PDGFR-beta are also highly expressed in melanocytic lesions [33]. Genomic profiling has revealed that AL and mucosal melanomas can have chromosomal amplification at chromosome 4q11spanning the PDFGRA locus [34, 35] that may contribute to increased PDGF signaling [36]. Finally, dominant-negative FGFR2 mutations have been reported in approximately 10% of nodular melanomas [37]. FGFR2 is a RTK that may mediate growth arrest in melanoma through interactions with stroma so its inactivation may promote tissue invasion.
PTEN is a phosphatase that regulates the activation of the serine/theronine kinases AKT1/2/3, which are global regulators of cell proliferation. PTEN is regulated as a tumor suppressor with complete PTEN loss (usually accompanied by genomic deletion) seen in 20-25% of melanomas (including those with BRAF mutation) [38-42], and is highly associated with uniform high-level AKT activation [43]. PTEN loss and concomitant AKT activation are both usually demonstrated by immunohistochemistry, with an anti-phosphoprotein antibody against an activation site on AKT (pS473) [40, 43]. Rare activation mutations in the AKT1 or AKT3 isoforms have been found in sun-exposed melanoma subtypes [44], and AKT overexpression may be associated with melanoma growth in situ [45, 46].
Variations in the incidence of different RTK pathway mutations in different geographic populations are evident [47], as well as variations in risk related to polymorphisms in other susceptibility loci [48-50]. Since most advanced-stage melanomas are resistant to existing adjuvant therapies, kinase inhibitors (KIs) have been tried in melanomas that demonstrate mutational activation of the kinases above. In single case reports or in small series, KIs have shown promising short-term responses that generally correlate with the presence of targetable RTK mutation in the tumor. For example, administration of sorafenib (a KI with activity against RAF, PDGFR, VEGF, and KIT) along with carboplatin and paclitaxel in a phase II trial has led to a partial response rate of 26% [51]. Results on the use of imatinib (Gleevec), a KI with high activity against KIT and PDGFR, have been disappointing with responses possibly correlated with either KIT mutation or high-level KIT protein expression (e.g. ≥75% of tumor cells) [52].
IV. MARKERS OF MELANOMA PROGRESSION IDENTIFIED BY GENE EXPRESSION STUDIES
Gene expression changes that occur during tumor progression can be due to chromosomal gains/losses resulting from cell cycle alterations (discussed below), activating mutations in pathways that modulate transcription factors (e.g. the RTK pathway mutation), or by epigenetic regulation. Melanoma at sun-exposed sites may more frequently demonstrate (UV-induced) genetic mutation whereas melanomas arising at non-sun-exposed sites may more frequently utilize epigenetic regulation but overlapping patterns are clearly seen.
This role of epigenetic regulation is clearly highlighted by silencing of multiple different tumor suppressor genes during melanoma progression. For example, the cell cycle regulator p16 is frequently silenced by promoter DNA CpG methylation (e.g. 32% of uveal melanoma) [53], as is the APC gene which regulates Wnt signaling in 10-20% of cases [54], and the kinase regulator RASSF1 in up to 50% of cases [55]. The DNA repair gene MGMT (O-6-methylguanine-DNA-methyltransferase) is silenced in approximately 20% of melanoma [56], and its inactivation corresponds with declines in the ability to repair DNA which may promote mutagenesis and potentiate the response to DNA-damaging chemotherapy [57].
Microarray gene expression profiling of primary and progressed melanoma and melanoma cell lines have revealed many of the coordinated changes in gene expression that correlate with clinical stage [36, 56, 58-65]. For example, early-stage melanomas often express high levels of the immune modulator CD24 and the transcription factor GATA3, whereas progressed melanomas exhibit upregulation of the melanoma antigen family A (MAGE) antigens of unknown function, and cell cycle regulators such as CDK2 [36]. The commonalities arising from these GEP studies of melanoma progression (Table 2) highlight several fundamental patterns of transcriptional dysregulation that may prove useful in individualizing therapy response and in developing novel treatment strategies.
Table 2.
Genes Involved in Melanoma Progression Identified by Gene Expression Profiling
| Gene | Function | Fold-Change | Comparison Group | References |
|---|---|---|---|---|
| Upregulated | ||||
| BIRC5 | Component of chromosome passenger complex that ensures chromosome alignment/segregation | ↑3-5X | primary → metastasis primary → metastasis |
[36, 64] |
| BUB | Mitotic kinase that functions in spindle checkpoint function | ↑4-11X | primary → metastasis nevi → melanoma |
[36, 59] |
| CDK2 | Kinase that regulates the G1-S transition | ↑3-9X | primary → metastasis nevi → melanoma |
[36] [65] |
| CHEK1 | Mitotic kinase that phosphorylates cdc25 at G2-M transition | nr | nevi → melanoma blood of metastatic cases | [60, 62] |
| CCNA2 (cyclin A) | Binds and activates CDC2 and CDK2 at the G1-S and G2-M transition | nr | nevi → melanoma | [60, 63] |
| MAGEA1 | Mediator of transformation through extracellular/adhesion signaling | ↑25X | primary → metastasis primary → metastasis |
[36, 61] |
| MAGEA2 | As above | ↑31X | primary → metastasis primary → metastasis |
[36, 61] |
| Downregulated | ||||
| MAP4 | Microtubule binding protein stabilizing the cyclin B/CDC2 kinase mitotic complex | ↓20X | nevi → melanoma | [36] |
| CDKN2A/p16 | Cyclin-dependent kinase inhibitor that regulates G1-S transition | nr | primary → metastasis primary → metastasis |
[58] [83] |
| CDKN1B/p27 | Inhibitor of cyclin E-CDK2 and cyclin D-CDK4 complexes at G1-S transition | nr | primary → metastasis primary → metastasis |
[67] [58] |
| SFN | Inhibitor of p53 function at G2-M transition | ↓24X | primary → metastasis primary → metastasis |
[36, 61] |
| FGFR3 | RTK stromal signals/differentiation | ↓8X | primary → metastasis | [36, 61] |
V. G1-S CHECKPOINT ALTERATIONS IN MELANOMA PROGRESSION
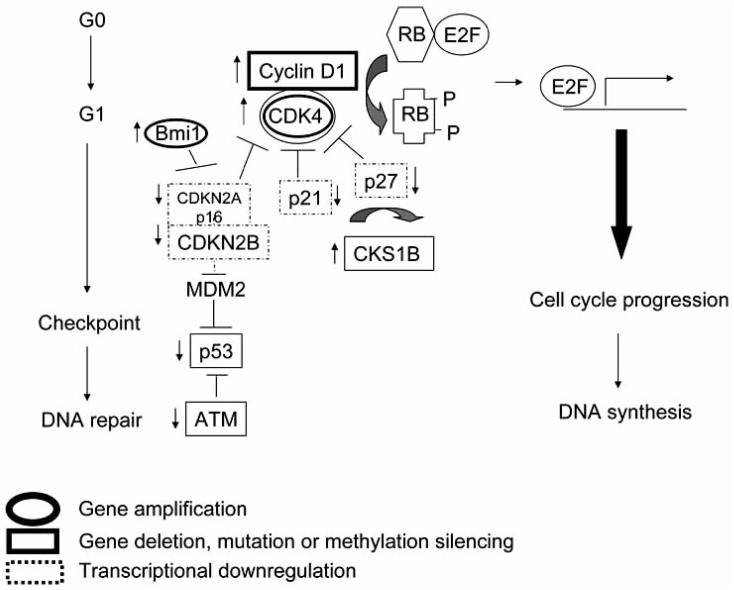
Genetic studies [4], genomic profiling [34], and GEP have all highlighted the multiple overlapping genetic and epigenetic alterations in the proteins regulating the G1-S transition in the cell cycle (Fig. 2). Among the 5-10% of melanomas with strong familial linkage or inherited germline defects, mutations in p16 (CDKN2A) [4, 5] and CDK4 [3] are common findings, as is loss of p27 [66, 67] and p16 [68, 69] expression during progression of sporadic melanomas. Indeed, loss of p16 appears to be common to most melanoma subtypes including superficial spreading, mucosal and nodular cases [38, 68-70]. Secondary genetic changes in melanoma also frequently involve the same genes, evidenced by the frequent genomic deletions at chr 12q14 spanning the CDK4 loci in AL melanoma [67], and chr 17p13 loss at the TP53 locus in chronic sun-damage melanomas [34].
Fig. (2).
Cell cycle dysregulation in melanoma progression. Complex patterns of transcriptional and epigenetic regulation of the cyclin-dependent kinases (CDKs), CDK inhibitors and the p53 axis have been demonstrated in microarray genomic and expression studies of melanoma progression. These include downregulation (↓) of inhibitors of CDK by DNA methylation silencing, deletion and transcriptional networks, and genomic amplification and microRNA regulation of CDKs and cyclins.
Multiple abnormalities in the G1-S proteins can occur simultaneously in the same melanoma, and occur in tandem with RTK alterations outlined above. All of these changes would be expected to mediate rapid progression through G1-S leading to propagation of unrepaired DNA errors. Although these effects are deleterious in terms of cancer progression, they may predict responsiveness to DNA-damaging chemotherapy such as alkylating agents, as discussed below.
VI. G2-M ALTERATIONS IN MELANOMA PROGRESSION
Another major cellular pathway that becomes dysregulated in melanoma progression is the G2-M mitotic transition. This stage of the cell cycle is regulated by a dynamic multi-protein spindle checkpoint complex that assures adequate centrosomal function and accurate chromosomal segregation. Upon activation, the centrosome divides to form spindle poles which function to guide chromosomal segregation. The progression from G2 to M is initiated at the centrosome by CDK1 and cyclin B if the checkpoint is adequately functioning. Spindle function can be negatively regulated by p21 and p27 which we have previously discussed as G1-S regulators that are frequently dysregulated in melanoma [71]. Other regulators of centrosomal/spindle pole function include the Aurora kinases and RASSF1A [71, 72], both of which been shown by us or others to be dysregulated during melanoma progression.
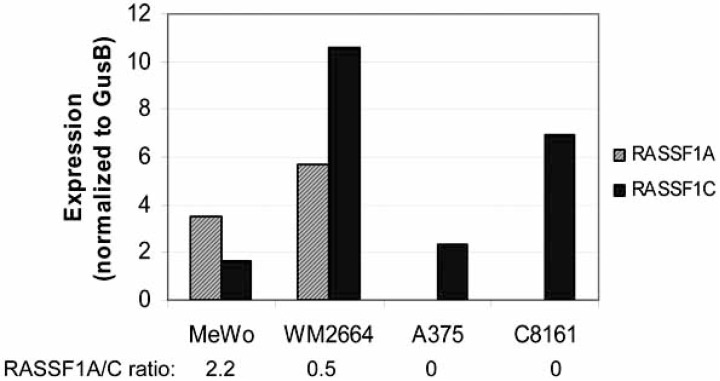
The RASSF1 gene, located at chr 3p21.3, has several different splice isoforms that encode proteins with SARAH, ras-association and diacylglycerol-binding domains. Although their functions have not been completely elucidated, RASSF1 proteins appear to bind and stabilize a number of different kinase complexes involved in apoptosis, proliferation, and genome maintenance [73, 74]. There are at least 5 RASSF1 splice isoforms transcribed from different promoters but we find that only the A and C isoforms are expressed in melanomas. We and others have shown that the RASSF1A splice isoform is differentially silenced by CpG DNA methylation during melanoma progression in 20-50% of primary tumors and in established melanoma cells lines [55, 75]. This silencing results in an imbalance between the amount of RASSF1A and the ubiquitously expressed RASSF1C isoforms (Fig. 3).
Fig. (3).
Variations in RASSF1 isoforms in melanoma cell lines. A. The MeWo line, established from a lymph node melanoma metastasis, shows higher RASS1A levels compared to RASSF1C. WM-2664, established from a cutaneous melanoma, shows higher RASSFC than RASSF1A. Lines A375 and C8161 lack RASSF1A expression due to promoter methylation silencing. Studies were performed by TaqMan reverse transcription (RQ)-PCR using a RASSF1A-specific primer-probe set (Applied Biosystems, Foster City, CA) with normalization to GUSB transcript levels.
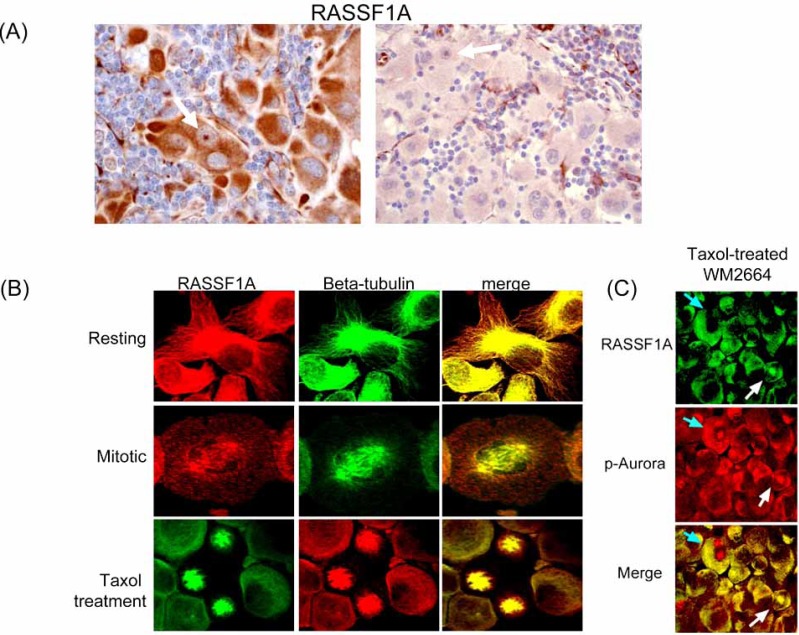
The best characterized function for RASSF1A is in complex stabilization of the mitotic spindle during one phase of the G2-S transition [72, 76]. But we have noted that differences in the levels of RASSF1A (due to varying levels of CpG methylation of its promoter) in melanoma cells leads to altered cellular localization patterns and likely different functions. For example, melanomas with very low RASSF1A expression show discrete nuclear positivity in only rare cycling tumor cells, whereas tumors with high RASSF1A expression show preferential cytoplasmic, membrane and nuclear localization patterns depending on tumor type (Fig. 4A and not shown).
Fig. (4).
Variable RASSF1A expression in melanoma. (A) High level RASSF1A expression in the cytoplasm of a melanoma metastatic to lymph nodes is contrasted with near absence of expression in another progressed melanoma. White arrows highlight the nuclear localization of RASSF1A seen in cycling cells. Immunohistochemical staining was performed on formalin-fixed paraffin-embedded tumor sections using a mouse monoclonal antibody (eB114-10H1. eBioscience, San Diego, CA) and the ABC avidin-biotin detection method. Activated endothelium within each tissue serves as a positive control. (B) RASSF1A in non-dividing cells is present in the cytoplasm in association with the actin-tubulin cytoskeleton. During the later stages of mitosis, RASSF1A transiently colocalizes with tubulin and other spindle components at the spindle poles. Use of mitotic spindle inhibitor (paclitaxel) results in trapping of RASSF1A in stalled mitotic spindle complexes. Confocal microscopy performed with RASSF1A polyclonal antisera (N-12, Santa Cruz Biotechnology, Santa Cruz, CA), and a beta-tubulin mouse monoclonal antibody (clone DM1A, Sigma). (C) In melanoma cell lines, RASSF1A and Aurora kinases colocalize at the mitotic spindle in a subset of cells. Confocal microscopy was performed using a pan-phospho-Aurora antisera (Thr288A/Thr232B/ Thr198C, Cell Signaling Technology, Beverly, MA) and a RASSF1A mouse monoclonal antibody (eB114-10H1, eBioscience).
During cell division in melanoma, RASSF1A shifts from its predominant localization with the microtubles in the cytoplasm to discrete locations within the mitotic spindle (Fig. 4B). These shifts are transient and dependent on the recruitment of other spindle components such as Aurora kinases (Fig. 4C). Melanoma cells treated with spindle toxins such as paclitaxel or vinblastine show trapping of RASSF1A in the altered mitotic spindle. Tumor cells with diminished RASSF1A have a greater tendency to develop chromosomal aberrations [77, 78].
VII. IDENTIFYING WHICH MELANOMAS MIGHT BENEFIT FROM CHEMOTHERAPY
Although profiling of growth factor pathway alterations may be useful in selecting patients for KI therapy, NRAS and BRAF mutation status have shown no or limited correlation with response to chemotherapy or immunotherapy. There is as yet too limited data on the correlative responses of melanomas with FGFR2, PTEN/AKT or KIT mutations. Therefore most studies have focused on identifying predictors of chemotherapeutic response.
The drugs typically used to treat melanoma include carboplatin and cisplatin, alkylating agents, and mitotic spindle poisons such as vinblastine and paclitaxel. Resistance to cisplatin in melanoma may be related to sequestration of the drug in melanosomes [79, 80], and resistance to alkylating agents may be mediated by expression of MGMT, which opposes their action [81]. However, since therapeutic activity of alkylating agents and DNA-damaging agents require tumor cell division, melanomas with a higher proliferative rate or those with genetic alterations in checkpoint function may be more likely to respond [82].
Since the spindle poisons (paclitaxel, vincristine, or vinblastine) are typically components of most multi-agent chemotherapy regimens, identification of predictors of response to this class of agents would be clinically useful. These drugs block cell division by interfering with microtubule function essential for chromosomal segregation and cytokinesis. Since abnormalities in mitotic regulators such as RASSF1A and Aurora kinases are common in melanoma they represent obvious candidate biomarkers. Indeed, in advanced stage melanoma, RASSF1A appears to correlate to some degree with response to chemotherapy. As a result, profiling of the activation state or the degree of mitotic spindle dysfunction using these markers shows promise in identifying those patients who would benefit most from spindle toxins. Additionally, strategies to restore loss of expression of spindle checkpoint proteins such as RASSF1A by use of demethylating agents (or more targeted methods) may be useful in reversing genetic instability associated with tumor progression.
REFERENCES
- 1.Rulyak SJ, Brentnall TA, Lynch HT, Austin MA. Characterization of the neoplastic phenotype in the familial atypical multiple-mole melanoma-pancreatic carcinoma syndrome. Cancer. 2003;98:798–804. doi: 10.1002/cncr.11562. [DOI] [PubMed] [Google Scholar]
- 2.Vasen HF, Gruis NA, Frants RR, van Der Velden PA, Hille ET, Bergman W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden) Int. J. Cancer. 2000;87:809–811. [PubMed] [Google Scholar]
- 3.Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 4.Hayward NK. Genetics of melanoma predisposition. Oncogene. 2003;22:3053–3062. doi: 10.1038/sj.onc.1206445. [DOI] [PubMed] [Google Scholar]
- 5.Ghiorzo P, Villaggio B, Sementa AR, Hansson J, Platz A, Nicolo G, Spina B, Canepa M, Palmer JM, Hayward NK, Bianchi-Scarra G. Expression and localization of mutant p16 proteins in melanocytic lesions from familial melanoma patients. Hum. Pathol. 2004;35:25–33. doi: 10.1016/j.humpath.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Terando A, Sabel MS, Sondak VK. Melanoma: adjuvant therapy and other treatment options. Curr. Treat. Options Oncol. 2003;4:187–199. doi: 10.1007/s11864-003-0020-0. [DOI] [PubMed] [Google Scholar]
- 7.Sabel MS, Sondak VK. Pros and cons of adjuvant interferon in the treatment of melanoma. Oncologist. 2003;8:451–458. doi: 10.1634/theoncologist.8-5-451. [DOI] [PubMed] [Google Scholar]
- 8.Anderson CM, Buzaid AC, Legha SS. Systemic treatments for advanced cutaneous melanoma. Oncology. 1995;9:1149–1158. [PubMed] [Google Scholar]
- 9.Wagner JD, Gordon MS, Chuang TY, Coleman JJ. 3rd. Current therapy of cutaneous melanoma. Plast. Reconstr. Surg. 2000;105:1774–1799. doi: 10.1097/00006534-200004050-00028. quiz 1800-1771. [DOI] [PubMed] [Google Scholar]
- 10.Chapman PB, Einhorn LH, Meyers ML, Saxman S, Destro AN, Panageas KS, Begg CB, Agarwala SS, Schuchter LM, Ernstoff MS, Houghton AN, Kirkwood JM. Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J. Clin. Oncol. 1999;17:2745–2751. doi: 10.1200/JCO.1999.17.9.2745. [DOI] [PubMed] [Google Scholar]
- 11.Mays SR, Nelson BR. Current therapy of cutaneous melanoma. Cutis. 1999;63:293–298. [PubMed] [Google Scholar]
- 12.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, Abrams J, Sznol M, Parkinson D, Hawkins M, Paradise C, Kunkel L, Rosenberg SA. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J. Clin. Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 13.Atkins MB, Kunkel L, Sznol M, Rosenberg SA. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J. 2000;6(1):S11–14. [PubMed] [Google Scholar]
- 14.Rosenberg SA, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, Seipp CA, Einhorn JH, White DE. Treatment of 283 consecutive patients with metastatic melanoma or renal cell cancer using high-dose bolus interleukin 2. JAMA. 1994;271:907–913. [PubMed] [Google Scholar]
- 15.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 16.Wong CW, Fan YS, Chan TL, Chan AS, Ho LC, Ma TK, Yuen ST, Leung SY. BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J. Clin. Pathol. 2005;58:640–644. doi: 10.1136/jcp.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poynter JN, Elder JT, Fullen DR, Nair RP, Soengas MS, Johnson TM, Redman B, Thomas NE, Gruber SB. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16:267–273. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha G, Potter L, Daforno P, Pringle JH. Cutaneous melanoma subtypes show different BRAF and NRAS mutation frequencies. Clin. Cancer Res. 2006;12:4499–4505. doi: 10.1158/1078-0432.CCR-05-2447. [DOI] [PubMed] [Google Scholar]
- 19.Viros A, Fridlyand J, Bauer J, Lasithiotakis K, Garbe C, Pinkel D, Bastian BC. Improving melanoma classification by integrating genetic and morphologic features. PLoS Med. 2008;5:e120. doi: 10.1371/journal.pmed.0050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdi AS, Palmedo G, Flaig MJ, Puchta U, Reckwerth A, Rutten A, Mentzel T, Hugel H, Hantschke M, Schmid-Wendtner MH, Kutzner H, Sander CA. Mutations of the BRAF gene in benign and malignant melanocytic lesions. J. Invest. Dermatol. 2003;121:1160–1162. doi: 10.1046/j.1523-1747.2003.12559.x. [DOI] [PubMed] [Google Scholar]
- 21.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. High frequency of BRAF mutations in nevi. Nat. Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 22.James MR, Dumeni T, Stark MS, Duffy DL, Montgomery GW, Martin NG, Hayward NK. Rapid screening of 4000 individuals for germ-line variations in the BRAF gene. Clin. Chem. 2006;52:1675–1678. doi: 10.1373/clinchem.2006.070169. [DOI] [PubMed] [Google Scholar]
- 23.Demunter A, Stas M, Degreef H, De Wolf-Peeters C, van den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J. Invest. Dermatol. 2001;117:1483–1489. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 24.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, Dias-Santagata D, Stubbs H, Lee DY, Singh A, Drew L, Haber DA, Settleman J. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J. Invest. Dermatol. 2007;127:179–182. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 26.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 27.Beadling C, Jacobson-Dunlop E, Hodi FS, Le C, Warrick A, Patterson J, Town A, Harlow A, Cruz F 3rd, Azar S, Rubin BP, Muller S, West R, Heinrich MC, Corless CL. KIT gene mutations and copy number in melanoma subtypes. Clin. Cancer Res. 2008;14:6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Zhou J, Yuen NK, Corless CL, Heinrich MC, Fletcher JA, Demetri GD, Widlund HR, Fisher DE, Hodi FS. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin. Cancer Res. 2008;14:7726–7732. doi: 10.1158/1078-0432.CCR-08-1144. [DOI] [PubMed] [Google Scholar]
- 29.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21:492–493. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 30.Jilaveanu L, Zito C, Lee SJ, Nathanson KL, Camp RL, Rimm DL, Flaherty KT, Kluger HM. Expression of sorafenib targets in melanoma patients treated with Carboplatin, Paclitaxel and sorafenib. Clin. Cancer Res. 2009;15:1076–1085. doi: 10.1158/1078-0432.CCR-08-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintas-Cardama A, Lazar AJ, Woodman SE, Kim K, Ross M, Hwu P. Complete response of stage IV anal mucosal melanoma expressing KIT Val560Asp to the multikinase inhibitor sorafenib. Nat. Clin. Pract. Oncol. 2008;5:737–740. doi: 10.1038/ncponc1251. [DOI] [PubMed] [Google Scholar]
- 32.Rakosy Z, Vizkeleti L, Ecsedi S, Voko Z, Begany A, Barok M, Krekk Z, Gallai M, Szentirmay Z, Adany R, Balazs M. EGFR gene copy number alterations in primary cutaneous malignant melanomas are associated with poor prognosis. Int. J. Cancer. 2007;121:1729–1737. doi: 10.1002/ijc.22928. [DOI] [PubMed] [Google Scholar]
- 33.Shen SS, Zhang PS, Eton O, Prieto VG. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J. Cutan. Pathol. 2003;30:539–547. doi: 10.1034/j.1600-0560.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 34.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am. J. Pathol. 2003;163:1765–1770. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer J, Bastian B. Genomic analysis of melanocytic neoplasia. Adv. Dermatol. 2005;21:81–99. doi: 10.1016/j.yadr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL, Bengston A, Yudt LM, Eliseenkova AV, Ma J, Curtin JA, Hyder P, Harper UL, Riedesel E, Mann GJ, Trent JM, Bastian BC, Meltzer PS, Mohammadi M, Pollock PM. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol. Cancer Res. 2009;7:41–54. doi: 10.1158/1541-7786.MCR-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, Frattini M, Pilotti S, Anichini A, Tragni G, Parmiani G, Pierotti MA, Rodolfo M. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004 doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 39.Abdel-Rahman MH, Yang Y, Zhou XP, Craig EL, Davidorf FH, Eng C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J. Clin. Oncol. 2006;24:288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 40.Slipicevic A, Holm R, Nguyen MT, Bohler PJ, Davidson B, Florenes VA. Expression of activated Akt and PTEN in malignant melanomas: relationship with clinical outcome. Am. J. Clin. Pathol. 2005;124:528–536. doi: 10.1309/YT58WWMTA6YR1PRV. [DOI] [PubMed] [Google Scholar]
- 41.Mirmohammadsadegh A, Marini A, Nambiar S, Hassan M, Tannapfel A, Ruzicka T, Hengge UR. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 42.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J. Invest. Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 43.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 44.Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 46.Govindarajan B, Sligh JE, Vincent BJ, Li M, Canter JA, Nickoloff BJ, Rodenburg RJ, Smeitink JA, Oberley L, Zhang Y, Slingerland J, Arnold RS, Lambeth JD, Cohen C, Hilenski L, Griendling K, Martinez-Diez M, Cuezva JM, Arbiser JL. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J. Clin. Invest. 2007;117:719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akslen LA, Puntervoll H, Bachmann IM, Straume O, Vuha-hula E, Kumar R, Molven A. Mutation analysis of the EGFR-NRAS-BRAF pathway in melanomas from black Africans and other subgroups of cutaneous melanoma. Melanoma Res. 2008;18:29–35. doi: 10.1097/CMR.0b013e3282f32517. [DOI] [PubMed] [Google Scholar]
- 48.Hacker E, Hayward NK. Germline MC1R variants and BRAF mutant melanoma. J. Invest. Dermatol. 2008;128:2354–2356. doi: 10.1038/jid.2008.236. [DOI] [PubMed] [Google Scholar]
- 49.Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Bressac-de Paillerets B, Chompret A, Ghiorzo P, Gruis N, Hansson J, Harland M, Hayward N, Holland EA, Mann GJ, Mantelli M, Nancarrow D, Platz A, Tucker MA. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J. Natl. Cancer Inst. 2002;94:894–903. doi: 10.1093/jnci/94.12.894. [DOI] [PubMed] [Google Scholar]
- 50.Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, Holland EA, Gillanders EM, Duffy DL, Maskiell JA, Jetann J, Ferguson M, Stephan DA, Cust AE, Whiteman D, Green A, Olsson H, Puig S, Ghiorzo P, Hansson J, Demenais F, Goldstein AM, Gruis NA, Elder DE, Bishop JN, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Martin NG, Trent JM, Mann GJ, Hayward NK. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flaherty KT, Schiller J, Schuchter LM, Liu G, Tuveson DA, Redlinger M, Lathia C, Xia C, Petrenciuc O, Hingorani SR, Jacobetz MA, Van Belle PA, Elder D, Brose MS, Weber BL, Albertini MR, O'Dwyer PJ. A phase I trial of the oral, multikinase inhibitor sorafenib in combination with carboplatin and paclitaxel. Clin. Cancer Res. 2008;14:4836–4842. doi: 10.1158/1078-0432.CCR-07-4123. [DOI] [PubMed] [Google Scholar]
- 52.Kim KB, Eton O, Davis DW, Frazier ML, McConkey DJ, Diwan AH, Papadopoulos NE, Bedikian AY, Camacho LH, Ross MI, Cormier JN, Gershenwald JE, Lee JE, Mansfield PF, Billings LA, Ng CS, Charnsangavej C, Bar-Eli M, Johnson MM, Murgo AJ, Prieto VG. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br. J. Cancer. 2008;99:734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Velden PA, Metzelaar-Blok JA, Bergman W, Monique H, Hurks H, Frants RR, Gruis NA, Jager MJ. Promoter hypermethylation: a common cause of reduced p16(INK4a) expression in uveal melanoma. Cancer Res. 2001;61:5303–5306. [PubMed] [Google Scholar]
- 54.Worm J, Christensen C, Gronbaek K, Tulchinsky E, Guldberg P. Genetic and epigenetic alterations of the APC gene in malignant melanoma. Oncogene. 2004;23:5215–5226. doi: 10.1038/sj.onc.1207647. [DOI] [PubMed] [Google Scholar]
- 55.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- 56.Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch W, Dammann R, Helmbold P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol. Histopathol. 2007;22:1005–1015. doi: 10.14670/HH-22.1005. [DOI] [PubMed] [Google Scholar]
- 57.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin. Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 58.Alonso SR, Ortiz P, Pollan M, Perez-Gomez B, Sanchez L, Acuna MJ, Pajares R, Martinez-Tello FJ, Hortelano CM, Piris MA, Rodriguez-Peralto JL. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am. J. Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller J.R 3rd, Allen RE, Singer MI, Leong SP, Ljung BM, Sagebiel RW, Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc. Natl. Acad. Sci. USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoek KS. DNA microarray analyses of melanoma gene expression: a decade in the mines. Pigment Cell Res. 2007;20:466–484. doi: 10.1111/j.1600-0749.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- 61.Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R, Kunz M. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin. Cancer Res. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 62.Mandruzzato S, Callegaro A, Turcatel G, Francescato S, Montesco MC, Chiarion-Sileni V, Mocellin S, Rossi CR, Bicciato S, Wang E, Marincola FM, Zanovello P. A gene expression signature associated with survival in metastatic melanoma. J. Transl. Med. 2006;4:50. doi: 10.1186/1479-5876-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nambiar S, Mirmohammadsadegh A, Hassan M, Mota R, Marini A, Alaoui A, Tannapfel A, Hegemann JH, Hengge UR. Identification and functional characterization of ASK/Dbf4, a novel cell survival gene in cutaneous melanoma with prognostic relevance. Carcinogenesis. 2007;28:2501–2510. doi: 10.1093/carcin/bgm197. [DOI] [PubMed] [Google Scholar]
- 64.Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seykora JT, Jih D, Elenitsas R, Horng WH, Elder DE. Gene expression profiling of melanocytic lesions. Am. J. Dermatopathol. 2003;25:6–11. doi: 10.1097/00000372-200302000-00002. [DOI] [PubMed] [Google Scholar]
- 66.Ivan D, Diwan AH, Esteva FJ, Prieto VG. Expression of cell cycle inhibitor p27Kip1 and its inactivator Jab1 in melanocytic lesions. Mod. Pathol. 2004;17:811–818. doi: 10.1038/modpathol.3800123. [DOI] [PubMed] [Google Scholar]
- 67.Woenckhaus C, Fenic I, Giebel J, Hauser S, Failing K, Woenckhaus J, Dittberner T, Poetsch M. Loss of heterozygosity at 12p13 and loss of p27KIP1 protein expression contribute to melanoma progression. Virchows Arch. 2004;445:491–497. doi: 10.1007/s00428-004-1049-6. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki N, Onda T, Yamamoto N, Katakura A, Mizoe JE, Shibahara T. Mutation of the p16/CDKN2 gene and loss of heterozygosity in malignant mucosal melanoma and adenoid cystic carcinoma of the head and neck. Int. J. Oncol. 2007;31:1061–1067. [PubMed] [Google Scholar]
- 69.Pavey SJ, Cummings MC, Whiteman DC, Castellano M, Walsh MD, Gabrielli BG, Green A, Hayward NK. Loss of p16 expression is associated with histological features of melanoma invasion. Melanoma Res. 2002;12:539–547. doi: 10.1097/00008390-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 70.Demirkan NC, Kesen Z, Akdag B, Larue L, Delmas V. The effect of the sun on expression of beta-catenin, p16 and cyclin d1 proteins in melanocytic lesions. Clin. Exp. Dermatol. 2007;32:733–739. doi: 10.1111/j.1365-2230.2007.02507.x. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Ji P, Liu J, Broaddus RR, Xue F, Zhang W. Centrosome-associated regulators of the G2/M checkpoint as targets for cancer therapy. Mol. Cancer. 2009;8:8. doi: 10.1186/1476-4598-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song MS, Chang JS, Song SJ, Yang TH, Lee H, Lim DS. The centrosomal protein RAS association domain family protein 1A (RASSF1A)-binding protein 1 regulates mitotic progression by recruiting RASSF1A to spindle poles. J. Biol. Chem. 2005;280:3920–3927. doi: 10.1074/jbc.M409115200. [DOI] [PubMed] [Google Scholar]
- 73.Whitehurst AW, Ram R, Shivakumar L, Gao B, Minna JD, White MA. The RASSF1A tumor suppressor restrains anaphase-promoting complex/cyclosome activity during the G1/S phase transition to promote cell cycle progression in human epithelial cells. Mol. Cell Biol. 2008;28:3190–3197. doi: 10.1128/MCB.02291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 75.Rother J, Bailey J, Alvarado G, Prieto V, Lazar A, Jones D, W-J H. Molecular dissection of the RAS/RAF/MAPK pathway in primary and metastatic melanoma. J. Clin. Oncol. 2006;24:8050. [Google Scholar]
- 76.Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26:7700–7708. doi: 10.1038/sj.onc.1210575. [DOI] [PubMed] [Google Scholar]
- 77.Haruta M, Matsumoto Y, Izumi H, Watanabe N, Fukuzawa M, Matsuura S, Kaneko Y. Combined BubR1 protein down-regulation and RASSF1A hypermethylation in Wilms tumors with diverse cytogenetic changes. Mol. Carcinog. 2008;47:660–666. doi: 10.1002/mc.20412. [DOI] [PubMed] [Google Scholar]
- 78.Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, Lee H, Choi N, Kim J, Kim H, Kim JW, Choi EJ, Kirschner MW, Lim DS. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat. Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- 79.Xie T, Nguyen T, Hupe M, Wei ML. Multidrug resistance decreases with mutations of melanosomal regulatory genes. Cancer Res. 2009;69:992–999. doi: 10.1158/0008-5472.CAN-08-0506. [DOI] [PubMed] [Google Scholar]
- 80.Chen KG, Valencia JC, Lai B, Zhang G, Paterson JK, Rouzaud F, Berens W, Wincovitch SM, Garfield SH, Leapman RD, Hearing VJ, Gottesman MM. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc. Natl. Acad. Sci. USA. 2006;103:9903–9907. doi: 10.1073/pnas.0600213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Augustine CK, Yoo JS, Potti A, Yoshimoto Y, Zipfel PA, Friedman HS, Nevins JR, Ali-Osman F, Tyler DS. Genomic and molecular profiling predicts response to temozolomide in melanoma. Clin. Cancer Res. 2009;15:502–510. doi: 10.1158/1078-0432.CCR-08-1916. [DOI] [PubMed] [Google Scholar]
- 82.Gao K, Lockwood WW, Li J, Lam W, Li G. Genomic analyses identify gene candidates for acquired irinotecan resistance in melanoma cells. Int. J. Oncol. 2008;32:1343–1349. doi: 10.3892/ijo_32_6_1343. [DOI] [PubMed] [Google Scholar]
- 83.Da Forno PD, Pringle JH, Hutchinson P, Osborn J, Huang Q, Potter L, Hancox RA, Fletcher A, Saldanha GS. WNT5A expression increases during melanoma progression and correlates with outcome. Clin. Cancer Res. 2008;14:5825–5832. doi: 10.1158/1078-0432.CCR-07-5104. [DOI] [PubMed] [Google Scholar]