Abstract
Higher plants not only provide human beings renewable food, building materials and energy, but also play the most important role in keeping a stable environment on earth. Plants differ from animals in many aspects, but the important is that plants are more easily influenced by environment than animals. Plants have a series of fine mechanisms for responding to environmental changes, which has been established during their long-period evolution and artificial domestication. The machinery related to molecular biology is the most important basis. The elucidation of it will extremely and purposefully promote the sustainable utilization of plant resources and make the best use of its current potential under different scales. This molecular mechanism at least includes drought signal recognition (input), signal transduction (many cascade biochemical reactions are involved in this process), signal output, signal responses and phenotype realization, which is a multi-dimension network system and contains many levels of gene expression and regulation. We will focus on the physiological and molecular adaptive machinery of plants under soil water stress and draw a possible blueprint for it. Meanwhile, the issues and perspectives are also discussed. We conclude that biological measures is the basic solution to solving various types of issues in relation to sustainable development and the plant measures is the eventual way.
Key Words: Higher plants, soil water stress, gene regulatory network, drought, anti-drought gene resources, signal, ion homeostasis, physiological mechanisms.
1. INTRODUCTION
Environmental stresses represent limiting factors for plant productivity on the globe. Drought is one of the important abiotic stresses, constraining global crop production and quality seriously and recent global climate change and increasingly erratic weather patterns in the future are likely to enhance this situation more seriously [1-11]. Abiotic stress factors mainly include temperature, salinity, drought, anaerobic, and mechanical stresses on plants. In most cases, soil water deficits directly result in drought, which is closely linked with natural rainfall [12-21]. Drought is a complex physical-chemical process, in which many biological macromolecules and small molecules are involved ,such as nucleic acids (DNA, RNA, microRNA), proteins, carbohydrates, lipids, hormones, ions, free radicals ,mineral elements [22-78]. In addition, drought is also related to salt stress, cold stress, high temperature stress, acid stress, alkaline stress, pathological reactions, senescence, growth, development, cell circle, UV-B damage, wounding, embryogenesis, flowering, signal transduction and so on [79-100], making the problem more complicated. The development and application of modern molecular biology have led to a better understanding of plant adaptations and responses to abiotic stress conditions. Genes responsible for adaptation processes to every example of given stress have been identified in Arabidopsis thaliana and other important model plants. The use of transgenic plants to overexpress or silence these genes is a powerful tool in determining if they are necessary or sufficient to induce stress tolerance. However, with stress tolerance being a quantitative character, it should not be expected that single genes confer a high level of corresponding tolerance [101-115].
Considering the interacting complexity (at least including water movement, solute transport, information exchange, ion homeostasis regulation, and other related physi-chemical changes) between plants and their surroundings, it is necessary to generalize the performance of physiological functions for higher plants under drought stress. In this article, related aspects of physiological and molecular responses of higher plants to drought stresses will be reviewed, mainly including the following parts: Plant Physiological Function Performance under Soil Water Stress, Plant Gene Regulatory Network System, A Model for Stress Signal Transduction Pathway in Higher Plants under abiotic Stresses, Pant Gene Regulatory Network System and Plant Drought Resistance Improvement. We then focus on the aspects of plant gene regulatory network system, which is the core controlling the interrelationship between plants and environment at the molecular level in a complex and coordinated manner.
2. PLANT PHYSIOLOGICAL FUNCTION PERFORMANCE IN FIELD UNDER SOIL WATER STRESS
Plants live in soil-plant-atmosphere continuum (SPAC) environment, and they have to coordinate the mechanisms of diverse types to respond to the above changing environment at any time for sustainable survival [1-9]. When abiotic-stress happened, the water in plants would be distributed again. And it could make the old leaf dead by the water movement from old leaf to new leaf, resulting in reduction of the yield of crops. Plant productivity realization is obtained eventually through physiological pathways at least at the level of individual and community [10-16]. One molecule, one kind of tissue or an organ can not produce any economic yield in terms of the need for human being [26-53]. Under the condition of ensuring plant survival, plants can produce corresponding yield.
Soil water is one of key factors influencing plant production and many reports have proved this clearly [17-25]. Loss of water in soil will lead to great reduction in plant production, which has been reflected from total grain yield of many countries in the world [54-58]. Soil water is also the important material for photosynthetic reactions that plants depend on to finish accumulation of photosynthetic products, which are impacted greatly by physiological pathways and environmental factors (such as soil water supply) [59-64]. The influence of water deficits for plant metabolism is very apparent, which is mainly restraining the anabolism by reducing the activity of synthase and strengthening the catabolism by increasing the activity of hydrolytic enzymes. This includes the reduction of protein, chlorophyll, DNA, RNA and plant growth hormone synthesis, which could destroy the normal metabolism and cause growth disorder. So, different soil water supplying will result in quite different physiological pathways, which directly determine the ability for plants to make photosynthetic products. Water deficits in soil environment also influence solute transport (ion and nutrient uptake of plants) to larger extent, which effects on photosynthetic reactions in plant chloroplasts in many ways [65-71]. This is the reason that ion homeostasis and redox state have been brought to attention [72-76].
The series of the above reactions and processes occurring at different biointerfaces is regulated and controlled by plant gene regulatory network system spatially and temporally on the basis of responding to plant developmental cue, through which plants can elegantly respond to the changing environment [77-82]. This network system has been formed by the interaction between plants and environment for a long time of evolution, which will continue to evolve with environmental succession [83-86]. From the angle of individual plant development, Plant Growth Grand Periodicity curve can reflect and show the above trend, displaying higher plasticity [87-90]. Besides, plant responses to soil water deficits (including nutrients) take a “slow-fast-slow” shaped curve in terms of main physio-biochemical indices change and this is in agreement with Plant Growth Grand Periodicity, which also illustrates this fact and wide plasticity for plants [36-53]. Surely, concerted expression of corresponding genes in plant gene regulatory network system makes it possible that we can see the phenotype and phenotype change under given temporal-spatial condition [91-96].
3. PLANT GENE REGULATORY NETWORK SYSTEM UNDER ABIOTIC STRESSES
Recent progress in molecular biology and bioinformatics (especially, DNA microarray technology), genomics, proteomics, metabolomics and transcriptomics) has provided insight into plant gene regulatory network system, which is mainly composed of inducible-genes (environmental factors and developmental cues), their expression programming and regulatory elements (cis-element and trans-element), corresponding biochemical pathways and diverse signal factors [97-103]. The genetic information for drought tolerance is expressed in many prokaryotes and lower eukaryotes, but only in very few higher plants. In higher plants, only seeds can survive for extended periods without water. Exceptional among higher plants is the small group of angiosperm plants termed 'resurrection plants' which can recover from complete dryness within one day of contact with water [116]. Under the condition of soil water deficits, related stress factors always result in overlapping responses, including anatomical, physiological, biochemical, molecular biological changes, which make plant gene regulatory network system more complicated and difficult to explore. Much information with respect to this topic is from the model plant, Arabidopsis thaliana. Main aspects will be illustrated below.
3.1. Typical Environmental Stress-responsive Transcriptional Elements
Plants can sense, process, respond to environmental stress and activate related-gene expression toincrease their resistance to stress. Environmental stress-inducible genes can be mainly divided into two types in terms of their protein products: one type of genes, whose coding products directly confer the function of plant cells to resist to environmental stress such as LEA protein, antifreezing protein, osmotic regulatory protein, enzymes for synthesizing betaine, proline and other osmoregulators; the other type of genes, whose coding products play an important role in regulating gene expression and signal transduction such as the transcriptional elements for sensing and transducing the protein kinases of MAP and CDP, bZIP, MYB and others [103-105]. Though these stress genes could be induced, they have not tissue specificity. At the same time, the protein of most different genes coding drought-resistance is rich in Gly, Pro and other hydrophilic amino acids, but the content of Trp and Cys is lower than others. Li et al. (1997) found that the content of Pro was 38 percentage in the 41.5kD drought-induced-protein in wheat seedlings, but it had not Trp and Cys [117].
Transcriptional elements are defined as the protein combining with the specialized DNA sequence of eukaryotic promoters or the protein having structural characteristics of known DNA-combining region, whose main function is to activate or suppress transcriptional effect of corresponding genes [61-69]. It is a critical factor for the transduction of stress signal. The transcriptional elements would be synthesized under the stress, and it could deliver and amplify the signal to regulate the genes expression to change the physiological function performance. There are two kinds of transcriptional elements according to their characters of expression: first, constitutive transcription elements, which could express normally in stress such as HvCBF2 in barley [118, 119]; the other is an inducible transcription element, whose expression mainly occurs in stress environment. Most transcription elements are the second type. It is proved that there are four function domains by protein analysis: DNA-binding domain, transcriptional regulatory domain (including activation and inhibitory domains), oligomerization site and nuclear localization signal. These domains can regulate the gene expression with the function domains of other transcriptional elements or promoter cis-acting elements in specific time [120]. But there are not sufficient evidences to prove how these transcription elements regulate the target gene expression and depict a legible stress-signal-responsive system.
Up to now, hundreds of transcriptional elements of environmental stress-responsive genes in higher plants have been isolated, which regulate and control the stress reaction related to drought, salinity, cold, pathogen and heat. In the genome of Arabidopsis and rice, they have about 1300-1500 genes for coding transcriptional elements, most of which have not been identified functionally. Recent study has shown that the transcriptional elements involved in plant stress responses mainly include four kinds: APETALA2/ EREBP, bZIP, WRKY, and MYB [70-79]. Some typical plant transcriptional elements have been summarized in Table 1 for reference.
Table 1.
Typical Transcriptional Elements Related to Abiotic Stresses in Plants and Crops.
| Plant Materials | Factors | Binding Sites/Factor Types |
|---|---|---|
| Arabidopsis thaliana | ABI5/AtDPBF | ABA response elements(ABREs)/bZIP |
| A.thaliana | AtDPBF2 | ABA response elements(ABREs)/bZIP |
| A.thaliana | AtDPBF3/AREB3 | ABA response elements(ABREs)/bZIP |
| A.thaliana | AtDPBF4 | ABA response elements(ABREs)/bZIP |
| A.thaliana | AtDPBF5/ABF3 | ABA response elements(ABREs)/bZIP |
| A.thaliana | ABF1 | ABA response elements(ABREs)/bZIP |
| A.thaliana | ABF2/AREB5 | ABA response elements(ABREs)/bZIP |
| A.thaliana | ABF4/AREB2 | ABA response elements(ABREs)/bZIP |
| A.thaliana | GBF3 | ABA response elements(ABREs)/bZIP |
| A.thaliana | AB53 | RY/sph elements/B3 domain proteins |
| A.thaliana | ATMTB2 | MTC |
| A.thaliana | ATHB6 | HD-Zip |
| A.thaliana | ATHB7 | HD-Zip |
| A.thaliana | ATHB12 | HD-Zip |
| A.thaliana | ABI4 | AP2 |
| Oryza | TRAB1 | ABA response elements(ABREs)/bZIP |
| Oryza | OsVPI | RY/sph elements/B3 domain proteins |
| Zea mays | VP1 | MYB |
| Triticum | EmBP-1 | ABA response elements(ABREs)/bZIP |
| Avena | AtVPI | RY/sph elements/B3 domain proteins |
| Helianthus | DPBF5,-2,-3 | ABA response elements(ABREs)/Bzip |
| Phaseolus | ROM2(repressor) | ABA response elements(ABREs)/Bzip |
| Phaseolus | PIARF | RY/sph elements/B3 domain proteins |
| Craterestinma | Cpvp1 | RY/sph elements/B3 domain proteins |
| Daucus | C-ABI3 | RY/sph elements/B3 domain proteins |
| Populus | PtABI3 | RY/sph elements/B3 domain proteins |
3.2. Complexity of Plant Gene Regulatory Network System: Specificity and Crosstalk
Many transcriptional element families participate in plant stress responses, each of which has many members with highly-conservative DNA-binding domain, composing a complicated, temporal-spatial network system for plant gene expression and regulation [81-86]. The specific amino acid sequence of DNA-binding domain decides the specificity of distinguishing and combining with cis-acting elements. Different members of TGA/OBF families have different DNA-binding specificity, protein-protein interaction and expressing profiles. Chromatin immunoprecipitation techniques indicated that tobacco TGA1a in vivo combined with xenobiotic-responsive promoters, but could not combine with PR promoter with as-1 cis-element [103-105]. Arabidopsis TGA2 could be responsive to SA signal, but not be responsive to xenobiotic stress signals.
Much analysis of genomic expression profiling by DNA microarray indicates that the mRNA coding transcriptional element genes in many plants are usually induced to express and accumulated [91, 98]. Most transcriptional element genes involved in plant stress responses have not only completely different expression profiles, but also some overlapping expression profiles, showing the complexity, specificity and crosstalk of plant gene regulatory network system [58-63]. In other words, one kind of stress may simultaneously activate many transcriptional elements and one transcriptional element may be activated by many types of plant stress responses. For instance, CBF3/DREB1a can be responsive rapidly to cold, at the same time, regulated by circadian clock [71-78], which reflects the functional complement between plant cold-responsive pathway and circadian clock-regulated circle in terms of CBF3/DREB1a functions [36, 54, 88, 94-105]. The gene, SbPRP derived from 2-week-old soybean seedlings, encodes 126 amino acids, which have a signal peptide in N-terminal. It is mainly in leaf and epicotyl, and its transcription is regulated by water-stress, salt-stress and plant hormone at the same time [121].
Shinozaki et al. (2003) thought that four signal pathways regulating the gene expression were involved in plant drought, cold and salinity responses, in which two were ABA-dependent (I and II), and two were non-ABA-dependent (III and IV). The gene expression depends on ABA accumulation in plants, which would change with the content of ABA. There are some conservative ABA responsive elements in these genes, whose characteristic sequence is PyACGTG (G/T)C. It can regulate the stress gene expression induced by ABA. Yamaguchi-Shinozaki et al. (1989) found two conservative cis-acting elements, motif 1(GTACGTGGC) and motif 2(CGG/CCGCGCT) after comparing four rab16 gene promoters of rice. Through analysis they found that motif 1 is the cis-acting factor to ABA reaction [122-125]. The gene expression of non-ABA-dependent pathway would be affected by drought and cold beside ABA. It means that ABA is unnecessary for its expression. Some studies indicated that it had a DREB transcriptional element in non-ABA-dependent pathway, which can distinguish the DRE element in the gene promoter. DRE/CRT is a cis-acting element in higher plants to respond the drought and cold stress, whose characteristic sequence is CCGAC. There are many elements associated with DRE found now. It is proved that the involvement of these elements in the stress response is without depending on ABA-dependent gene expression [126-129].
The process of stress signal sensing and transducing, transcriptional regulating, and functional expressing was existent in these pathways [106-115]. Zhu T (2003) and Zhu JK (2000, 2003) concluded that molecular mechanism of plant stress responses (drought and salinity) included three main steps, i.e. stress signal input, transducing process, and regulatory product output through the study of Arabidopsis drought and salinity for many years. Results of many genetic mutants and key intermediate molecules from his lab supported his view powerfully. Recent related anti-drought data (dynamic change of anti-oxidative enzymes and soil water stress threshold) from my lab also proved the point [58-66]. From plant developmental context, plant responses to environmental stresses have a universal law, which has been reflected completely by Plant Growth Grand Periodicity curve. Our study on dynamic changing of wheat anti-oxidative enzymes under soil water deficit have indicated that wheat with different genotypes responded to soil water stress by taking a“ slow- rapid- slow” characteristic curve during wheat life cycle. This is the physiological basis for water-saving agriculture and dry land farming, which also provides substantial evidence for the above viewpoint [34, 37, 46, 49, 50, 91, 95, 103, 105-115].
4. A POSSIBLE MODEL FOR STRESS SIGNAL TRANSDUCTION PATHWAY IN HIGHER PLANTS UNDER ABIOTIC STRESSES
Animals must change their behavior to fit the living environment fluctuates. The same case happens on higher plants and sessile higher plants must also change behavior to increase fitness as the local environment fluctuates. A stronger spatial dimension network underlies signal transduction; for instance, and higher plants must be able to detect gradients in signals (such as light) and resources (such as nitrate and water). Higher plant development itself also is decidedly polar [7-11]. The spatial dimension is satisfied in many ways. Higher plant cells place receptors, channels, G proteins, and kinases, in specific membranes. Some signaling protein complexes are permanent, such as relatively stable and perhaps hardwired COP9 signalosome. Other signaling protein complexes are likely to be ephemeral and formed immediately as a result of signaling [12-16]. There are at least 300 receptor kinases in Arabidopsis, and most of them are membrane bound. Incompatibility and disease defense signal transduction use receptor kinases. After ligand binding and autophosphorylation, such kinases may act as nucleation sites for the construction of ephemeral signaling complexes that contain many proteins [21-25].
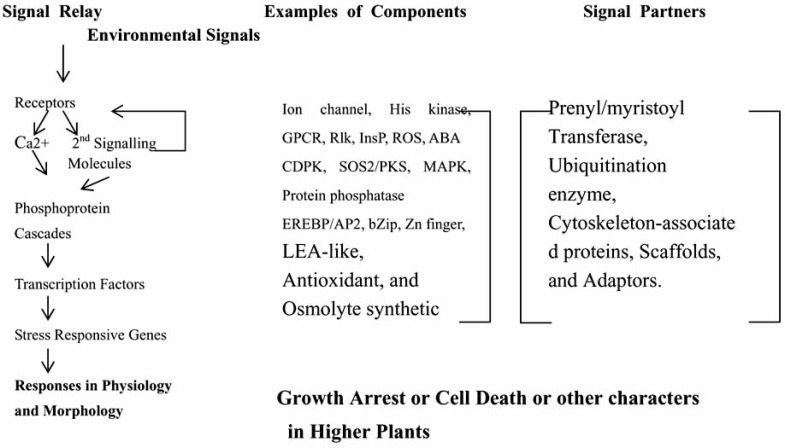
Although there are some differences in different higher plants, a common signal model for stress transduction pathway exist in higher plants [104-115] (Fig.1). This model begins with the perception of signals from environments, followed by the generation of second messengers (such as inositol phosphates and reactive oxygen species). Second messnengers can modulate intracellular Ca2+ levels, often initiating a protein phosphorylation cascade that finally targets proteins directly involved in cellular protection or transcription factors controlling specific sets of stress-regulated genes. The products of these genes may participate in the production of regulatory molecules like the plant hormones abscisic acid (ABA), ethylene, and salicylic acid (SA). Some of these regulatory molecules can, in turn, initiate a second round of circulation.
Fig. (1).
A framework for the signal transduction of abiotic stresses in higher plants.
5. PANT GENE REGULATORY NETWORK SYSTEM AND PLANT DROUGHT RESISTANCE IMPROVEMENT
Pant Gene Regulatory Network System is very complex. More information is obtained from the model plant, Arabidopsis thaliana. The transcriptional elements induced by stress signals decide which gene expression should be increased and the function of different transcriptional elements in plants is synergetic and complementary for each other. It is important to remember the fact that some transcriptional elements may regulate several metabolic pathways and one metabolic pathway may need orchestrated regulation from some transcriptional elements, which is the nature of plant gene regulatory network system [100-115]. So, in some cases, only introducing a transcriptional element can not obtain targeted phenotype and may lead to metabolic unbalance in plants. In addition, because of coordinated evolution of transcriptional elements and their regulating metabolic pathways the genetically-modifying strategy for the same transcriptional element could produce different phenotypes in different plant species. Besides, some transcriptional elements not only regulate metabolic pathways, but also influence transport and allocation of secondary metabolites. Plant secondary metabolism plays an important role in plant responding to environmental stresses. Long-step progress has taken place in terms of introducing transcriptional elements to regulate targeted pathways. These issues need deeper exploration to establish an efficient genetically-modifying system by transcriptional elements and their network system for improving plant stress resistance and global eco- environment and feeding the world [53-63]. Potential genes mediating resistance to soil water stress and related abiotic stress in plants have been listed in Table 2 for reference.
Table 2.
Potential Genes Mediating Resistance to Soil Water Stress and Related Abiotic Stress in Plants
| Gene | Gene Action | Species | Phenotype |
|---|---|---|---|
| adc | Polyamine synthesis | Rice | Drought resistance |
| Apo-Inv | Apoplastic invertase | Tobacco | Salt tolerance, high “osmotic pressure” |
| AtGolS2 | Galactinol and raffinose accumulation | Arabidopsis | Reduced transpiration |
| AtHAL3 | Phosphoprotein phosphatase | Tobacco | Improved salt, osmotic and Lithium tolerance of cell cultures |
| AtHAL3a | Phosphoprotein phosphatase | Arabidopsis | Regulate salinity and osmotic tolerance and plant growth |
| ATP-PRT | ATP-phosphoribosyltransferase | Alyssum | His accumulation and Nickel tolerance |
| AtTPS1 | trehalose-6-phosphate synthase | Tobacco | Drought resistance; sustained photosyntehsis |
| BADH-1 | Betaine aldehyde dehydrogenase | Carrot | Salinity tolerance |
| BADH-1 | Betaine aldehyde dehydrogenase | Rice | Cd tolerance |
| BADH-1 | Betaine aldehyde dehydrogenase | Maize | Salinity tolerance |
| BADH-1 | Betaine aldehyde dehydrogenase | Tobacco | Heat tolerance in photosynthesis |
| BADH-1 | Betaine aldehyde dehydrogenase | Tobacco | Salinity tolerance |
| BADH-1 | Betaine aldehyde dehydrogenase | Tomato | Maintenance of osmotic potential |
| betA | Choline dehydrogenase (glycinebetaine synthesis) | Maize | Drought resistance at seedling stage and high yieldafter drought |
| betA | Choline dehydrogenase (glycinebetaine synthesis) | Tobacco | Increased tolerance to salinity stress |
| CHIT33, CHIT42 | Endochitinase synthesis | Tobacco | Salt and metal toxicity resistance (& disease) |
| CMO | Choline monooxygenase (glycine betaine synthesis) | Tobacco | Better in vito growth under salinity and osmotic (PEG6000) stress |
| codA | Choline oxidase (glycine betaine synthesis) | Arabidopsis | Increased stress tolerance |
| codA | Choline oxidase (glycine betaine synthesis) | Arabidopsis | Salt tolerance in terms of reproduction |
| codA | Choline oxidase (glycine betaine synthesis) | Arabidopsis | Seedlings tolerant to salinity stress and increasedgermination under cold |
| codA | Choline oxidase (glycine betaine synthesis) | Brassica juncea | Tolerance to stress induced photoinhibition |
| codA | Choline oxidase (glycine betaine synthesis) | Rice | Increased tolerance to salinity and cold |
| codA | Choline oxidase (glycine betaine synthesis) | Rice | Recovery from a week long salt stress |
| codA | Choline oxidase (glycine betaine synthesis) | Tobacco | Freezing toleance |
| codA | Choline oxidase (glycine betaine synthesis) | Tomato | Chilling tolerance in yield and oxidative stress tolerance |
| codA | Choline oxidase (glycine betaine synthesis) | Tomato | Chilling tolerance |
| COR15a | Cold induced gene | Arabidopsis | Increased freezing tolerance |
| COX | Choline oxidase (glycine betaine synthesis) | Rice | Salt and 'stress' tolerance |
| Ect A...ect C | Edtoin accumulation in chloroplasts | Tobacco | Salt and cold tolerance |
| GS2 | Chloroplastic glutamine synthetase | Rice | Increased salinity resistance and chilling tolerance |
| IMT1 | Myo-inositol o-methyltransferase (D-ononitol synthesis) | Tobacco | Better CO2 fixation under salinity stress. Better recovery after drought stress. |
| LWR1, LWR2 | Solute accumulation (proline) | Arabidopsis | Growth, osmotic adjustment, water status |
| M6PR | Mannose-6-phosphate reductase | Arabidopsis | Mannitol accumulation under salt stress leading tosalt tolerance |
| M6PR | Mannose-6-phosphate reductase | Arabidopsis | Mannitol accumulation and salt tolerance due to chloroplast protection |
| mt1D | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Arabidopsis | Increased germination under salinity stress |
| mt1D | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Petunia | Chilling tolerance |
| mt1D | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Tobacco | Increased plant height and fresh weight under salinity stress |
| mt1D | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Tobacco | No contribution to sustained growth under salinityand drought stress. |
| mt1D | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Wheat | Drought and salinity tolerance of calli and plants |
| mt1D & GutD | Mannitol-1-phosphate dehydrogenase & glucitol-6-phosphate dehydrogenase | loblolly pine | High salt tolerance due to mannitol and glucitol accumulation |
| mtlD | Mannitol-1-phosphate dehydrogenase (mannitol synthesis) | Populus tomentosa | Salinity tolerance |
| Osm1 …Osm4 | Osmotin protein accumulation | Tobacco | Drought and salt tolerance in plant water status andproline accumulation |
| Osm1 ...Osm4 | Osmotin protein accumulation | Strawberry | Proline accumulation & salt tolerance |
| Osmyb4 | Cold induced transcription factor | Arabidopsis | Accumulation of compatible solutes |
| Osmyb4 | Specifically cold inducible | Tobacco | Freezing and Chilling tolerance |
| Osmyb4 | Cold induced transcription factor | Tomato | Drought but not cold resistance |
| OsP5CS2 | Highly homologous to P5CS | Rice | Cold and salinity tolerance |
| otsA | Trehalose-6-phosphate synthase (trehalose synthesis) | Tobacco | Increased leaf dry weight and photosynthetic activityunder drought. Increased carbohydrate accumulation. |
| otsB | Trehalose-6-phosphate synthase (trehalose synthesis) | Tobacco | Increased leaf dry weight and photosynthetic activityunder drought. Increased carbohydrate accumulation. |
| P5CR | Pyrroline carboxylate reductase (proline accumulation) | Soybean | Antioxidants activity under stress |
| P5CR | Pyrroline carboxylate reductase (proline accumulation) | Soybean | Amino acid accumulation |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) (tomato) | Citrus | Osmotic adjustment and drought resistance |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Petunia | Drought resistance and high proline |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Potato | Salinity tolerance |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Rice | Increased biomass production under drought and salinity stress |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Rice | Reduced oxidative stress under osmotic stress |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Rice | Resistance to water and sainity stress |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Soybean | Resistance to osmotic stress and heat |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) (tomato) | Soybean | Drought resistance, high RWC, high proline |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) (tomato) | Sugarcane | Drought resistance via antioxidant role of proline |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Tobacco | Increased biomass production and enhance flower development under salinity stress |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Tobacco | Freezing tolerance |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) | Wheat | Drought resistance due to antioxidative action |
| P5CS | Pyrroline carboxylate synthase (proline synthesis) (tomato) | Yeast | Reduced growth under none-stress and some promoted growth under mild stress |
| pdc1 | Pyruvate decarboxylase overexpression | Rice | Increased submergence tolerance |
| pdc1; pdc2 | Pyruvate decarboxylase overexpression | Arabidopsis | Hypoxic stress survival |
| PPO | Polyphenol oxidases suppression | Tomato | Drought resistance |
| SAMDC | S-adenosylmethioninedecarboxylase (polyamine synthesis) | Rice | Better seedling growth under a 2 day NaCl stress |
| SAMDC | S-adenosylmethioninedecarboxylase (polyamine synthesis) | Tobacco | drought, salinity, Verticillium and Fusarium wiltsresistance |
| SMT | selenocysteine methyltransferase | Arabidopsis, Indian Mustard | Selenium hyperaccumulation tolerance |
| SPE | Spermidine synthase | Arabidopsis | Chilling, freezing, salinity, drought hyperosmosis |
| spe1-1; spe2-1 | Spermidine non-accumulating | Arabidopsis | Decreased salt tolerance |
| SST/FFT | Fructan accumulation | Potato | Reduced proline accumulation at low water status |
| TaCRT | Ca2+-binding protein | Tobacco | Better water status, WUE and membrane stability |
| TPP1 | Trehalose synthesis | Rice | Salt and cold tolerance |
| TPS; TPP | Trehalose synthesis | Arabidopsis | Drought, freezing, salt and heat tolerance |
| TPS1 | Trehalose synthesis | Tomato | Drought, salt and oxidative stress tolerance |
| TPS1 | Trehalose synthesis | Potato | Delayed wilting under drought |
| TPS1 & TPS2 | Trehalose synthesis | Tobacco | Maintenance of water status under drought stress |
| TPSP | Trehalose synthesis | Rice | Drought, salt and cold tolerance expressed by chlorophyll fluorescence |
| WCOR15 | Cold induced gene | Tobacco | Increased freezing tolerance |
The plant stress resistance mainly depends on varying proteins directly. There are many kinds of anti-proteins upon water stress, but we must point out that, in evolutionary terms, the possibility of producing specific proteins normally is very low except seriously drought-stress. Changing the relative content of different proteins in plants is the main way for resisting the stress, which is reflected in many plants such as NADP-malic enzymes in wheat and rice. It has an internal molecular basic of the drought-resistance in the drought-resistance species [127-130].
6. CONCLUDING REMARKS AND PERSPECTIVES
Plants have more refine mechanisms to regulate themselves from molecular level to ecosystem to respond to environmental changing. For instance, there are many coding-protein genes downstream only for osmotic regulation in abiotic stress resistance (Table 3). Plants are always in the state of passiveness for confronting environmental succession and the related issue is more complicated, which is the main cause that plants are behind animals in the study of most fields [52-56].
Table 3.
Some Examples of the Osmotic Regulating Genes Downstream in Abiotic Resistance
| Components | Metabolic Functions | Gene/Proteins |
|---|---|---|
| ROS scavenging | Increase in ROS scavenging enzymes | GP, PHGPX |
| Chaperones | Heat-/cold-/salt-shock proteins; protein folding | |
| Hsp,Csp,Ssp,DnaJ | ||
| Fructan | Osmoprotection | SacB |
| Trehalose | Osmoprotection | |
| Tps;Tpp,trehalase | ||
| Glycine betaine | Protein protection and carbon sink | codA |
| Proline | Substrate for mitochondrial respiration; redox control | P5CS/P5CR |
| Ectoine | Osmoprotectant | EctA,BC |
| K+-transporters | High affinity K+ uptake | Hkt1,Hak1 |
| K+-channels | Low affinity or dual affinity K+ uptake | Akt1, Akt |
| H2O channel proteins | Membrane cycling control | TIP |
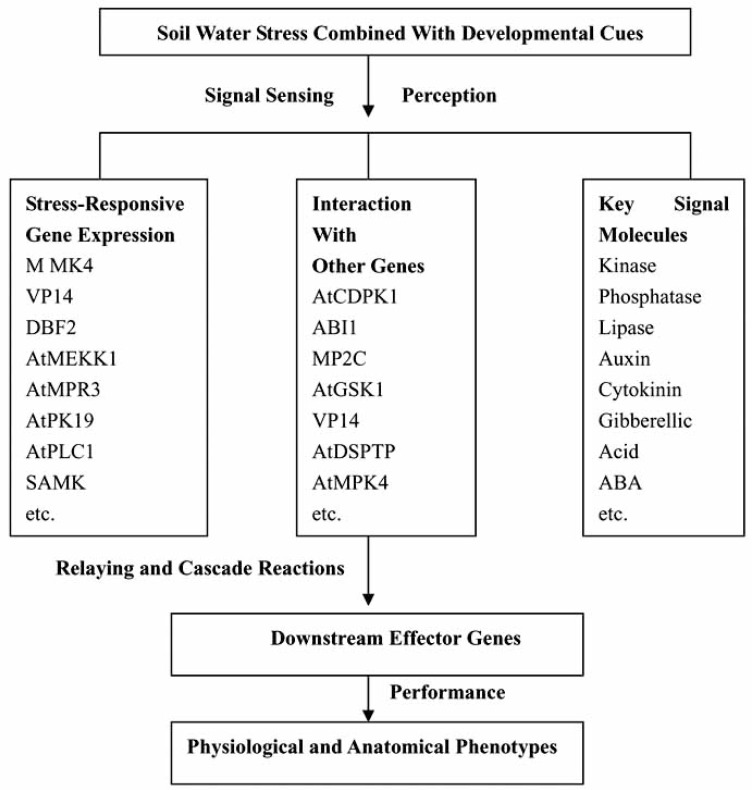
Charting plant gene regulatory network system under soil water deficits is a great challenge. Nowadays, there are indeed many favorable conditions for charting this blueprint, including much available data from Arabidopsis, rice, grass, yeast and fruit fly, but the range of tested plants is very much limited, many stress-responsive genes have not been unified in terms of their refine functions, and many genes participating in environmental stresses are interacted and overlapped, which have led to incorrect placing of key genes (gene effectors) and signal molecules in the whole plant gene regulatory network system. Besides, much data are from under condition of one type of stresses. It is a fact that plants always confront more than two kinds of individual environmental stresses or their combination simultaneously in field [91-115]. Although drawing this dimensional plant gene regulatory network system with great details and complete pathways is impossible currently, the basic draft for this blueprint could be summarized in Fig. (2). This draft was established in combination with recent advance in this hot topic and from the context of development, which will provide instructions for further investigation and insights into understanding of plant refine plasticity for abiotic environmental stresses.
Fig. (2).
The basic draft for plant gene regulatory network system.
In a word, precise elucidation of plant gene regulatory network system under abiotic stresses is of importance to molecularly engineering plant resistance, because of which many excellent scientists world-wide have been engaged in this frontier field, resulting in a long-step progress. There are also many issues remained to be solved and needed to make efforts. Scope of tested plants needs to be extended; comprehensive study on a combination of environmental stress factors in laboratories and in field should be given much attention; system development viewpoint and computer simulation analysis method should be also applied. The combination of molecular biology, biotechnology and plant physiology (especially in field) is also the key. With accumulation of data from being extended plant range, plant gene regulatory network system under environmental stresses will be clearer and clearer.
ACKNOWLEDGEMENTS
One Hundred-Talent Plan of Chinese Academy of Sciences, Shao M.-A.’s Innovation Team Project of Education Ministry of China (Changjiang Scholar Innovation Team Group) and Northwest A&F University, International Cooperative Partner Plan of Chinese Academy of Sciences (To Shao M.-A.), the Cooperative&Instructive Foundation of State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau (10501-HZ), and the Award and Open Foundation of State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau (10501-194) (To Shao H-B). Our thanks are given to Dr. Professor Shao M.- A. for his support and leadership. Our sincere thanks also extended to Editors, Dr. Christian Neri and Afsheen Nawab for their critical comments on and helpful suggestions for this invited submission, respectively.
REFERENCES
- 1.Amtmann A. Learning from evolution: Thellugiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2009;2:3–12. doi: 10.1093/mp/ssn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ. Evaluation of seven function- known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant. 2009;2:73–83. doi: 10.1093/mp/ssn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuberosa R, Salvi S. Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci. 2006;11:405–412. doi: 10.1016/j.tplants.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang ZY, Li FM, Xiong YC, Xu BC. Soil-water threshold range of chemical signals and drought tolerance was mediated by ROS homeostasis in winter wheat during progressive soil drying. J. Plant Growth Regul. 2008;27:309–319. [Google Scholar]
- 5.Chaves MM, Oliveria MM. Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J. Exp. Bot. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- 6.Deng XP, Shan L, Zhang HP, Turner NC. Improving agricultural water use efficiency in arid and semiarid areas of China. Agric. Water Manag. 2006;80:23–40. [Google Scholar]
- 7.Wang HB, Liu DC, Sun JZ, Zhang M. Asparagine synthetase gene TaASN1 from wheat is up-regulated by salt stress,osmotic stress and ABA. J. Plant Physiol. 2005;162:81–89. doi: 10.1016/j.jplph.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Nadeau JA. Stomatal development: new signals and fate determinants. Curr. Opin. Plant Biol. 2009;12:29–35. doi: 10.1016/j.pbi.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson FC, Skeffington AW, Gardner MJ, Web AAR. Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 2009;69:419–427. doi: 10.1007/s11103-008-9407-4. [DOI] [PubMed] [Google Scholar]
- 10.Bressan R, Bohnert H, Zhu JK. Abiotic stress tolerance:from gene discovery in model organisms to crop improvement. Mol. Plant. 2009;2:1–2. doi: 10.1093/mp/ssn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Secenji M, Bebes A, Hideg E, Gyorgyey J. Transcriptional changes in ascorbate- glutathione cycle under drought conditions. Acta Biol. Szeged. 2008;52:93–94. [Google Scholar]
- 12.Materna SC, Davidson EH. Logic of gene regulatory networks. Curr. Opin. Biotech. 2007;18:351–54. doi: 10.1016/j.copbio.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldgur Y, Rom S, Ghirlando R, Shkolnik D, Shadrin N, Konrad Z, Bar-Zvi D. Desiccation and zinc binding induce transition of tomato abiscisic acid stress ripening 1, a water stress-and salt stress-regulated plant-specific protein, from unfolded to folded state. Plant Physiol. 2007;143:617–628. doi: 10.1104/pp.106.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu KM, Wang L, Xu YY, Chen N, Ma QB, Li F, Chong K. Overexpression of OsCOIN,a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought,and enhanced praline level in rice. Planta. 2007;226:1007–1016. doi: 10.1007/s00425-007-0548-5. [DOI] [PubMed] [Google Scholar]
- 15.Wan SQ, Norby RJ, Ledford J, Weltzin JF. Responses of soil respiration to elevated CO2, air warming, and changing soil water availability in a model old-field grassland. Global Change Biol. 2007;13:2411–2424. [Google Scholar]
- 16.Chow B, McCourt P. Hormone signaling from a developmental context. J. Exp. Bot. 2004;55:247–251. doi: 10.1093/jxb/erh032. [DOI] [PubMed] [Google Scholar]
- 17.Wang PT, Song CP. Guard-cell signaling for hydrogen peroxide and abiscisic acid. New Phytol. 2008;178:703–718. doi: 10.1111/j.1469-8137.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 18.Penfield S. Temperature perception and signal transduction in plants. New Phytol. 2008;178:1–14. doi: 10.1111/j.1469-8137.2008.02478.x. [DOI] [PubMed] [Google Scholar]
- 19.McDowell N, Pockman WT, Allen CD, Breshears DD, Kolb T, Plaut J, Sperry J, West A, Williams DG, Yepez EA. Mechanisms of plant survival and mortality during drought:why do some plants survive while others succumb to drought? New Phytol. 2008;178:719–739. doi: 10.1111/j.1469-8137.2008.02436.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim MC, Chung WS, Yun D-J, Cho MJ. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant. 2009;2:13–21. doi: 10.1093/mp/ssn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 2004;55:2343–2351. doi: 10.1093/jxb/erh276. [DOI] [PubMed] [Google Scholar]
- 22.Chen XJ, Guo ZJ. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. Int. J. Mol. Sci. 2008;9:2601–2613. doi: 10.3390/ijms9122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weckwerth W. Integration of metabolomics and proteomics in molecular plant physiology- coping with the complexity by data-dimensionality reduction. Physiol. Plant. 2008;132:176–189. doi: 10.1111/j.1399-3054.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhu HY, Choi HK, Cook DR, Shoemaker RC. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005;137:1189–1196. doi: 10.1104/pp.104.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, Krishnan A, Nataraja K, Udayakumar M, Pereira A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. PNAS. 2007;104:15270–15275. doi: 10.1073/pnas.0707294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monaghan P, Metcalfe NB, Torres R. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol. Lett. 2009;12:75–92. doi: 10.1111/j.1461-0248.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu XA, Vance Baird WmV. Differential expression of genes regulated in response to drought or salinity stress in sunflower. Crop Sci. 2003;43:678–687. [Google Scholar]
- 28.Chae L, Sudat S, Dudoit S, Zhu T, Luan S. Diverse transcriptional programs associated with environmental stress and hormones in the Arabidopsis receptor-like kinase gene family. Mol. Plant. 2009;2:84–107. doi: 10.1093/mp/ssn083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shyur L-F, Yang N-S. Metabolomics for phytomedicine research and drug development. Curr. Opin. Chem. Biol. 2008;12:66–71. doi: 10.1016/j.cbpa.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 30.Gao WR, Wang XS, Liu QY, Peng H, Chen C, Li JG, Li JG, Zhang JS, Hu SN, Ma H. Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.) Biochem. Biophy. Res. Comm. 2008;376:578–583. doi: 10.1016/j.bbrc.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 31.Xiao BZ, Huang YM, Tang N, Xiong LZ. Overexpression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 32.Shih M-D, Hoekstra FA, Hsing Y-I. Late embryogenesis abundant proteins. Adv. Bot. Res. 2008;48:212–255. [Google Scholar]
- 33.Morris PC. MAP kinase signal transduction pathways in plants. New Phytol. 2001;151:67–89. doi: 10.1046/j.1469-8137.2001.00167.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu XG, Yue YL, Li B, Nie YL, Li W, Wu WH, Ma LG. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abiscisic acid. Science. 2007;10 doi: 10.1126/science.1135882. 1126/Science. 1135882. [DOI] [PubMed] [Google Scholar]
- 35.Kearney M, Porter W. Mechanistic niche modeling:combing physiological and spatial data to predict species’ ranges. Ecol. Lett. 2009;12:1–17. doi: 10.1111/j.1461-0248.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 36.Shao HB, Liang ZS, Shao MA. Osmotic regulation of 10 wheat (Triticum aestivum L.) genotypes at soil water deficits. Biointerf. 2006;47:132–139. doi: 10.1016/j.colsurfb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Shao HB, Liang ZS, Shao MA. Changes of anti-oxidative enzymes and MDA content under soil water deficits among 10 wheat (Triticum aestivum L.) genotypes at maturation stage. Biointerf. 2005;45:7–13. doi: 10.1016/j.colsurfb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Shao HB, Liang ZS, Shao MA. Dynamic changes of anti-oxidative enzymes of 10 wheat genotypes at soil water deficits. Biointerf. 2005;42:187–195. doi: 10.1016/j.colsurfb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Shao HB, Liang ZS, Shao MA, Wang BC. Changes of anti-oxidative enzymes and membrane peroxidation for soil water deficits among 10 wheat genotypes at seedling. Biointerf. 2005;42:107–113. doi: 10.1016/j.colsurfb.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Shao HB, Chu LY. Plant molecular biology in China: Opportunities and challenges. Plant Mol. Biol. Rep. 2005;23:345–358. [Google Scholar]
- 41.Shao HB, Liang ZS, Shao MA. LEA proteins in higher plants: Structure, function, gene expression and regulation. Biointerf. 2005;45:131–135. doi: 10.1016/j.colsurfb.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 42.Shao HB, Liang ZS, Shao MA, Sun Q, Hu ZM. Investigation on dynamic changes of photosynthetic characteristics of 10 wheat (Triticum aestivum L.) genotypes during two vegetative-growth stages at water deficits. Biointerf. 2005;43:221–227. doi: 10.1016/j.colsurfb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Chu LY, Shao HB, Li MY. Molecular mechanism of phytochrome signal transduction in higher plants. Biointerf. 2005;45:154–161. doi: 10.1016/j.colsurfb.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Shao HB, Chen XY, Chu LY, Zhao XN, Yuan YB, Zhao CX, Hu ZM. Investigation on the relationship of Proline with wheat anti-drought under soil water deficits. Biointerf. 2006;53:113–119. doi: 10.1016/j.colsurfb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Shao HB, Jiang SY, Li FM, Chu LY, Zhao CX, Shao MA, Zhao XN , Li F. Some advances in plant stress physiology and their implications in the systems biology era. Biointerf. 2007;54:33–36. doi: 10.1016/j.colsurfb.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H, Zhang ZB, Shao HB, Xu P, Foulkes MJ. Genetic Correlation and Path Analysis of Transpiration Efficiency for Wheat Flag Leaves. Environ. Exp. Bot. 2008;64:128–134. [Google Scholar]
- 47.Shao HB, Chu LY, Shao MA. Calcium as a versatile Plant Signal Transducer under Soil Water Stress. BioEssays. 2008;30:634–641. doi: 10.1002/bies.20770. [DOI] [PubMed] [Google Scholar]
- 48.Shao HB, Chu LY, Shao MA, Zhao CX. Advances in functional regulation mechanisms of plant aquaporins: Their diversity, gene expression, localization, structure and roles in plant soil-water relations. Mol. Membr. Biol. 2008;25:179–191. doi: 10.1080/09687680801914508. [DOI] [PubMed] [Google Scholar]
- 49.Shao HB, Chu LY, Shao MA, Li SQ, Yao JC. Improving Plant Resistance to abiotic Stresses by the global Calcium Signal System. Biotech. Adv. 2008;26:503–510. doi: 10.1016/j.biotechadv.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Shao HB, Li B, Wang B.C, Tang K, Liang Y.L. A study on differentially expressed gene screening of Chrysanthemum plants under sound stress. C.R. Biol. 2008;331:329–333. doi: 10.1016/j.crvi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Shao HB, Chu LY, Jaleel CA, Zhao CX. Water-deficit stress-induced anatomical changes in higher plants. C. R. Biol. 2008;331:215–225. doi: 10.1016/j.crvi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Shao HB, Chu LY, Lu ZH, Kang CM. Primary antioxidant free radical scavenging and redox signaling pathways in higher plant cells. Int. J. Biol. Sci. 2008;4:8–14. doi: 10.7150/ijbs.4.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao HB, Chu LY, Shao MA. Understanding molecular mechanisms for improving phytoremediation of heavy metal-contaminated soils. Crit. Rev. Biotech. 2009;29 doi: 10.3109/07388550903208057. (In Press) [DOI] [PubMed] [Google Scholar]
- 54.Rubio V, Bustos R, Irigoyen ML, Cardona-Lopez X, Rojas-Triana M, Paz-Ares J. Plant hormones and nutrient signalling. Plant Mol. Biol. 2009;69:361–373. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- 55.Bradshaw AD. Unravelling phenotypic plasticity-why should we both? New Phytol. 2006;170:644–648. doi: 10.1111/j.1469-8137.2006.01761.x. [DOI] [PubMed] [Google Scholar]
- 56.Apel K, Hirt A. Reactive oxygen species: Metabolism, Oxidative Stress, and Signal Transduction. Ann. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 57.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrew DB, Martin U, Elizabeth A. Photosynthesis, productivity and yield of maize are not affected by open-air elevation of CO2 concentration in the absence of drought. Plant Physiol. 2006;140:779–790. doi: 10.1104/pp.105.073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andjelkovic V, Thompson R. Changes in gene expression in maize kernel in response to water and salt. Plant Cell Rep. 2006;25:71–79. doi: 10.1007/s00299-005-0037-x. [DOI] [PubMed] [Google Scholar]
- 60.Angela H. The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytol. 2004;162:9–24. [Google Scholar]
- 61.Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- 62.Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM. Allelopathy and exotic plant invasion: from molecules and genes to species interaction. Science. 2003;301:1377–1380. doi: 10.1126/science.1083245. [DOI] [PubMed] [Google Scholar]
- 63.Bauer P, Bereczkey Z. Gene networks involved in iron acquisition strategies in plants. Agronomie. 2003;23:447–454. [Google Scholar]
- 64.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 65.Brooker RW. Plant-plant interactions and environmental change. New Phytol. 2006;171:271–284. doi: 10.1111/j.1469-8137.2006.01752.x. [DOI] [PubMed] [Google Scholar]
- 66.Boudsocq M, Lauriere C. Osmotic signaling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138:1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyer JS. Plant productivity and environment potential for increasing crop plant productivity, genotypic selection. Science . 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 68.Chen Z, Gallie DR. The Ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell. 2004;16:1143–1162. doi: 10.1105/tpc.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Capell T, Bassie L, Christou P. Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. PNAS. 2004;101:9909–9914. doi: 10.1073/pnas.0306974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaves MM, Maroco J, Pereira J. Understanding plant responses to drought—from genes to the whole plant. Funct. Plant Biol. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- 71.Cushman JC, Bohnert J. Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 2000;3:117–124. doi: 10.1016/s1369-5266(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 72.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 73.Condon AG, Richards RA, Rebetzke GJ, Farquhar GD. Breeding for high water-use efficiency. J. Exp.Bot. 2004;55:2447–2460. doi: 10.1093/jxb/erh277. [DOI] [PubMed] [Google Scholar]
- 74.Dhanda S, Sethi GS, Behl RK. Indices of drought tolerance in wheat genotypes at early stages of plant growth. J. Agron. Crop. Sci. 2004;190:6–12. [Google Scholar]
- 75.De Ronde JA, Laurie RN, Caetano T. Comparative study between transgenic and non-transgenic soybean lines proved transgenic lines to be more drought tolerant. Euphytica. 2004;138:123–132. [Google Scholar]
- 76.Doelle HW. Biotechnology and human development in developing countries. Electro. J. Biotech. 2002;2:1–10. [Google Scholar]
- 77.Franklin KA, Whitelam GC. Light signals, phytochromes and cross-talk with environmental cues. J. Exp. Bot. 2004;55:271–276. doi: 10.1093/jxb/erh026. [DOI] [PubMed] [Google Scholar]
- 78.Foyer CH, Noctor G. Oxidant and antioxidant signaling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005;28:1056–1071. [Google Scholar]
- 79.Gregory PJ. Roots, rhizosphere and soils: the route to a better understanding of soil science? Eur. J. Soil Sci. 2006;57:2–12. [Google Scholar]
- 80.Hsiao TC. Plant response to water stress. Ann. Rev. Plant Physiol. 1973;24:519–534. [Google Scholar]
- 81.Halliwell B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006;141:312–322. doi: 10.1104/pp.106.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hodge A. The plastic plants: root responses to heterogeneous supplies of nutrients. New Phytol. 2004;162:9–24. [Google Scholar]
- 83.How GT, Brunner AM. An evolving approach to understanding plant adaptation. New Phytol. 2005;167:1–5. doi: 10.1111/j.1469-8137.2005.01469.x. [DOI] [PubMed] [Google Scholar]
- 84.Liu XA, Vance Baird WM. Dentification of a novel gene, HAABRC5, from Helianthus annuus (Asteraceae) that is upregulated in response to drought, salinity and abscisic acid. Am. J. Bot. 2004;91:184–191. doi: 10.3732/ajb.91.2.184. [DOI] [PubMed] [Google Scholar]
- 85.Lenski RL, Barrick JE, Ofria C. Balancing robustness and evolvability. PLoS Biol. 2006;4:e428. doi: 10.1371/journal.pbio.0040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li JX, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science . 2000;287:300–302. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- 87.Kitano H. Looking beyond the details: a rise in system-oriented approaches in genetics and molecular biology. Curr. Genet. 2002;41:1–10. doi: 10.1007/s00294-002-0285-z. [DOI] [PubMed] [Google Scholar]
- 88.Kasuga M, Liu Q, Miura S. Improving plant drought, salt and freezing tolerance by gene transfer of a single-inducible transcription factor. Nat. Biotech. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 89.Kreps JA, Wu YJ, Chang HS. Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kennedy GC, Wilson IW. Plant functional genomics: opportunities in microarray databases and data mining. Funct. Plant Biol. 2004;31:295–314. doi: 10.1071/FP03216. [DOI] [PubMed] [Google Scholar]
- 91.Mark T, Antony B. Abiotic stress tolerance in grasses. From model plants to crop plants. Plant Physiol. 2005;137:791–793. doi: 10.1104/pp.104.900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–252. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 93.Mittler R. Abiotic stress,the field environment and stress combination. Trends Plant Sc. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 94.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:855–862. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 95.Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase path-ways in plant stress signaling. Trends Plant Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 96.Pitzschke A, Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2005;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poethig RS. Life with 25,000 genes. Genome Res. 2001;11:313–316. doi: 10.1101/gr.180001. [DOI] [PubMed] [Google Scholar]
- 98.Riechmann JL, Heard J, Martin G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science . 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 99.Shou X, Bordallo P, Wang K. Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 2004;55:1013–1019. doi: 10.1093/jxb/erh129. [DOI] [PubMed] [Google Scholar]
- 100.Somerville C, Dangl J. Plant Biology in 2010. Science. 2000;290:2077–2078. doi: 10.1126/science.290.5499.2077. [DOI] [PubMed] [Google Scholar]
- 101.Shinozaki K, Dennis ES. Cell signaling and gene regulation global analyses of signal transduction and gene expression profiles. Curr. Opin. Plant Biol. 2003;6:405–409. doi: 10.1016/s1369-5266(03)00093-1. [DOI] [PubMed] [Google Scholar]
- 102.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 103.Sakuma Y, Maruyama K, Osakabe Y. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shulaev V, Oliver DJ. Metabolic and proteomic markers for oxidative stress. New tools for reactive oxygen species research. Plant Physiol. 2006;141:367–372. doi: 10.1104/pp.106.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suzuki N, Rizhsky L, Liang HJ, Shuman J, Mittler R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein Bridging Factor 1c(MBF1C) Plant Physiol. 2005;139:1313–1322. doi: 10.1104/pp.105.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vasil IK. The science and politics of plant biotechnology-a personal perspective. Nat. Biotech. 2003;21:849–851. doi: 10.1038/nbt0803-849. [DOI] [PubMed] [Google Scholar]
- 107.Wang WX, Vinocur P, Altman A. Plant responses to drought,salinity and extreme temperatures:towards genetic engineering for stress tolerance. Planta. 2003; 218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 108.Wu YS, Tang KX. MAP Kinase cascades responding to environmental stress in plants. Acta Bot. Sin. 2004;46:127–136. [Google Scholar]
- 109.Wei GH, Liu DP, Liang CC. Charting gene regulatory networks: strategies, challenges and perspectives. Biochem. J. 2004;381:1–12. doi: 10.1042/BJ20040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weitz JS, Benfey PN, Wingreen NS. Evolution, interactions and biological networks. PLoS Biol. 2007;5:e11. doi: 10.1371/journal.pbio.0050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiong LM, Wang RG, Mao GH, Koczan JM. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abiscisic acid. Plant Physiol. 2006;142:1065–1074. doi: 10.1104/pp.106.084632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu T. Global analysis of gene expression using GeneChip microarrays. Curr. Opin. Plant Biol. 2003;6:418–425. doi: 10.1016/s1369-5266(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 113.Zhu JK. Salt and drought stress signal transduction in plants. Ann. Rev. Plant Biol. 2000;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu JK. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 115.Zhang JX, Kirham MB. Drought stress-induced changes in activities of superoxide dismutase, catalase and peroxidase in wheat species. Plant Cell Physiol. 1994;35:785–791. [Google Scholar]
- 116.Bartels D, Schneider K, Terstappen G, Piatkowski D ; Salamini, E Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta. 1990;18:27–34. doi: 10.1007/BF00202321. [DOI] [PubMed] [Google Scholar]
- 117.Li NY, Gao JF. Effect of Osmotic Stress on Isoelectric Point and Subun it Composition of Proteins in Shoots of Wheat. Acta Univ. Agric. Boreali-occidentalis. 1997;25:6–11. [Google Scholar]
- 118.Xue GP. The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low temperature responsive genes in barely is modulated by temperature. Plant J. 2003;33:373–338. doi: 10.1046/j.1365-313x.2003.01630.x. [DOI] [PubMed] [Google Scholar]
- 119.Luo SN, Yang GS, Shi XH, Lu XY, Xu P. On the Application of Transcription Factor to Plant Stress Resistance. J. Hunan Agri. Uni. 2005;31:219–223. [Google Scholar]
- 120.He CY, Wu XL, Dong FY, Du BX, Zhang JS, Chen SY. Isolation and characterization of a new defense gene from soybean. Sci. China Ser. C. 2001;4:409–420. doi: 10.1007/BF02879608. [DOI] [PubMed] [Google Scholar]
- 121.Yamaguchi-Shinozaki K, Mundy J, Chua NH. Four tightly linked rab genes are differentially expressed in rice. Plant Mol. Biol. 1989;14:29–39. doi: 10.1007/BF00015652. [DOI] [PubMed] [Google Scholar]
- 122.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 123.Downton WJS, Loveys BR. Abscisic acid content and osmotic relations of salt- stressed grapevine leaves. Aust. J. Plant Physiol. 1981;8:443–453. [Google Scholar]
- 124.Guiltinan MJ, Marcotte WR, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science . 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 125.Xiang X, Fu JR. The Expression and Regulation of ABA-responsive Genes and The Relation With Stress. Chin. Bull. Bot. 1998;15:11–16. [Google Scholar]
- 126.Becker D, Hoth S, Ache P, Paran LN, Naros PE. Regulation of the ABA-sensitive Arabidopsis potassium channel gene GORK in response to water stress. FEBS Lett. 2003;554:119–126. doi: 10.1016/s0014-5793(03)01118-9. [DOI] [PubMed] [Google Scholar]
- 127.Thomashow MF. In: Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. Meyerowitz EM, Somervlle CR, editors. New York: Cold Spring Harbor Laboratory press; 1994. pp. 807–834. [Google Scholar]
- 128.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness t o drought, lowtemperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu MJ, Liu GR, Yang XJ, Wang LJ. The study of drought induced protein of winter wheat cultivars. J. Agri. Uni. Hebei. 2002;25:11–15. [Google Scholar]
- 130.Ren DT, Zhao SL. Effects of Water Stress on Protein Metabolism of Flag Leaves of Spring Wheat Growing in Semi-arid Region. Acta Agron. Sin. 1997;23:468–473. [Google Scholar]