Abstract
A novel technique for biomolecular detection in microliter droplets floating on the surface of high density oil is presented. Each droplet was captured and manipulated dielectrophoretically and was used as a site for a microscopic bioassay based on agglutination of antibody-conjugated particles. The results were read out by the pattern of unagglomerated gold nanoparticles collected on the droplet surface. Two formats of bioassays, namely gold only agglutination and gold and latex agglutination, were investigated experimentally by varying analyte concentration, particle size and concentration, number of antigen binding sites per particle, time for incubation, and rate of particle collection on the droplet surface. The microbioassays performance was also evaluated with ricin antibodies and compared to the ricin assays in field use. It is estimated that the droplet based assays require 100× smaller sample volume and are ten times more sensitive, though they require longer times to complete. The experiments were interpreted by modeling the kinetics of particle agglutination and mass transfer processes inside the droplets. The incubation time and antigen concentration values calculated by the model correlate well with the experimental results. The results could allow for development of efficient immunoassays on a chip requiring even smaller sample volumes.
INTRODUCTION
The last five decades have brought forward significant development in the immunological techniques for biomolecular detection and identification.1, 2 Many of the immunoassays for clinical diagnostics and detection of chemical and biological agents are based on particle agglutination principles.3 They are used in detection of various proteins such as immunoglobulin, toxins, and hormones present in blood serum.2, 4, 5, 6, 7, 8, 9 Microscale devices are commonly used in conjunction with immunological methods to process multiple samples in an efficient and rapid manner. Microfluidic operation in small volumes reduces the time needed for analysis of a sample. The volume of analyte solution may be critical in applications such as biodefense and forensic diagnostics, where only limited sample amounts are available.
The typical immunoagglutination assays are based on polystyrene latex microspheres with antibody molecules bound to their surface.3, 4, 5, 6, 7, 8, 9, 10 An aqueous suspension of these microspheres is mixed with a sample containing antigen molecules from whole blood, serum, urine, etc. The antigen molecules bind two antibody molecules situated on different microspheres and cause agglutination (aggregation) of latex microspheres. Several techniques such as nephelometry and spectrophotometry could be employed to determine and quantify the aggregation state of the latex particles. The immunoagglutination methods, however, are not readily compatible with conventional microfluidic devices with channels due to problems with mixing, clogging of the channels by particles or aggregates, protein fouling, high pressure heads generated by viscous fluid flow, and long result read-out times.11, 12 Some of these problems can be addressed by “digital microfluidics”—moving droplets on solid surface using electrowetting.13, 14, 15 This technique, however, still may encounter problems with contact angle hysteresis, contact line pinning of droplets, and fouling. Complex optical detection methods would be required to read the results of agglutination assays in the sessile droplets.
In this article a new type of immunoassay based on an alternative droplet microfluidic technique is explored and characterized. It is based on a fluidic chip, where freely suspended droplets are entrapped and transported by dielectrophoresis without any contact with the solid surfaces.16, 17, 18 The microdroplets are suspended on the surface of perfluorinated hydrocarbon and serve as self-contained microscopic containers and reactors for performing and reading out assays for biological detection. The electric fields that hold and guide the droplets and particles are applied through arrays of electrodes submerged in the oil (Fig. 1). The droplet technique does not encounter the problems of high pressure head, channel clogging, protein fouling, and waste disposal existent in conventional microfluidic devices.
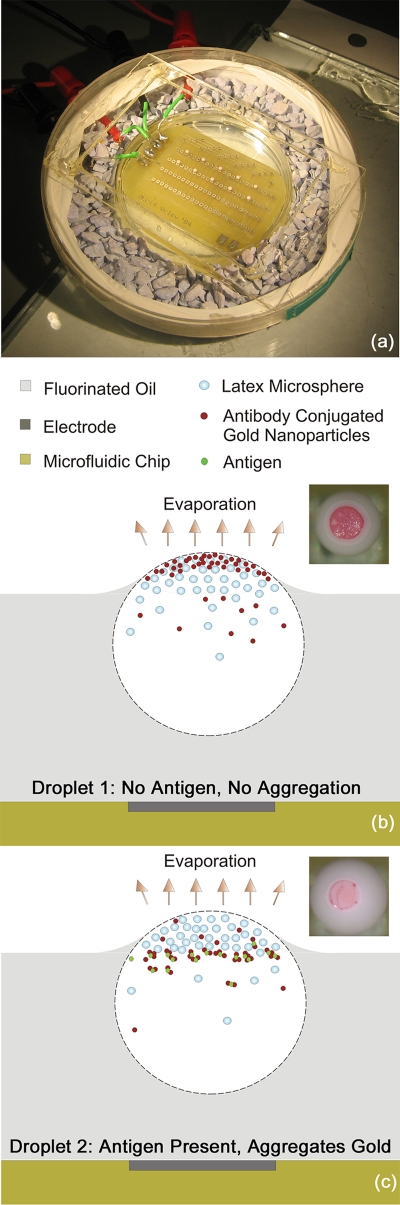
Figure 1.
(a) Experimental setup with evaporating droplets on a DEP chip. (b) Schematics and optical micrograph from above of evaporating droplet without antigen. (c) Schematics and micrograph of gold nanoparticle aggregation in a droplet containing antigen.
A detailed experimental study of the liquid flow and particle distribution, combined with simulation of the heat and mass transfer, inside single floating microdroplets was recently completed.19 Evaporation from the exposed portion of droplets protruding through the oil leads to internal water circulation, mixing, and microseparation of the particles in the top part of the droplets. The internal circulation is driven by Marangoni flow. Finite element simulations for hydrodynamic flows inside the droplet were in good correlation with experimental observations. Various chemical reactions and materials synthesis processes can be performed in these microcontainers.19 Here we show how such “droplet engineering” could find applications in novel bioassays.
Principles of the immunological bioassays in microdroplets
The evaporation process of the droplets can be used for on-chip detection of antibody-antigen driven agglutination. In the earlier demonstration of the principle, we mixed aqueous suspensions of 0.32 μm latex particles and of 40 nm gold nanoparticles coated with goat anti-rabbit IgG.19 One part of the suspension was kept as is, and another part was mixed and incubated with rabbit IgG (the antigen for the IgG bound on the gold nanoparticles). 1.0 μL droplets of each suspension, (Droplet 1 no antigen—“negative control” droplet, Droplet 2 with antigen—“test droplet”) were deposited on the F-oil, entrapped by the electric field and observed during drying under the microscope (Fig. 1). As the droplets began to dry, a dark gold nanoparticle “eyeball” spot appeared on the top surface of the negative control droplet without rabbit IgG [Fig. 1b, inset]. Deposits of gold nanoparticles in the droplet with rabbit IgG, however, were not visible on the surface [Fig. 1c, inset].
The differences in the particle collection pattern in the top part of droplets arises because the gold nanoparticles in the test droplet bind to other gold nanoparticles particles via antibody-antigen interaction, forming large clusters as a result of this agglutination process. The gold nanoparticles in the negative control droplet, on the other hand, do not agglutinate in the absence of antigen and remain freely dispersed. The agglutinated gold nanoparticles in the test droplet cannot pass through the interstices between the latex particles collected on the top section of the droplet showing a positive result. The unbound free nanoparticles in the negative control droplet are dragged to the surface and form the darker spot indicating negative result. Thus microseparation inside the droplets allows direct and easy distinguishing of the aggregation state of the suspended particles affected by biomolecular binding. This process was developed further and investigated in depth in our present study to enable the development of sensitive biological microbioassays.
Formats of immunoagglutination bioassays studied
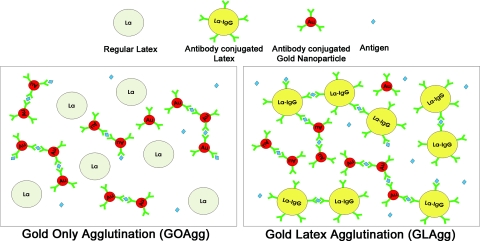
Two types of assays, schematically shown in Fig. 2, were developed and limit of detection (LOD) experiments were performed. Both assays use gold nanoparticles functionalized with antibodies for the targeted biological or chemical molecule. The difference between the two formats lies in the types of latex particles present in the droplets. The first assay reported here is Gold Only Agglutination (GOAgg) which uses nonfunctionalized latex microspheres. The functionalized gold nanoparticles agglutinate in the presence of an antigen, forming clusters within the bulk of the droplet. The second assay format, coded here as Gold and Latex Agglutination (GLAgg) is based on similar detection principles, but in this case both the gold nanoparticles and the latex spheres are conjugated with immunoglobulin. The antigen leads to agglutination of all particles, including the gold nanoparticles, the latex spheres and cross-agglutination between the gold and latex particles. The detection is carried out by the microseparation procedure and a positive result is detected by the absence of gold nanoparticle ring or spot on the droplet top.
Figure 2.
Schematics of the immunorecognition and agglutination processes taking place in the two microbioassay formats studied.
There are important differences between the above mentioned formats of microbioassays. The GOAgg format is less expensive and simpler to implement and read. These assays, however, may be easily oversaturated with antigen, resulting in false negative results. Oversaturation occurs when antigen concentration in the droplets is enough to bind to all antibodies at a ratio of at least 1:1, as a result of which cross-linking of the particles becomes impossible. This ratio was found to be higher in the experiments owing to the slow diffusion and orientation constraints in binding of the particles. The GLAgg format, on the other hand, requires two types of functionalized particles, and thus is a bit more expensive and complex. However, it may be less prone to oversaturation because of the higher number of antigen binding sites available on both latex and gold nanoparticles.
In the following sections we present the experimental data and evaluate the microbioassays, GOAgg and GLAgg, using the goat anti-rabbit immunoglobulin and rabbit immunoglobulin pair. The performance of assays was assessed in terms of reliability, sample volume, limit of detection, incubation time, particle size, and concentration detection range. The microbioassays are also characterized using antibodies and antigens supplied by Critical Reagents Program (CRP), US Department of Defense (DOD) for a realistic biological defense application—detection of ricin. The parameters of the microbioassays developed using particle agglutination techniques were compared with the ones of common hand-held and laboratory CRP assays. In the second part of the article, results are presented from the theoretical model of the kinetics of particle agglutination and correlated with experimental results.
EXPERIMENTAL PROCEDURES
Materials
The detection in microbioassay droplets is based on gold nanoparticles penetrating through cavities in the latex particles cap. Calculations for the geometry of the cavity formed between the spheres in a hexagonally close-packed crystal show that the minimal opening size is ∼15% of the diameter of microspheres. The diameter of latex particles should be such that the aggregated 40 nm gold nanoparticles can not pass through the interstices of the latex microspheres. Hence, polystyrene latex microspheres of 0.32 μm diameter were chosen to detect the presence of antigen in the microbioassay droplet. Aqueous surfactant-free sulfate-stabilized 0.32 μm polystyrene latex microspheres were purchased from Interfacial Dynamics Corp. (Portland, OR, USA). Goat anti-rabbit IgG (H&L)—Flouresbrite™ Carboxylate YG Beads were purchased from Polysciences Incorporation (Warrington, PA, USA). The microspheres were centrifuged at 1100 g for 10 min with Marathon micro-A centrifuge (Fisher Scientific, USA) and washed with deionized (D.I.) water. The collected microspheres were resuspended in D.I. water and sonicated (Branson Ultrasonics Corp., CT, USA). The D.I. water used was obtained from Millipore RiOs 16 reverse osmosis water purification systems (Bedford, MA, USA).
An inert, high density perfluorinated oil, FC-70, was purchased from Sigma-Aldrich (St. Louis, MO, USA). 40 nm gold nanoparticles were obtained from British Biocell International (Cardiff, UK). 40 nm goat anti-rabbit IgG conjugated gold particles were purchased from EY Labs (San Mateo, CA, USA). Bovine serum albumin (BSA) was purchased from Sigma-Aldrich. Rabbit IgG plasma was purchased from Calbiochem (San Diego, CA, USA). Ricin antigen (ricin A-chain) and ricin antibody (goat anti-ricin toxin) were supplied by CRP, DOD. Standard hand held assays (HHA) for the detection of ricin were also obtained through CRP. These assays operate on immunochromatographic principle.1
Experimental Setup
The DEP chip used to capture microdroplets carries arrays of electrodes situated on a circuit board.20 The square waves of frequency 800 Hz and amplitude of 700 V applied to the electrodes were generated using a FG-7002C Sweep∕Function generator (EZ Digital Company Limited, Korea) and a Piezo Driver∕Amplifier (Model PZD 700, Trek Incorporation, USA). The electrode chip was immersed in 4.5 mL high density fluorinated oil (FC-70) contained in a small Petri dish (Millipore Co., MA, USA). The Petri dish was in turn kept inside a bigger chamber containing desiccant to enhance evaporation of droplets (Fig. 1).
Microseparation of particles due to evaporation in the droplets was continuously monitored from top using SZ61 0.7–4.5× zoom stereomicroscope (Olympus America Inc., NY, USA). Their images were captured at regular intervals using DSC-V1 Cyber-Shot digital camera (SONY, Japan) coupled with the microscope. Characterization of the droplet geometry was done using Olympus BX-61 optical microscope (Olympus America Inc., NY, USA). Images of the droplets were taken using high resolution DP70 digital CCD microscope camera (Olympus America Inc., NY, USA).
Methods
Water droplets of volume 1.0 μL containing the microspheres, functionalized gold nanoparticles and antigen were dispensed onto the oil surface using ultramicropipette (Eppendorf North America Inc., NY, USA). The droplets for GOAgg microbioassay were prepared by washing the latex particles twice with 0.01 M PBS and centrifuging them at 3000 g for 20 min. The supernatant was decanted, the latex particles were sonicated and then mixed with 0.01 M PBS containing 2 mg∕mL BSA and incubated for 30 min. BSA was routinely added to the solutions to prevent any specific adsorption of antigens on the surface of latex microspheres during the microbioassays.21 In the next step the microspheres were again washed with PBS and then centrifuged to remove unadsorbed immunoglobulin in the solution. Subsequently, a solution containing 0.2 mg∕mL BSA and 0.1 wt % Tween-20 in 0.01 M PBS (referred to further as “PBSA”) was added with sonication to adjust final latex concentration to 15 wt %.
The latex solution was then mixed in 1:1 volume ratio with goat anti-rabbit IgG conjugated suspension containing 0.04 wt % of 40 nm gold particles. 10.0 μL aliquots of this latex∕gold mix were taken and increasing concentrations of antigen (Negative control—no antigen, 1.0 μg∕mL, 10.0 μg∕mL, and 10.0 μg∕mL) were added to each. To study the effect of incubation time, several sets of 10.0 μL aliquots of latex and antibody coated gold were prepared. Aliquots of each set were then mixed with antigen concentration varying from 0 to 10.0 μg∕mL. These assays were incubated for times ranging from 5 min to 45 min.
Latex solutions for the GLAgg assay were prepared using goat anti-rabbit IgG coated Flouresbrite™ Carboxylate YG Beads of 1.03 μm diameter. These antibody-coated particles were pretreated by the same procedures as described above for latex in GOAgg assay to adjust latex concentration to 2.6 wt %. The latex suspension was then mixed in 1:1 volume ratio with 0.04 wt % suspension of antibody-conjugated 40 nm gold nanoparticles. The latex∕gold particle suspension was divided into 10.0 μL aliquots and increasing concentrations of antigen were added before incubation and deposition of 1.0 μL droplets on the chip.
The effect of gold nanoparticles and Tween-20 on the evaporation rate of droplets was examined in order to characterize the drying process leading to detection. This was done with sets of droplets, which had similar contents except for the presence of gold nanoparticles and Tween-20. The preparation procedure was the same as for droplets in GOAgg microbioassays. Two sets of 1.0 μL droplets were compared. The droplets in the first set contained 15 wt % latex and a mixture of latex and 0.04 wt % gold nanoparticles in PBSA. The droplets in the second set had the same particles, but 0.05 wt % Tween-20 was added to all samples. The droplets were entrapped on the DEP chip and their diameter was measured with time using high magnification optical microscopy to compare the rate of evaporation.
RESULTS AND DISCUSSION
Gold only agglutination microbioassay
In the initial set of experiments it was verified that the assay functions were as expected. A detailed study of the effects of the major experimental parameters was then performed. The results can be summarized as follows.
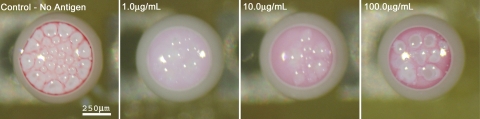
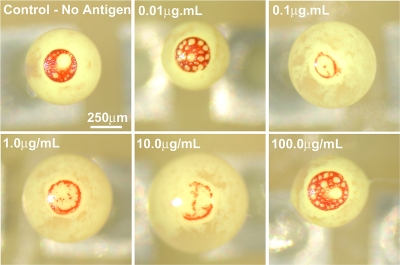
Effect of antigen concentration: The suspension containing latex and gold nanoparticles and varying concentration of antigen was preincubated for 30 min. Images of droplets evaporating on F-oil surface were then taken at regular intervals. After 12 min of drying time, the droplets showed a clear difference in the collection pattern of colloidal gold on top (Fig. 3). The gold nanoparticles in negative control droplets were able to pass through the interstices between latex microspheres and collect on top. No nanoparticle aggregation had taken place owing to the absence of antigen. The droplet with 1.0 μg∕mL antigen concentration showed the least amount of gold nanoparticle collection on top. This points out that the gold nanoparticles had agglutinated strongly and formed aggregates large enough to get entrapped in the cavities between the latex particles.
Figure 3.
Optical micrographs of droplets in a GOAgg assay. 1.0 μg∕mL antigen concentration shows the least amount of gold nanoparticles collection on top for 30 min incubation time. The white areas in the top of the droplets are the dense latex particle phases. The gold nanoparticles reaching the top are easily observed because of their intensively red color.
The assay droplets containing 10.0 μg∕mL antigen displayed more gold nanoparticles collected in its top portion than the 1.0 μg∕mL antigen droplets. This can be explained with effective over saturation of the antigen-binding antibody sites on the gold nanoparticles. The binding process occurs when free antibody on one particle gets in contact with an antigen bound to an antibody on another particle. The binding does not take place when both antibodies on the two particles are saturated with antigen. The collision of heavily antigen-covered gold particle with another antigen-saturated nanoparticle site does not lead to aggregation. A larger number of gold nanoparticles remained unaggregated, passing through the interstices of collected latex particles cap and migrating to droplet top.
The droplet with 100.0 μg∕mL antigen showed maximal amount of gold nanoparticle collection. The concentration of antigen in this droplet was high enough to saturate all, or nearly all, antigen binding sites on the surface of the colloidal gold. Thus, the gold particles did not aggregate and collected in the top portion of the droplet. Notably, the color of the gold nanoparticles collected here differs from the one of the negative control, displaying a more bluish tint. This can be explained by the partial aggregation of the nanoparticles before full surface saturation takes place. The plasmon absorption band of colloidal gold depends on the effective size of the nanoparticles and as the nanoparticles aggregate they show a red shift in the absorption spectra.22 In practice, the difference in the color could not be a parameter reliable enough to distinguish the negative control droplets from the over saturated ones. Thus, over saturation (in this case occurring at an antigen concentration ∼100× higher than the optimal one) could lead to error in the readout of these assays.
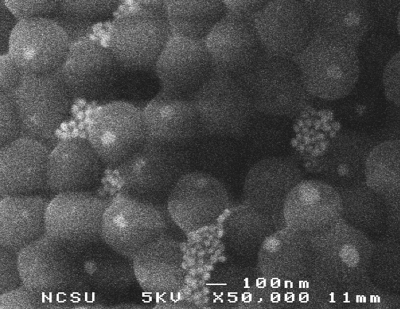
Scanning electron microscopy observations of a dried microbioassay droplet confirm the assumption of hexagonal closed packing of latex particles in the top portion of droplets. Micrographs of the bottom side of the particle aggregate were taken after flipping it over an SEM grid, illustrating how the agglutinated gold nanoparticle clusters get captured in between the interstices of the latex particles (Fig. 4).
Figure 4.
Scanning electron micrograph showing clusters of aggregated gold nanoparticles trapped in the interstices between latex particles in the bottom portion of droplet cap.
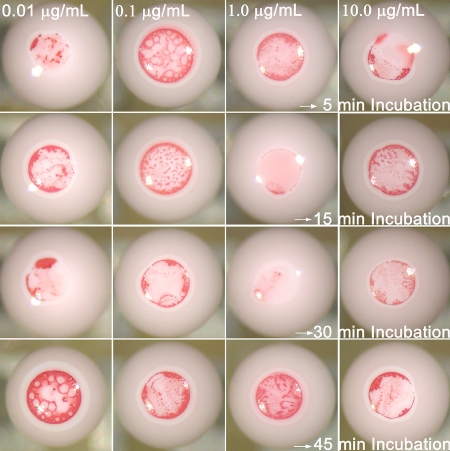
Effect of incubation time: The influence of incubation time (before depositing and drying the droplets) on the performance of microbioassays was evaluated using the GOAgg system (Fig. 5). Short incubation times (<5 min) did not result in visible pattern that can be interpreted for successful antigen detection. The gold nanoparticles and antigen molecules do not undergo enough effective collisions at such short times. The gold nanoparticles get pushed to the droplet top by the evaporation flux before they had formed big enough clusters to be caught in the latex particles pores. The microbioassays show differentiable pattern for 15 min of incubation time. The smallest amount of gold nanoparticles coming to the top is registered after 30 min of incubation, indicating that this is about the optimal incubation time, during which the major fraction of the Au nanoparticles have been included into aggregates large enough to prevent them from reaching the top surface during the evaporation.
Figure 5.
Optical micrographs of GOAgg microbioassay for varying incubation times (vertical direction) and varying concentration (horizontal direction).
Surprisingly, a larger fraction of the gold nanoparticle collection for the assays was consistently observed for 45 min of incubation, in comparison to the ones performed at smaller incubation times (see bottom row in Fig. 5). The difference between the positive and negative control assays becomes hard to visualize. Thus, the assays seemed to deteriorate and free particles were released from the aggregates formed. It has been hypothesized that the detachment is caused by the thermal motion of the gold nanoparticles and the presence of a large pool of free surfactant (Tween-20) in the medium. The antibodies are physically adsorbed on the gold surface and can be pulled off partially during the thermal fluctuations on the large agglutinated gold particles. Once an antibody gets pulled off partially from the nanoparticle surface, the surfactant molecules present in the droplet compete to adsorb at their place and prevent immunoglobulin readsorption. The danger of “over-incubating” the assays is significant for practical applications and will be investigated in the future due to its complex origins.
Gold and latex agglutination microbioassay
The GLAgg agglutination process involves more complex interactions in comparison to the GOAgg microbioassay. This assay includes agglutination of both the antibody conjugated latex microspheres and the antibody conjugated gold particles (Fig. 2). Aliquots of 10.0 μL latex and gold nanoparticle suspension were incubated for 15 min and 55 min to allow for completion of the agglutination processes. The droplets were then dispensed on the DEP liquid-liquid chip to follow the microseparation due to evaporation. A similar gold nanoparticles collection pattern on top was observed for a wider range of antigen concentrations (0.1 μg∕mL∼10.0 μg∕mL), regardless of the time during which droplets were incubated (Fig. 6). This was in contrast to GOAgg microassays, which only performed optimally for 30 min incubation at a concentration of 1.0 μg∕mL. In addition to the lack of dark red spot, the latex particles do not collect effectively on the surface due to the formation of loose latex aggregates.
Figure 6.
Optical micrographs of droplets with GLAgg assays at increasing antigen concentrations.
In contrast to the GOAgg microbioassays, the GLAgg systems were not sensitive to the size of the latex particles. GOAgg assays with spheres 0.78 μm and larger were not successful because the large size interstices in the latex cap allowed even aggregated gold nanoparticles to pass through. The GLAgg assays worked successfully with particles of 1.03 μm in size, because the gold nanoparticles are prevented from reaching the surface by binding rather than filtering in the cavities. A gold particle with antibody sites covered with antigen rising to the droplet surface can become attached to the latex particles collected in the top portion of the droplets via their antigen free antibody sites.
The comparison between the two assay formats leads us to the conclusion that the GLAgg microbioassays are less affected by incubation time and less responsive to analyte concentration. There are more antigen binding sites that can adsorb more antibodies before saturating. In addition, the gold particles in GLAgg microbioassays have low probability of making it to the top surface of latex particles cap even if the cavities between the particles are larger than that in the GOAgg assays.
In summary, the results point out that the rapid and reliable detection in these assays depends on the balance between incubation time and analyte concentration. GOAgg assays are simpler and are, in principle, more sensitive. The GLAgg microbioassays can give faster results and appear less prone to oversaturation in comparison to GOAgg format due to the presence of larger amount of antigen binding sites available on both types of particles.
Gold only agglutination microbioassay with ricin antibody
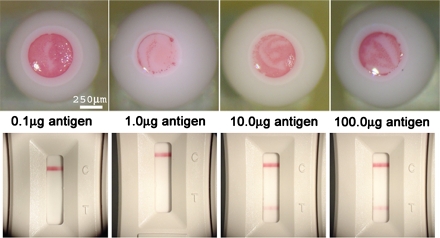
Assays based on goat anti-rabbit antibodies are a research standard, but they might not be a realistic enough simulation for practical toxin antigens. For this purpose, experiments were done with GOAgg bioassays based on gold nanoparticles coated with ricin antibody. The antibody was conjugated to the colloidal gold using the protocol given by Beesley 23, 24, 25, 26. The experiment was then conducted by the same protocol as for GOAgg microbioassays. This experiment also allowed us to evaluate the performance of the droplet assays to the one of the standard DOD hand held assays (HHA) operating on immunochromatographic principles 1, 27, 28. The ricin droplet microbioassays showed minimum gold nanoparticles collection for 1.0 μg∕mL concentration at 30 min incubation time (Fig. 7). This correlates well with the results of GOAgg microbioassays made with goat anti-rabbit IgG conjugated gold. The hand held assays needed at least 10 μg∕mL to yield positive results.
Figure 7.
Comparison between optical micrographs of droplet GOAgg assay with ricin as an antigen (top) and conventional HHA for ricin. 30 min incubation.
A summary of the evaluation of the ricin droplet microbioassays and the conventional HHA is presented in Table 1. The droplet based assays take 3× as much time as HHA to produce detection results. However, they have 10× lower limit of detection (LOD) and are also 100× more efficient in utilizing sample volume. These advantages make them suitable for analysis of biotoxin agents and forensic samples of microscopic volumes and low concentrations. Assays based on other antibodies for ricin with higher sensitivity have been reported previously 28, 29, 30. However, these assays use larger sample volumes in comparison to the droplet based microbioassay.
Table 1.
Summary comparison between HHAs and droplet based microbioassays.
| Parameter | Hand held assay | Droplet based assay |
|---|---|---|
| Incubation time | ∼15 min | ∼45 min |
| Volume of sample | >100 μL | <1.0 μL |
| Lower limit of detection | 10.0 μg∕mL | 1.0 μg∕mL |
Factors affecting evaporation rate of droplets
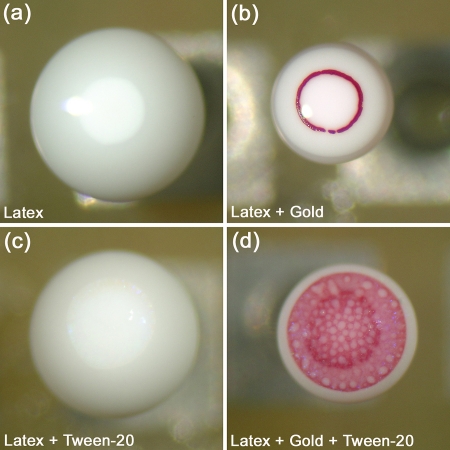
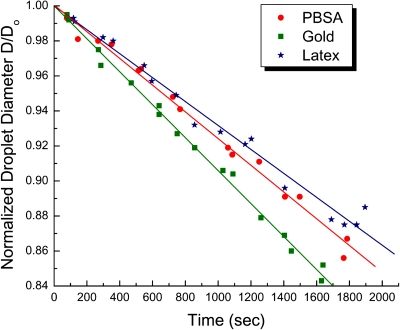
The speed of microseparation of particles in the top section of the droplets is controlled by evaporation. The microbioassays can provide faster results if the evaporation rate of droplets is increased. In order to facilitate the future improvement and optimization of the droplet assays, we investigated the effect of gold nanoparticle concentration, presence of Tween-20, background protein (BSA) concentration, and electric field intensity to the evaporation rate. Four sets of drying droplets were compared to examine the effect of Tween-20 and Au nanoparticles on evaporation rate (Fig. 8). The protocol for these experiments is detailed in the Methods section. The data indicates that the presence of surfactant assures a slower but more uniform drying of the droplets [compare Fig. 8a with Fig. 8c, Fig. 8b with Fig. 8d]. On the other hand, the gold nanoparticles strongly increased the evaporation rate in comparison to the surfactant [compare Fig. 8a with Fig. 8b, Fig. 8c with Fig. 8d].
Figure 8.
Micrographs of typical droplets, illustrating the effect of presence of gold nanoparticles and Tween-20 on the drying rate of droplets. (a) latex only, (b) latex and gold nanoparticles, (c) latex and Tween-20, and (d) latex, gold nanoparticles and Tween-20. All droplets were allowed to evaporate for 65 minutes.
The gold nanoparticle effect on evaporation was examined quantitatively by measuring the diameter with time for three types of droplets (Fig. 9). The concentration of gold nanoparticles was kept the same as in GOAgg droplet bioassays. The normalized droplet diameter in all cases decreased approximately linearly with time. The difference in slope points out that droplets containing nanoparticles were evaporating faster in comparison to the PBSA and latex droplets. This supports the conclusion drawn from Fig. 8 that the presence of gold nanoparticles increases the evaporation rate of droplets. The higher evaporation rate in the presence of nanoparticles is possibly a consequence of the deformation and corrugation of the surface by the layer of particles collected and pressed against it from the water side and the resulting higher area of evaporation. The results in general point out that the concentration of nonionic surfactant and nanoparticles should be sustained constant in order to compare the results of the various assays.
Figure 9.
Droplet diameter variation with evaporation time for droplets containing different ingredients.
Since the droplets are attracted to the underlying electrodes where the electric field intensity is high, it was also speculated that higher field intensities can pull them down toward the electrodes and thus control the degree to which they protrude from the surfaces. This could change the size of the meniscus and the top open area, where evaporation takes place. Droplets containing PBS were observed while varying the magnitude of electric field intensity in the operating range of 50,000 V∕cm∼80,000 V∕cm. Contrary to the hypothesis, however, it was found that changing electric field intensity does not affect the droplet meniscus size in the system.
The major factor in assay performance recognized so far is the dynamics of particle agglutination. Section 4 evaluates the particle aggregation dynamics for GOAgg microbioassay on the basis of modified agglutination theory and kinetic models available in the literature.
MODEL OF PARTICLE AGGLUTINATION DYNAMICS
The optimization of droplet microbioassays requires fast aggregation of antibody-coated particles to produce rapid detection results. A particle agglutination model to explain the binding dynamics of the biologically functionalized particles in the microbioassays was developed. Several assumptions and modifications were made to existing theories to make this model simple yet versatile enough. The particles are approximately spherical, so it can be assumed that the process is similar to reaction between spheres for which the rate constant can be expressed as
| (1) |
where kD=4πDR is the diffusion rate constant (R is the sum of the radii of reacting spheres, D is the relative diffusion constant) and kR is the reaction constant, which characterizes the binding of the biomolecules on the particle surfaces 31. Antigen-antibody binding reactions are specific and their rate is known to be rapid in comparison to the rate of diffusion 32. However, for certain system geometries the binding process may be reaction limited.
The ratio of the reaction control to the diffusion control in the binding process can be estimated by the Damkohler number. It is defined as
| (2) |
where R is the radius of the gold nanoparticles in cm, kf is the maximum forward reaction rate in ml∕(mol-s) considering orientation and other rate limiting factors, Γo is the surface concentration of antibody sites on the gold nanoparticles in mol∕cm2 and DB is the diffusion constant for antigen molecules in cm2∕s. For the system Da=0.6, which suggests that it is reaction limited. This reaction control, however, switches to diffusion control after a certain time interval (δ) which is defined as33
| (3) |
δ is on the order of few milliseconds, which signifies that the aggregation (agglutination) process in our system is effectively diffusion controlled. Equation 1 for the aggregation kinetics then simplifies to
| (4) |
where kD is given by the Smoluchowski theory,33, 34rA and rB are the radius of reacting spheres. The diffusion constants of the spheres can be related to their radii by the Stokes-Einstein equation
| (5) |
where η is the fluid viscosity, T is absolute temperature and kB is the Boltzmann constant. Equation 4 can be rewritten as
| (6) |
where koe=8kBT∕3η is the universal rate constant for particles of equal radius and fr=(rA+rB)2∕4rArB is the geometrical factor.
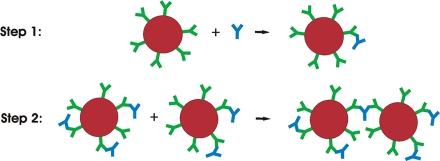
The aggregation process in the GOAgg microbioassay system takes place in two steps. The first step includes binding between antigen molecules and antibodies conjugated to gold particles. In the second step the antigen bound to an antibody site on one gold particle binds with another free antibody site on another gold particle and binds the two gold particles together (Fig. 10).
Figure 10.
Schematics of two step model for aggregation of gold nanoparticles in the presence of antigen.
The diffusion rate constant in Eq. 6 takes into account only the translational diffusion of reacting spheres. It is accurate only when the surface of the sphere is completely covered with reactive sites and all collisions are fully effective. However, the gold nanoparticles used had an average of 10–12 antigen binding sites per particle available for biomolecular reaction. Even if the reactive sites of the particles come into contact during collision, they might not aggregate because of unfavorable orientation. Apart from rotational diffusion, steric factors and reactive site area need to be considered for the calculation of rate constant. For the first step of aggregation after steric factors are taken into account, the reaction rate constant is given as
| (7) |
where D=DA+DB, R=rA+rB, rB is the radius of the smaller reactant and rA the radius of the larger particle.33 For the second step we consider two spheres A and B, having reactive sites described by parameters θA and θB, respectively. θ is the ratio of the radius of reactive site on the particle and the particle radius itself. The diffusion rate constant in this case is given by35
| (8) |
In our system rA=rB, hence θA=θB=rS∕rA, where rs is the radius of the reactive site, which corresponds to the radius of the area occupied by the IgG onto the surface of gold. The rate constant for the second step of aggregation can then be expressed as
| (9) |
Agglutination of two antibody-coated gold nanoparticles requires that an antibody site with bound antigen on one gold particle collides with an antigen-free antibody site on another gold nanoparticle. The rate of agglutination depends on the concentration of antigen, which in turn controls the number of antigen-bound and antigen-free antibody sites on the gold nanoparticles. The concentration of single gold nanoparticles not only changes via collisions with other single gold nanoparticles but also via collisions with bigger aggregates (doublets, triplets, etc.). For spherical particles of the same size, the concentration of any aggregate can be calculated using the theory of Smoluchowski as
| (10) |
where
| (11) |
is the initial concentration of antibody conjugated gold nanoparticles (monomers), τ is the characteristic half-time of aggregation and kD is the diffusion rate constant. The value of j varies as 1,2,3,… corresponding respectively to monomers, dimers, trimers, etc.34, 35, 36 The theory assumes that the diffusion rate constant is the same for dimers, trimers, and higher order aggregates. This is in contrast to what Eq. 9 suggests. However, this simplification does not affect our evaluations, as we are interested only in the formation of aggregates of second order (dimers).
To account for the probability of a successful binding collision leading to agglutination, the diffusion rate constant should include a collision frequency factor.37, 38 The collision frequency factor P can be estimated as follows:
| (12) |
where ϕ is the number of antibody sites with bound antigen molecules and (1−ϕ) is the number of antibody sites not bound to antigen molecules. After taking the collision frequency into account, we can calculate the corrected half time for aggregation as
| (13) |
For a given value of the diffusion rate constant and initial concentration, the half time is minimal at maximal probability. The collision frequency factor has a maximum at the value of ϕ=0.5 for 50% coverage of antibody sites by antigen on each particle. Immunoglobulins are Y-shaped bi-functional structures, so each antibody site can bind two antigen molecules.39 As mentioned before, on average there are six antibody sites on every gold nanoparticle, leading to a total of 12 antigen binding sites available on each nanoparticle. For maximum collision efficiency factor, the number of antigen binding sites covered is six.
The values of the parameters used in the estimates are listed in the Appendix. The diffusion rate constant for the second process (particle collision) is two orders of magnitude lower than for the first one, suggesting that it is the rate limiting step for binding kinetics. After 3 τ2=44.6 min, 94% of the gold nanoparticles in the solution form at least a dimer. The time of 44.6 min is close to the one observed in the experiments (30 min incubation followed by 20 min of drying). For 3.09×1017 particle∕m3 of gold nanoparticles, the concentration of antigen must be 1.854×1018 particle∕m3, which corresponds to an optimal antigen concentration of 0.5 μg∕mL. This is close to the value of 1.0 μg∕mL that we established in the experimental results.
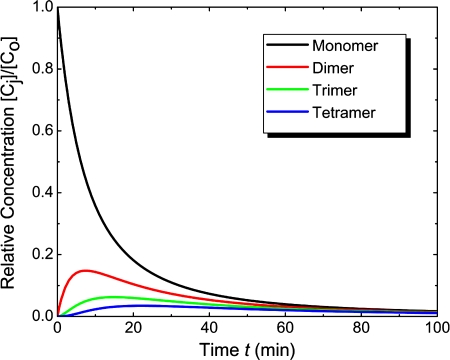
The concentration of each type of gold nanoparticle aggregate (relative to initial antibody-coated gold nanoparticles concentration) is plotted with respect to time in Fig. 11. As predicted by the aggregation half-time τ, the concentration of unaggregated antibody-coated gold nanoparticles (black curve) goes down to 6% of its initial value in about 45 min. For comparison, the time evolution of the concentration of aggregates like dimers (red curve), trimers (green curve), tetramers (blue curve), and other higher order aggregates was plotted.
Figure 11.
Concentration profiles of different types of aggregates relative to initial gold nanoparticle concentration [Eqs. 10, 13].
A comparison between the experimental results and the theoretically calculated values is given in Table 2. The agreement between experiments and theory suggests that the model can be used to predict the behavior of the microbioassays for any change in system parameters. This can be used for calculating the optimal particle concentration and minimal incubation times in designing future microbioassays, both in droplet on a chip or other formats. The applicability of the model to the case of ricin was verified on the basis of the similarity in size and correspondingly in diffusion rates.40, 41
Table 2.
Comparison between experimental results and theoretical values for droplet microbioassays.
| Parameter | Experimental value | Evaluated by theory |
|---|---|---|
| Incubation time | ∼50 min | 45 min |
| (30 min incubation, 20 min drying) | ||
| Optimum antigen concentration | 1.0 μg∕mL | 0.5 μg∕mL |
CONCLUDING REMARKS
It was shown that microliter droplets captured by DEP can be used as containers for microscopic bioassays. The detection is based on agglutination of antibody functionalized particles in the presence of antigen. Two microbioassay formats based on the type of functionalized particles, GOAgg and GLAgg were demonstrated. The experiments prove that both assay formats can be used to detect antibodies as expected. GOAgg assay has a lower limit of detection since only gold nanoparticles have binding sites available to consume antigen molecules, but it requires longer time for detection and is oversaturated more easily. However, the antigen concentration threshold for oversaturation is higher than expected for both microbioassays, probably because of the slow mass transfer processes.
The performance of the microbioassays was described as a function of several parameters including sample size, particle size, analyte concentration, limit of detection, incubation time, and rate of evaporation. The results from the droplet microbioassays were compared with the ones from HHA obtained from CRP, DOD in terms of incubation time, sample volume, and lower limit of detection (Table 1). The HHA needed 3× less time for result read-out. On the other hand the lower limit of detection for GOAgg assays was also found to be 10× better, 1.0 μg∕mL as opposed to 10.0 μg∕mL needed for HHA. The microbioassays consumed 100× less sample volume than HHA’s. Efficient usage of the sample makes it a viable immuno-detection method for biological defense applications, with a tradeoff in terms of testing time.
The results were matched against a model of particle aggregation kinetics developed on the basis of the kinetic theory of agglutination by Smoluchowski using rate constants provided in the literature. The calculations for the aggregation time of particles using this model were in good correlation with the experimental values (Table 2). The calculated antigen concentration was of the same order as the ones observed in experiments. The quantification of the agglutination and detection process can in the future be improved by measurement of the amount of nanoparticles on the droplet surfaces by image processing. By identifying the experimental conditions conducive to efficient detection in the assays and developing a model that could predict the kinetic response we make possible the further development of efficient microbioassays on a chip.
ACKNOWLEDGMENTS
This research was supported by the U.S. Army Research Office. The authors are thankful to Stephen Lee and the DOD CRP program for providing realistic antibodies and antigens. The authors thank Suk Tai Chang for valuable discussions and Brian Prevo for assistance with SEM imaging.
APPENDIX: LIST OF CONSTANTS AND VARIABLES USED IN THEORETICAL CALCULATIONS
Concentration of gold nanoparticles CA=3.09×1017 particle∕m3.
Molecular weight of immunoglobulin MIgG=160 kDa.30
Antigen concentration CB=(CNA)∕(MIgG) where C is antigen concentration in μg∕mL.
CB=3.764×1018 particle∕m3 for C=1.0 μg∕mL.
Radius of gold nanoparticles rA=2.0×10−8 m.
Average radius of IgG (see Ref. 42) rB=3.5×10−9 m.
Diffusion rate constant for first step kD1=1.87×10−18 m3∕s [Eq. 7].
Diffusion rate constant for second step kD2=1.45×10−20 m3∕s [Eq. 9].
Half-time constant for first step of aggregation τ1=0.564 s.
Half-time constant for second step of aggregation τ2=891.2 s=14.86 min.
References
- Baylis C. L., in Detecting Pathogens in Food, edited by McMeekin T. A. (CRC LLC, Boca Raton, FL, 2004), p. 217. [Google Scholar]
- Bangs L. B., Pure Appl. Chem. 68, 1873 (1996). [Google Scholar]
- Andreotti P. E., Ludwig G. V., Peruski A. H., Tuite J. J., Morse S. S., and L. F.Peruski, Jr., BioTechniques 35, 850 (2003). [DOI] [PubMed] [Google Scholar]
- Wang Z. G., Shang H., and Lee G. U., Langmuir 10.1021/la052902p 22, 6723 (2006). [DOI] [PubMed] [Google Scholar]
- Molina-Bolivar J. A., and Galisteo-Gonzalez F., J. Macromol. Sci., Polym. Rev. C45, 59 (2005). [Google Scholar]
- Bangs L. B. and Kenny M. T., Ind. Res. 18, 46 (1976). [Google Scholar]
- Tovar G. E. M. and Weber A., Dekker Encyclopedia of Nanoscience and Nanotechnology, edited by Schwarz J. A., Contescu C. I., and Putyera K. (Marcel Dekker, New York, 2004), p. 277. [Google Scholar]
- Lowe C. R., Hin B. F. Y. Y., Cullen D. C., Evans S. E., Stephens L. D. G., and Maynard P., J. Chromatogr. 510, 347 (1990). [DOI] [PubMed] [Google Scholar]
- Meulenberg E. P., Mulder W. H., and Stoks P. G., Environ. Sci. Technol. 10.1021/es00003a001 29, 553 (1995). [DOI] [PubMed] [Google Scholar]
- Bangs L. B., J. Clin. Immunoassay 13, 127 (1990). [Google Scholar]
- Stone H. A. and Kim S., AIChE J. 10.1002/aic.690470602 47, 1250 (2001). [DOI] [Google Scholar]
- Stone H. A., Stroock A. D., and Ajdari A., Annu. Rev. Fluid Mech. 10.1146/annurev.fluid.36.050802.122124 36, 381 (2004). [DOI] [Google Scholar]
- Cho S. K., Moon H., and Kim C. J., J. Microelectromech. Syst. 10.1109/JMEMS.2002.807467 12, 70 (2003). [DOI] [Google Scholar]
- Srinivasan V., Pamula V. K., and Fair R. B., Lab Chip 10.1039/b403341h 4, 310 (2004). [DOI] [PubMed] [Google Scholar]
- Su F., Chakrabarty K., and Fair R. B., IEEE Trans. Comput.-Aided Des. 10.1109/TCAD.2005.855956 25, 211 (2006). [DOI] [Google Scholar]
- Velev O. D. and Bhatt K. H., Soft Mater. 10.1039/b605052b 2, 738 (2006). [DOI] [Google Scholar]
- Velev O. D., Prevo B. G., and Bhatt K. H., Nature 10.1038/426515a 426, 515 (2003). [DOI] [PubMed] [Google Scholar]
- Velev O. D., Bhatt K. H., Prevo B. G., and Lumsdon S. O., Abstr. Pap. - Am. Chem. Soc. 226, U479 (2003). [Google Scholar]
- Chang S. T. and Velev O. D., Langmuir 10.1021/la052695t 22, 1459 (2006). [DOI] [PubMed] [Google Scholar]
- Millman J. R., Bhatt K. H., Prevo B. G., and Velev O. D., Nat. Mater. 10.1038/nmat1270 4, 98 (2005). [DOI] [PubMed] [Google Scholar]
- Bangs L. B., Am. Clin. Lab. 9, 16 (1990). [PubMed] [Google Scholar]
- Turkevich J., Gold Bull. (Geneva) 18, 125 (1985). [Google Scholar]
- Malamud D. and Drysdale J. W., Anal. Biochem. 10.1016/0003-2697(78)90790-X 86, 620 (1978). [DOI] [PubMed] [Google Scholar]
- Righetti P. G. and Caravaggio T., J. Chromatogr. A 127, 1 (1976). [DOI] [PubMed] [Google Scholar]
- Roth J., Techniques in Immunocytochemistry, edited by Bullock G. R. and Petrusz P. (Academic, London, 1982), p. 107. [Google Scholar]
- Beesley J. E., Colloidal Gold: A New Perspective for Cytochemical Marking (Royal Microscopical Society, Oxford, England, 1989), p. 10. [Google Scholar]
- Sun X.-L., Zhao X.-L., Tang J., Zhou J., and Chu F. S., Int. J. Food Microbiol. 99, 185 (2005). [DOI] [PubMed] [Google Scholar]
- Shyu R. H., Shyu H. F., Liu H. W., and Tang S. S., Toxicon 10.1016/S0041-0101(01)00193-3 40, 255 (2002). [DOI] [PubMed] [Google Scholar]
- Lubelli C., Chatgilialoglu A., Bolognesi A., Strocchi P., Colombatti M., and Stirpe F., Anal. Biochem. 355, 102 (2006). [DOI] [PubMed] [Google Scholar]
- Shyu H. F., Chiao D. J., Liu H. W., and Tang S. S., Hybridoma Hybridomics 21, 69 (2002). [DOI] [PubMed] [Google Scholar]
- Solc K. and Stockmayer W. H., J. Chem. Phys. 10.1063/1.1675283 54, 2981 (1971). [DOI] [Google Scholar]
- Dmitriev D. A., Massino Y. S., and Segal O. L., J. Immunol. Methods 280, 183 (2003). [DOI] [PubMed] [Google Scholar]
- Stenberg M. and Nygren H., J. Immunol. Methods 113, 3 (1988). [DOI] [PubMed] [Google Scholar]
- Evans D. F. and Wennerstrom H., The Colloidal Domain: Where Physics, Chemistry, Biology, and Technology Meet (Wiley, New York, 1999), pp. 417–428. [Google Scholar]
- Berg O. G. and Von Hippel P. H., Annu. Rev. Biophys. Biophys. Chem. 10.1146/annurev.bb.14.060185.001023 14, 131 (1985). [DOI] [PubMed] [Google Scholar]
- Schurr J., J. Phys. Chem. 10.1021/j100558a026 80, 1934 (1976). [DOI] [Google Scholar]
- Lamer V. K., Discuss. Faraday Soc. 10.1039/df9664200248 42, 248 (1966). [DOI] [Google Scholar]
- Hogg R., J. Colloid Interface Sci. 10.1016/0021-9797(84)90215-7 102, 232 (1984). [DOI] [Google Scholar]
- Quesada M., Puig J., Delgado J. M., Peula J. M., Molina J. A., and Hidalgo Alvarez R., Colloids Surf., B 10.1016/S0927-7765(96)01329-X 8, 303 (1997). [DOI] [Google Scholar]
- Radius of ricin is ∼2.5 nm as compared to 3.5 nm for IgG. The diffusion rate for the first step is roughly 118× larger than the rate of diffusion for second step. The antibody used to detect ricin is also an immunoglobulin attached onto the gold nanoparticle surface. Hence our assumption of second step to be the rate limiting is still valid and the model explains the dynamics of ricin GOAgg assays.
- Rutenber E., Katzin B. J., Ernst S., Collins E. J., Mlsna D., Ready M. P., and Robertus J. D., Proteins 10.1002/prot.340100308 10, 240 (1991). [DOI] [PubMed] [Google Scholar]
- Handley D. A., Colloidal Gold: Principles, Methods and Applications, edited by Hayat M. A. (Academic, San Diego, 1989), pp. 1. [Google Scholar]