SUMMARY
Dietary restriction increases life span and slows the onset of age-associated disease in organisms from yeast to mammals. In humans, several age-related diseases are associated with aberrant protein folding or aggregation, including neurodegenerative disorders such as Alzheimer’s, Parkinson’s, and Huntington’s diseases. We report here that dietary restriction dramatically suppresses age-associated paralysis in three nematode models of proteotoxicity. Similar to its longevity-enhancing properties, dietary restriction protects against proteotoxicity by a mechanism distinct from reduced insulin/IGF-1-like signaling. Instead, the heat shock transcription factor, hsf-1, is required for enhanced thermotolerance, suppression of proteotoxicity, and life span extension by dietary restriction. These findings demonstrate that dietary restriction confers a general protective effect against proteotoxicity and promotes longevity by a mechanism involving hsf-1.
Keywords: C. elegans, dietary restriction, proteotoxicity, longevity, hsf1, proteotoxicity
INTRODUCTION
Dietary restriction is known to increase life span and retard a variety of age-associated pathologies in rodents and invertebrate organisms (Weindruch & Walford 1988; Masoro 2005). Among the diseases beneficially impacted by DR in mice are cancer, diabetes, and cardiovascular disease. In addition, DR has been shown to improve outcome in mouse models of neurodegeneration (Martin et al. 2006). The mechanism by which DR confers these benefits remains unknown, but is of high interest.
DR can be studied in the nematode Caenorhabditis elegans using either environmental or genetic approaches. In the laboratory, C. elegans are typically maintained on a nutrient agar nematode growth medium (Youngman et al. 1992) and provided E. coli OP50 as their dietary food source. Under these standard growth conditions, life span extension from bacterial food restriction is maximized when animals are maintained on nutrient-agar plates without bacterial food after early adulthood (BD, bacterial food deprivation; also referred to previously as dietary restriction through food deprivation or dietary deprivation) (Kaeberlein et al. 2006; Lee et al. 2006). While BD is similar in concept to an alternative method for DR in C. elegans, axenic growth (Vanfleteren & Braeckman 1999), BD is instituted post-developmentally and utilizes standard C. elegans growth conditions.
An alternative method of DR in C. elegans is the use of mutations that reduce food intake, such as loss of function alleles of eat-2. eat-2 mutants have defects in pharyngeal pumping (Avery 1993) and an adult life span 20–40% longer than wild type N2 animals (Lakowski & Hekimi 1998). Life span extension from BD is non-additive with the long-lived eat-2(ad465) allele (Kaeberlein et al. 2006; Lee et al. 2006), indicating that BD and mutation of eat-2 are likely to promote longevity via similar or overlapping mechanisms.
Although the mechanism by which DR increases longevity in C. elegans is unknown, DR is thought to act through a pathway that is genetically distinct from insulin/IGF-1-like signaling (IIS). Mutations that decrease IIS, such as loss of function alleles of insulin-like receptor daf-2 (Kenyon et al. 1993; Kimura et al. 1997) or the PI3-kinase age-1 (Friedman & Johnson 1988a; Friedman & Johnson 1988b; Morris et al. 1996), increase longevity via a mechanism that is dependent on the FOXO-family transcription factor daf-16 (Lin et al. 1997; Ogg et al. 1997). In contrast, life span extension from DR (whether accomplished by BD, axenic growth, or mutation of eat-2), does not require daf-16 and is additive with mutation of daf-2 (Lakowski & Hekimi 1998; Houthoofd et al. 2002; Kaeberlein et al. 2006; Lee et al. 2006). More recently, two transcription factors, skn-1 and pha-4, have been shown to be necessary for life span extension from DR (Bishop & Guarente 2007; Panowski et al. 2007); however, it remains unclear how these factors respond to DR and what the relevant downstream targets for life span extension might be.
We have used C. elegans to explore the effect of DR on disease processes associated with protein misfolding or aggregation (proteotoxicity). Several transgenic models of proteotoxicity have been developed and characterized in worms. These models show pleiotropic phenotypes (Link 2001; Brignull et al. 2007), including defective posterior mechanosensation (Parker et al. 2001; Parker et al. 2005), impaired ubiquitin-proteasomal function (Khan et al. 2006), decreased nose touch response (Faber et al. 1999), or age-associated paralysis (Link 1995; Satyal et al. 2000; Morley et al. 2002; Link et al. 2006). Here we report that both genetic and environmental models of DR are potent suppressors of proteotoxicity and we identify a novel role for the heat shock transcription factor, hsf-1, as a key mediator of DR in C. elegans
RESULTS
DR confers protection against polyglutamine proteotoxicity
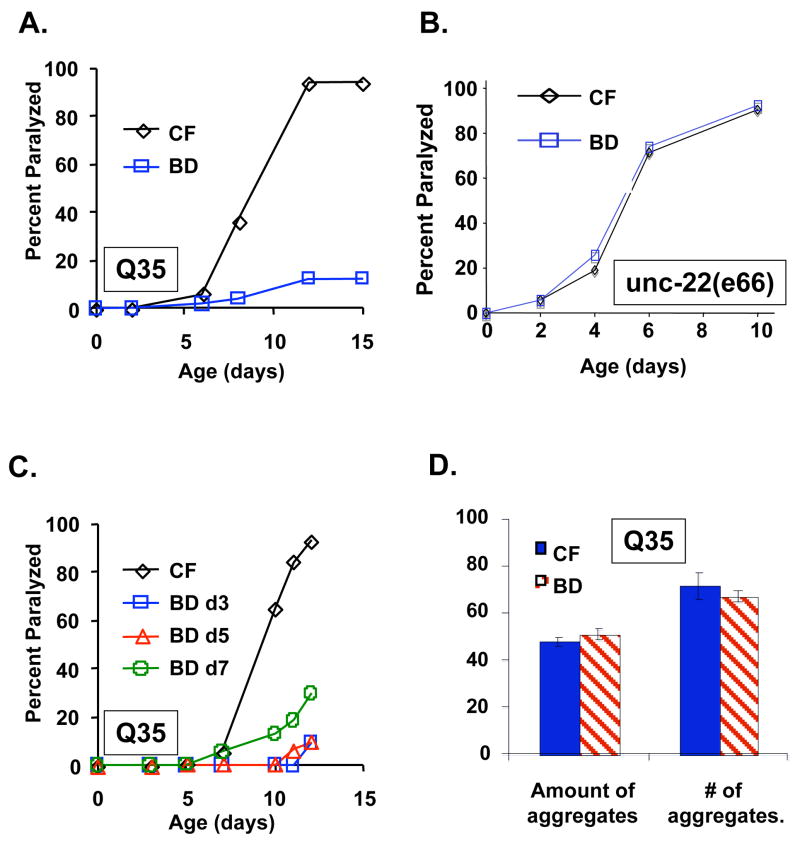
We first examined the effects of DR on proteotoxicity using a nematode model of polyglutamine disease. Huntington’s disease and several other neurodegenerative disorders are caused by polyglutamine tract expansions, though the mechanism of disease progression remains unknown (Bonini & La Spada 2005). For our studies, we utilized a nematode model of Huntington’s disease in which a tract of 35 consecutive glutamine residues is fused to YFP (Q35YFP) and expressed in the body wall muscles (Morley et al. 2002). This tract length has been shown to be at the threshold for age-related toxicity in C. elegans (Morley et al. 2002). As previously reported (Morley et al. 2002), we observed an age-dependent paralysis phenotype in Q35YFP animals fed a control diet of abundant UV-killed E. coli OP50 (Figure 1A). The life span of these animals was relatively normal (Supplemental Figure 1A), suggesting that the proteotoxicity caused by the Q35YFP protein, while sufficient to cause paralysis, does not necessarily limit longevity. When placed on a BD regimen at the 2nd day of adulthood, the survival of Q35YFP animals was enhanced in a manner similar to wild-type controls (Supplemental Figure 1A). Strikingly, Q35YFP animals on BD were resistant to proteotoxicity (p = 6.2 × 10−12) and remained largely paralysis free (Figure 2A; Supplemental video files).
Figure 1. Bacterial food deprivation suppresses proteotoxicity in a nematode model of polyglutamine disease.
(A) Age associated paralysis caused by expression Q35YFP (Q35) is significantly reduced in BD animals relative to control fed (CF) animals (p=6.2 × 10−12). (B) The age-associated paralysis of unc-22(e66) mutants is unaltered in BD animals relative to control fed (CF) animals. (C) BD significantly reduces polyglutamine-associated paralysis when initiated at day 3 (p=8.3 × 10−12), day 5 (p = 7.6 × 10−14), or day 7 (p=5.1 × 10−5) of adulthood. (D) BD does not significantly reduce the formation of polyglutamine aggregates. The total amount of aggregates, as measured by the integral of the fluorescent area per animal or the number of discrete aggregates visible as fluorescent spots, of BD animals relative to CF animals were counted. Error bars are standard error of the mean.
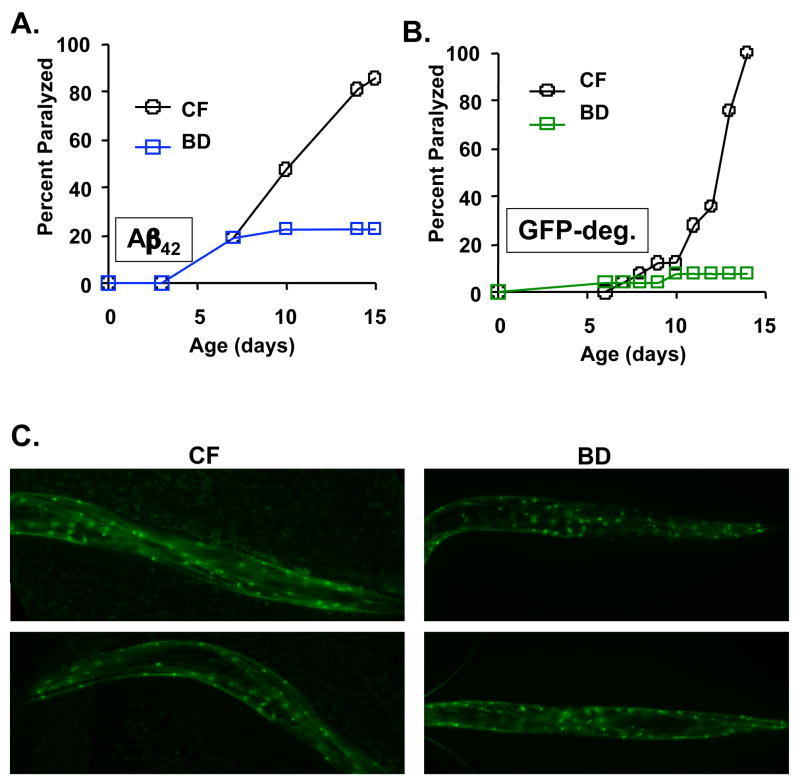
Figure 2. BD suppresses proteotoxicity in a nematode model of Alzheimer’s disease.
(A) Relative to control fed (CF) animals BD significantly reduces age-associated paralysis in animals expressing Aβ42 (p=1.4 × 10−7). (B) BD significantly reduces age-associated paralysis in animals expressing the aggregation prone GFP-degron peptide (GFP-deg), relative to CF animals (p=1.0 × 10−14). (C) By fluorescence microscopy, the steady-state levels of the GFP-degron peptide are not reduced in BD animals as compared to control fed (CF) animals.
To determine whether BD confers general protection against muscular defects independently of the Q35YFP transgene, we examined the effects of BD in two mutant backgrounds that display and age-associate defect in movement similar to that of Q35YFP: unc-52(e444) and unc-22(e66) (Moerman & Baillie 1979; Waterston et al. 1980; Rogalski et al. 1995). In both cases, BD failed to improve movement or suppress paralysis, relative to control fed isogenic animals (Figure 1B; Supplemental video files). Thus, we conclude that the suppression of paralysis by BD is unlikely to be the result of a general improvement in muscle function or increase in movement.
We have previously reported that BD increases life span to a similar extent whether initiated early in adulthood (day 2) or post-reproductively (day 10) (Kaeberlein et al. 2006). Accordingly, we wished to determine whether suppression of polyglutamine toxicity is observed by initiating BD at ages later than 2 days of adulthood. Consistent with the effect of BD on longevity, BD initiated at day 3, 5, or 7 of adulthood significantly reduced age-associated paralysis in Q35YFP animals (Figure 1C; p = 8.3 × 10−12 day 3, 7.6 × 10−14 day 5, 5.1 × 10−5 day 7). BD was not sufficient to reverse paralysis, however. Out of more than 50 individual paralyzed animals examined, none regained movement when transferred from a control fed diet to BD. These observations can be explained by a model in which BD confers protection against polyglutamine toxicity until a threshold level of toxicity results in paralysis, after which BD is unable to reverse the accumulated cellular damage.
The formation of insoluble aggregates is a hallmark of human polyglutamine diseases such as Huntington’s disease (Borrell-Pages et al. 2006). Whether aggregates are causal in disease progression remains controversial, however. To address this question, we quantified the abundance of aggregates in aged animals fed either a control diet or maintained on BD from the 2nd day of adulthood. Although paralysis was dramatically reduced in 14 day old BD animals relative to control fed animals, there was no significant difference in either the number or size of Q35YFP-aggregates between the two groups (Figure 1D; See also Supplemental Videos and Supplemental Figure 2). Consistent with this, the relative amount of Q35YFP protein, when normalized to total protein or to actin (0.96 ± 0.1), was not significantly different in BD animals relative to control fed animals at day 9 of adulthood (Supplemental Figure 2C). Thus, BD appears to protect against proteotoxicity without significantly altering the age-associated accumulation of polyglutamine aggregates.
DR is a general suppressor of proteotoxicity
In order to determine whether the suppression of proteotoxicity conferred by BD would be specific to polyglutamine toxicity, we next examined a transgenic model of Alzheimer’s disease. A 42 amino acid amyloid beta peptide (Aβ42) is expressed in body wall muscle cells of these animals under control of the unc-54 promoter (Link 1995). The Aβ42 peptide is a cleavage product of amyloid precursor protein, implicated in Alzheimer’s disease (Koo et al. 1999). Similar to the Q35YFP animals, BD was sufficient to largely suppress age-associated paralysis caused by Aβ42-induced proteotoxicity (Figure 2A; p = 1.4 × 10−7). BD also increased both median and maximum survival of Aβ42 animals (Supplemental Figure 1B; p = 8.7 × 10−4). In multiple independent experiments we observed that BD caused a notable increase in age-independent early mortality. This may indicate that amyloid beta toxicity (but not polyglutamine toxicity) causes frailty in response to food restriction among a percentage of the population. Interestingly, the subset of animals that died early also tended to show early paralysis, while the survivors remained largely paralysis free. Further studies will be necessary to address the mechanism behind this observation.
Given that BD was effective at suppressing proteotoxicity in two different nematode models of human disease, we wished to further characterize the generality of this phenomenon. Recently, an aggregation prone form of GFP (GFP-degron) was described that causes progressive paralysis when expressed transgenically in C. elegans from a myo-3 promoter (Link et al. 2006). As observed in the Q35YFP and Aβ42 models, BD significantly suppressed paralysis in animals expressing the GFP-degron peptide (Figure 2B; p = 1.0 × 10−14). We also assayed wether the levels of aggregated GFP:degron was decreased in worms subjected to BD. As with the Q35YFP animals, age-matched GFP:degron animals did not display a notable loss of the aggregated protein by microscopy (Figure 2C).
Genetic models of DR suppress proteotoxicity
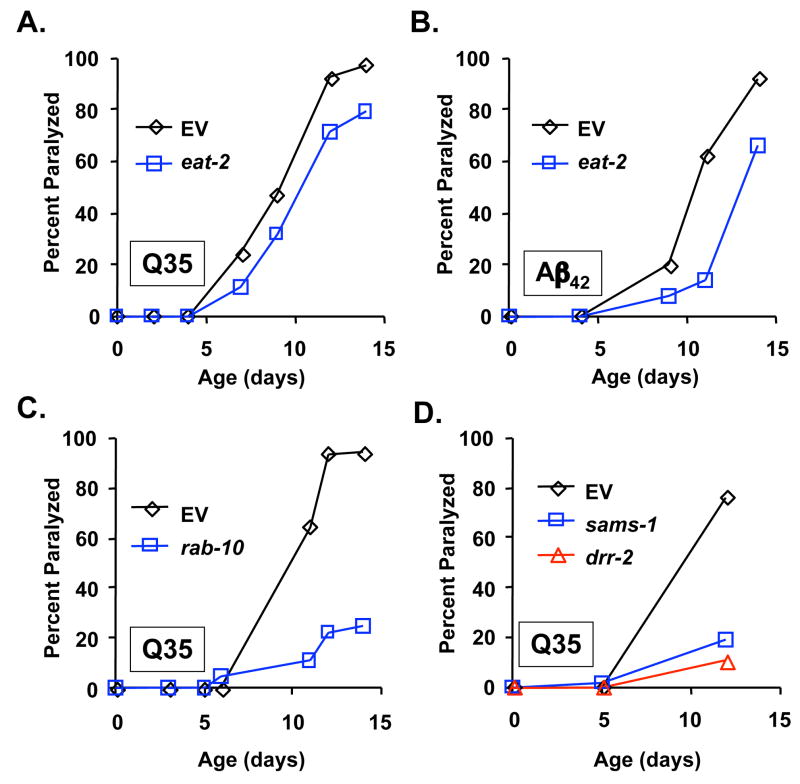
Although BD is unquestionably a form of food (dietary) restriction, it remains possible that BD could have different or additional properties that are not shared by other methods of DR described in the nematode. To address this possibility, we examined whether decreased eat-2 function could suppress proteotoxicity. RNAi inhibition of eat-2 resulted in a significant decrease in paralysis in both the Q35YFP (Figure 3A; p = 1.0 × 10−3) and Aβ42 (Figure 3B; p = 1.0 × 10−5) models. Suppression of proteotoxicity by inhibition of eat-2 did not appear to be as effective as BD, however. This is similar to the relative effects of BD and mutation of eat-2 on life span (Kaeberlein et al. 2006; Lee et al. 2006), and may suggest that BD represents a level of DR more optimal for longevity and suppression of proteotoxicity. Alternatively, BD may have additional properties that influence aging and proteotoxicity other than those shared by both BD and mutation of eat-2.
Figure 3. Genetic models of dietary restriction suppress proteotoxicity.
(A) RNAi knock-down of eat-2 significantly reduces paralysis in Q35YFP animals, relative to animals maintained on empty vector bacteria (p=1.0 × 10−3). (B) RNAi knock-down of eat-2 significantly reduces paralysis in Aβ42 animals, relative to animals maintained on EV bacteria (p=1.0 × 10−5). (C) RNAi knock-down of rab-10 significantly reduces paralysis in Q35YFP animals, relative to animals maintained on empty vector bacteria (p=1.3 × 10−11). (D) RNAi knock-down of sams-1 or drr-2 significantly reduces paralysis in Q35YFP animals, relative to animals maintained on empty vector bacteria (p=1.3 × 10−14 for sams-1 and p < 10−15 for drr-2).
We next examined genetic models of DR identified from a genome-wide RNAi screen for increased life span (Hansen et al. 2005). RNAi knock-down of rab-10, sams-1, or drr-2 increases life span through a daf-16-independent mechanism, but fails to further increase the life span of eat-2(ad1116) animals (Hansen et al. 2005). Each of these genes are transcriptionally down-regulated in eat-2 mutants, suggesting that they function downstream of food consumption to modulate life span (Hansen et al. 2005). Paralysis was dramatically reduced in Q35YFP animals in response to RNAi inhibition of rab-10 (Fig 3C; p = 1.3 × 10−11), sams-1 (Fig 3D; p = 1.3 × 10−14) or drr-2 (Fig 3D; p < 1 × 10−15). Suppression of proteotoxicity in these three genetic models of DR was comparable in magnitude to BD. Therefore, we conclude that both genetic and environmental models of DR confer a substantial protective effect against proteotoxicity in C. elegans.
BD suppresses proteotoxicity through a mechanism distinct from IIS
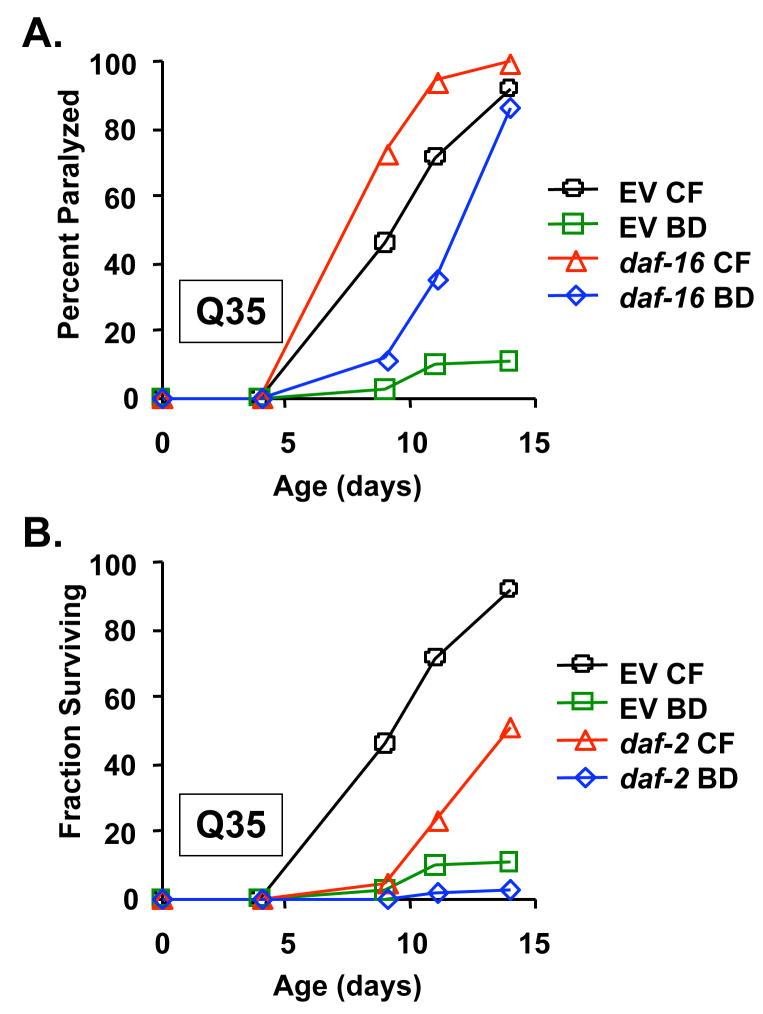
IIS has been previously found to modulate resistance to proteotoxicity in both Aβ42 and Q35YFP animals (Hsu et al. 2003; Cohen et al. 2006). We therefore wished to determine the relationship, if any, between DR and IIS in these models. As previously reported for Aβ42 animals (Cohen et al. 2006), we observed that RNAi knock-down of the FOXO-family transcription factor, daf-16, accelerated paralysis in Q35YFP animals (Figure 4A; p = 8.0 × 10−3), while RNAi knock-down of the insulin-like receptor, daf-2, delayed paralysis (Figure 4B; p = 7.6 × 10−7). For these experiments, animals were maintained on RNAi bacteria from egg until the second day of adulthood, then transferred to either empty vector RNAi bacteria or BD. Interestingly, RNAi knock-down of daf-16 did not block the effectiveness of BD (Figure 4A; p = 4.5 × 10−9). When combined with RNAi knock-down of daf-2, BD conferred an even greater suppression of paralysis than was observed in BD animals fed empty vector RNAi (Figure 4B; p = 1.0 × 10−14). Identical epistasis interactions were observed between IIS and BD in Aβ42 animals (Supplemental Figure 3). These data recapitulate the genetic relationship between DR and IIS with respect to longevity: DR can increase life span by a mechanism that is independent of daf-16 and additive with mutation of daf-2 (Lakowski & Hekimi 1998; Houthoofd et al. 2003; Kaeberlein et al. 2006; Lee et al. 2006). Thus, BD and reduced IIS appear to modulate both longevity and proteotoxicity through distinct genetic pathways.
Figure 4. BD suppresses proteotoxicity by a mechanism distinct from insulin/IGF-1 like signaling.
(A) RNAi inhibition of daf-16 significantly accelerates paralysis in Q35YFP animals relative to growth on empty vector bacteria (8.0 × 10−3), but does not prevent suppression of paralysis by BD (p=4.5 × 10−9). (B) RNAi inhibition of daf-2 significantly reduces paralysis in Q35YFP animals relative to growth on EV bacteria (p=7.6 × 10−7). BD further reduces paralysis of daf-2 RNAi treated animals (p=1.0 × 10−14).
DR protects against proteotoxicity through an hsf-1 dependent mechanism
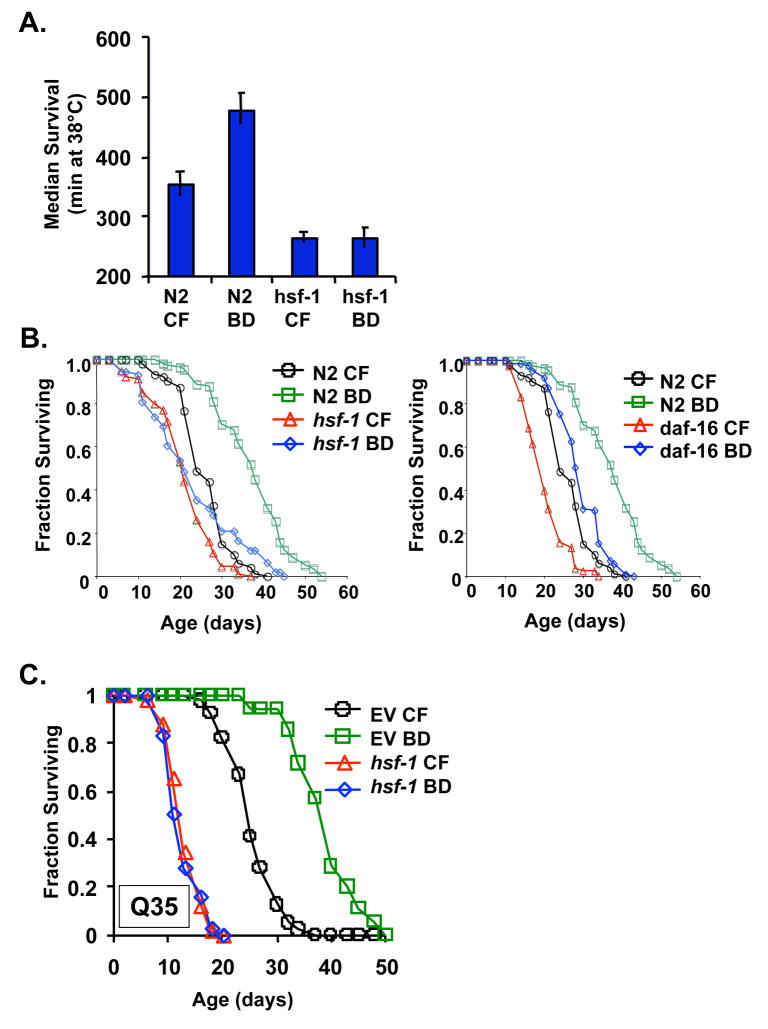
The heat shock transcription factor, hsf-1, regulates expression of many different heat-inducible target genes such as small heat shock proteins, and has been implicated in modulating both longevity (Garigan et al. 2002; Hsu et al. 2003; Morley & Morimoto 2004) and proteotoxicity (Hsu et al. 2003; Cohen et al. 2006). In order to determine whether hsf-1 is required for phenotypes associated with DR, we subjected hsf-1(sy441) animals to BD and monitored thermotolerance and longevity (Kaeberlein et al. 2006; Lee et al. 2006). The hsf-1(sy441) allele contains a point mutation in the hsf-1 transactivation domain that prevents induction of hsf-1 target genes in response to heat stress (Hajdu-Cronin et al. 2004). Unlike the case in N2 animals, BD failed to result in enhanced thermotolerance in hsf-1(sy441) animals (Figure 5A). The effect of BD on the longevity of hsf-1(sy441) animals was complex: BD failed to significantly increase median life span (p = 0.34); however, maximum life span appeared to be increased (Figure 5B). The inability of BD to increase the median life span of hsf-1(sy441) animals is notably different than the increase in both median and maximum life span of daf-16(mu86) animals in response to BD (Figure 5B; p = 3.9 × 10−19).
Figure 5. Heat shock factor 1 is required for enhanced thermotolerance and increased life span in response to BD.
(A) Animals carrying a mutant allele of hsf-1, hsf-1(sy441), fail to show increased survival at 38°C when subjected to BD. Error bars are standard error of the mean. (B) BD fails to increase median life span of hsf-1(sy441) animals (p=0.34) but significantly increases the life span of daf-16(mu86) animals (C) RNAi inhibition of hsf-1 prevents life span extension from BD.
There are two possibilities to explain the observation that hsf-1 mutation prevents median, but not maximum, life span extension from BD. First, it may indicate that life span extension from BD involves two different mechanisms, one of which acts through hsf-1 and one of which is hsf-1-independent. Alternatively, the residual hsf-1 function present in hsf-1(sy441) animals may be sufficient to partially mediate BD-associated life span extension. Consistent with this latter possibility, RNAi knock-down of hsf-1 completely prevented both median and maximum life span extension by BD (Figure 5C). Thus, mutation of the hsf-1 transactivation domain prevents induction of thermotolerance and median life span extension in response to BD, and RNAi knock-down of hsf-1 completely abrogates median and maximum life span extension by BD.
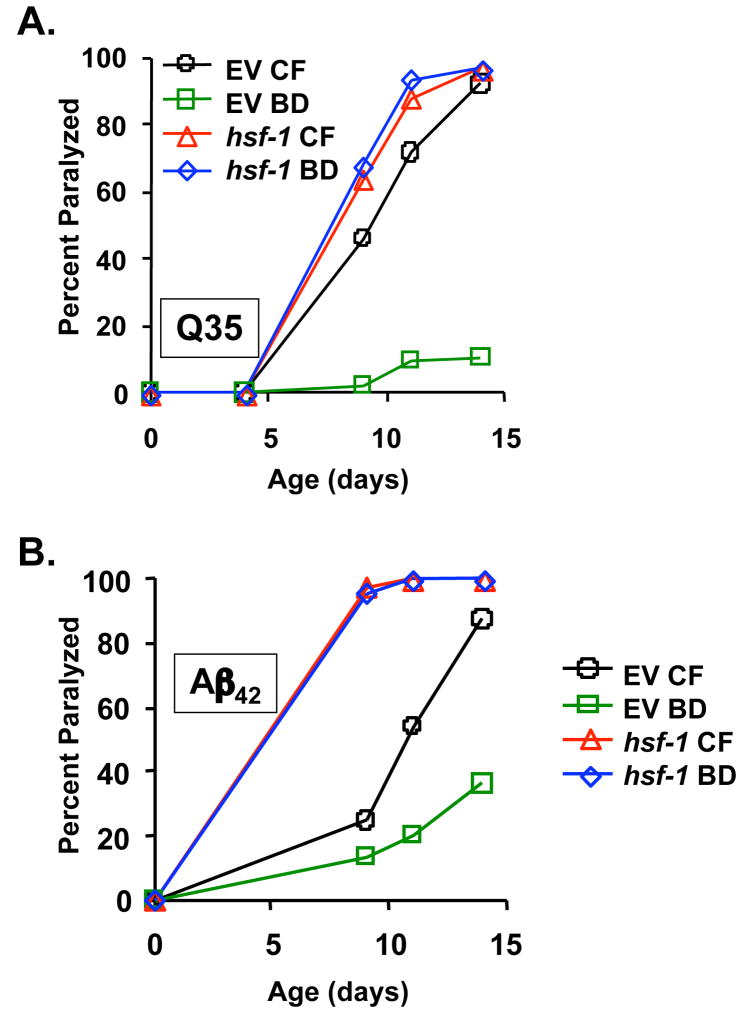
Finally, we asked whether RNAi knock-down of hsf-1 would affect suppression of proteotoxicity by BD. Q35YFP animals were maintained on hsf-1 RNAi from egg until the 2nd day of adulthood then transferred to either empty vector bacteria or BD. The onset of paralysis was accelerated when hsf-1 function was reduced by RNAi, relative to animals maintained on empty vector RNAi from egg (Figure 6A; p = 1.1 × 10−11). Strikingly, BD failed to significantly suppress paralysis in Q35YFP animals where hsf-1 was inhibited (p = 0.5). RNAi knock-down of hsf-1 had a similar effect on paralysis in Aβ42 animals (Fig 6b; p = 0.1). Thus, we conclude that hsf-1 activity is required both for life span extension and for the general suppression of proteotoxicity associated with BD.
Figure 6. Heat shock factor 1 is required for suppression of proteotoxicity in response to BD.
(A) BD significantly reduces paralysis of Q35YFP animals grown on empty vector bacteria (p=1.1 × 10−11), but does not reduce paralysis of Q35YFP animals grown on hsf-1 RNAi (p=0.51). (B) BD significantly reduces paralysis of Aβ42 animals grown on empty vector bacteria (p=8.9 × 10−14), but does not reduce paralysis of Aβ42 animals grown on hsf-1 RNAi (p=0.1).
DISCUSSION
Relationship between DR, IIS, and hsf-1
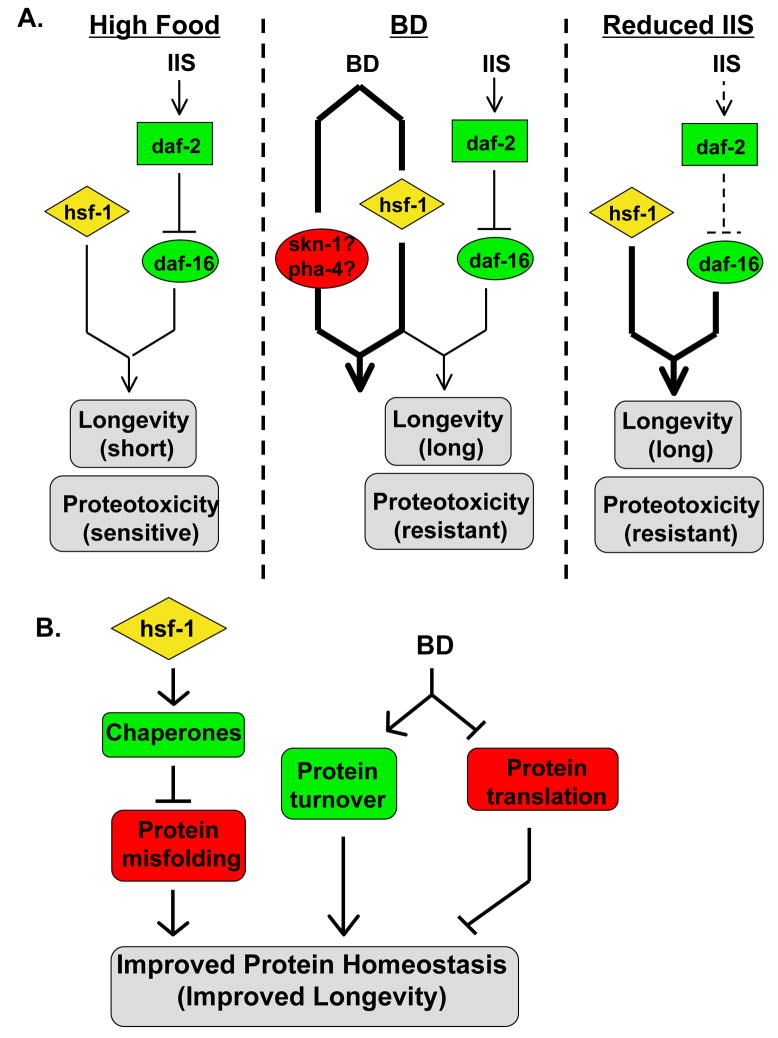
DR and reduced IIS result in several shared phenotypes, including enhanced thermotolerance, enhanced resistance to proteotoxicity, and increased longevity. In the case of reduced IIS, each of these phenotypes is known to be dependent on both hsf-1 and daf-16, which function to co-regulate the expression of a subset of IIS-responsive genes (Hsu et al. 2003; Murphy et al. 2003). In contrast, the effects of DR require hsf-1, but daf-16 is expendable. A model consistent with these observations is one in which BD promotes the activity of a transcription factor which functions in a manner similar to daf-16 to regulate target genes co-operatively with hsf-1 (Figure 7A). In this model, BD would not act solely by inducing hsf-1 activity or expression, but hsf-1 would function together with another transcription factor. Thus, hsf-1 activity would be necessary, but not sufficient, to recapitulate many of the phenotypes associated with BD, consistent with the observation that daf-16 is required for life span extension from overexpression of hsf-1 in animals fed a normal bacterial diet (Hsu et al. 2003; Morley & Morimoto 2004).
Figure 7. Models for the role of hsf-1 in determining longevity and sensitivity to proteotoxicity.
(A). The effect of BD is mediated by hsf-1. Under conditions of abundant food, the dietary restriction pathway is inactive and hsf-1 activity and insulin/IGF-1-like signaling (IIS) are at normal levels. This leads to short life span and sensitivity to proteotoxicity. Under DR conditions, hsf-1 is activated along with other DR-responsive proteins (perhaps skn-1 and pha-4) to increase life span and suppress proteotoxicity. In mutants with reduced IIS, the DR pathway is inactive, but life span and resistance to proteotoxicity are increased through activation of daf-16. (B) BD and hsf-1 function in parallel to alter protein homeostasis. hsf-1 activity upregulates chaperone activity, which decreases the amount of misfolded proteins in vivo. At the same time, BD might both increase the rate of protein turnover and decrease the synthesis of new protein. These three factors would combine to improve protein homeostasis, decreasing the rate of aging.
Recently, the daf-16 homolog pha-4 was found to be required for life span extension in eat-2 mutants (Panowski et al. 2007). Expression of pha-4 is normally low during adulthood, but is increased in eat-2 mutants (Panowski et al. 2007). Unlike hsf-1 (Hsu et al. 2003), however, overexpression of pha-4 is not sufficient to increase life span in control fed animals (Panowski et al. 2007). Thus, it could be that in well-fed animals hsf-1 activity is limiting for longevity, but in response to nutrient deprivation both hsf-1 and pha-4 are activated and promote longevity. Since skn-1 is also required for life span extension from DR (Bishop & Guarente 2007), it may be the case that hsf-1 and skn-1 act to co-regulate genes important for DR or that skn-1 acts upstream of hsf-1 and pha-4 in response to DR. Similar to the case for pha-4, increased expression of skn-1 is not sufficient to increase the life span of control fed animals (Bishop & Guarente 2007). Skn-1 is important for regulating gene expression in response to oxidative stress (An & Blackwell 2003; Inoue et al. 2005), but no role for pha-4 or skn-1 in modulating the response to proteotoxic stress has been described.
An alternative, but not mutually exclusive, possibility is that BD and hsf-1 function in parallel to promote longevity by improving protein homeostasis (Figure 7B). This hypothesis is attractive, since hsf-1 is known to regulate expression of chaperones, and BD could contribute to protein homeostasis by enhancing protein degradation or by modulating mRNA translation (or both). In C. elegans, starvation causes rapid turnover of muscle proteins and induces autophagy (Fostel et al. 2003; Jia & Levine 2007), suggesting that turnover of damaged proteins is likely to be enhanced by BD. This model also explains that apparent correlation between longevity and resistance to proteotoxicity. For example, a recent RNAi screen identified several genes that play a role in both longevity determination and the formation of polyglutamine aggregates (Curran & Ruvkun 2007). We have also observed that RNAi of several previously reported longevity factors (Dillin et al. 2002; Lee et al. 2003; Hamilton et al. 2005; Hansen et al. 2005; Hansen et al. 2007; Pan et al. 2007) also suppresses paralysis in Q35YFP and Aβ42 animals (KAS, CD, DC and MK, unpublished data). Further studies will be necessary to clarify the precise relationship between hsf-1 and DR, and the importance of protein homeostasis as a determining factor for longevity.
Conclusion
The data presented here demonstrate a novel and previously unsuspected role for hsf-1 as a key mediator of DR. This finding is noteworthy, as hsf-1 orthologs are highly conserved from yeast to humans, as are many of the known hsf-1 targets. Although it remains unclear whether hsf-1 is activated by DR in mammals, DR enhances Hsf1 binding activity in aging rats (Heydari et al. 1996), indicating that the link between DR and hsf-1 activity may be conserved. Activation of hsf-1 or specific hsf-1 target genes may prove therapeutic toward proteotoxic insults leading to neurodegeneration and other age-associated pathologies in people and may also mimic the longevity enhancing effects of DR.
EXPERIMENTAL PROCEDURES
Strains and genetics
Standard procedures for C. elegans strain maintenance and manipulation were used. Experiments were conducted at 20°C unless otherwise noted in the text. Nematode strains were obtained from the Caenorhabditis Genetics Center, or the laboratories of Dr. Richard Morimoto (Northwestern University, Chicago, IL), Dr. Chris Link (University of Colorado, Boulder, CO) or Dr. Jim Thomas (University of Washington, Seattle, WA). The strains used in this study are listed in Supplemental Table 1.
Bacterial Food Deprivation
BD experiments were carried out essentially as described previously (Kaeberlein et al. 2006). In brief, adult hermaphrodites were allowed to lay eggs on nematode growth medium (Youngman et al.) containing UV-killed E. coli OP50. At L4, worms were transferred to fresh NGM + UV-killed OP50 supplemented with 50 μM 5-fluorodeoxyuridine (FUDR) to prevent further eggs from hatching. Unless otherwise stated, worms were then transferred to experimental media at the 2nd day of adulthood: NGM + UV-killed OP50 + 50μM FUDR (control fed) or NGM + 50μM FUDR (Parker et al.). For the experiment shown in Figure 1C, adult worms were transferred to experimental media on the days indicated in the figure legend. OP50 were UV-killed by irradiation with a Stratagene UV Stratalinker 2400 on maximal energy setting. UV-killing was periodically verified by streaking UV-irradiated bacteria onto LB plates and confirming absence of growth.
RNA interference combined with BD
RNA interference (RNAi) experiments were conducted using feeding protocols according to standard procedures. Unless otherwise stated, RNAi feeding strains were obtained from the Ahringer RNAi library (Kamath et al. 2003); library clones were sequenced for verification. RNAi plates consisted of NGM supplemented with 1mM β-D-isothiogalactopyranoside (IPTG) and 25μg/ml carbenicillin. Unless otherwise indicated, worms were raised on RNAi bacteria from egg. At L4, they were then transferred to plates containing freshly-seeded RNAi bacteria plus 50μM FUDR. To combine RNAi with BD, worms were maintained on the indicated RNAi bacteria until the 2nd day of adulthood, at which point they were transferred to experimental media: RNAi plates + 50μM FUDR seeded with empty vector control bacteria (control fed) or RNAi plates + 50μM FUDR (Parker et al.). The daf-2 RNAi construct used for these studies was provided to us by J. McElwee.
Paralysis and life span quantification
The paralysis of worms expressing any of the indicated transgenes (Q35-YFP, Aβ1-42, or GFP::degron) was determined by visual analysis. Worms were scored as paralyzed if they were unable to make forward progress on the NGM surface in response to both plate-tapping and tail-prodding. If paralyzed, worms were scored as alive if nose-tapping resulted in nose movement. If no nose movement could be seen, worms were scored as dead.
Thermotolerance
Animals were maintained at 20°C on either control fed or BD media as described until the 6th day of adulthood (4 days without food for the BD group). The morning of the 6th day of adulthood, plates (each containing approximately 20 animals) were transferred to 38°C. Viability was determined periodically by removing one plate at a time and assaying for movement in response to nose-prodding.
Fluorescence microscopy and quantification of Q35YFP aggregates
Movie files were generated using Canon Powershot S31S digital camera connected to the eyepiece of a Zeiss SteREO Lumar.V12 microscope equipped with a broad-range GFP filter. Still GFP images were obtained with a Biorad MRC-1024 LSCM (Hercules, CA). This package included a computer assisted Zeiss inverted microscope and 15mW Krypton/Argon laser (ILT laser, Aachen Germany) emitting 3 three excitation lines at 488 nm, 568 nm, and 647nm. The 488 nm line was used to excite the green fluorescent protein. The emitted green fluorescence was filtered through a 522 nm DF 32 band pass filter. Unless otherwise stated, a 10x objective lens was used for fluorescence measurements. Identical settings were used for all worms analyzed (laser power at 1%, gain @1500, Iris@4.0 uM, scan time@488 lines/sec). The worms were paralyzed with 25mM sodium azide and placed on a glass slide under a cover slip to flatten the worm somewhat and optimize viewing. The microscope was focused at the position of maximum fluorescence for each worm. The images were analyzed using Image J 1.341.5.0_07 software [ available from NIH website (http://rsb.info.nih.gov/ij/java)] and a Macintosh computer with OS X operating system. The “count objects” command of Image J was used to count the number of fluorescent spots in the images. A constant minimum intensity threshold was set at 75/255 pixels, and a minimum spot size of 3 pixels/spot were used for all worms analyzed.
Quantification of Q35YFP expression by Immunoblot Analysis
Worms were prepared for immunoblot analysis using the control fed and BD protocols described above. At day 9 of adulthood, 1000 worms per group were harvested in 1ml M9 buffer and concentrated by low speed centrifugation. Worms were washed in M9 twice, then resuspended in PBS at a concentration of 1 worm per microliter buffer. They were homogenized in a glass homogenizer and spun at 3000rpm for 3 minutes; the supernatant was collected as “total lysate.” Samples were subjected to SDS PAGE analysis followed by immunoblot, according to standard protocols. Q35-YFP was detected with a GFP-specific antibody (Roche Applied Science, Indiana, USA); protein loading was controlled for using a β-actin specific antibody (Abcam, Massachusetts, USA). The relative abundance of Q35YFP protein and actin was determined by quantifying the band intensities after probing with the GFP-specific antibody or the β-actin specific antibody, respectively. When normalized to actin, the relative abundance of Q35YFP in control fed to BD animals was 0.96 ± 0.1, based on three biological replicates (independent experiments and independent blots).
Statistical analysis and replication
A Wilcoxon Rank-Sum test (MATLAB ‘ranksum’ function; Mann-Whitney U Test) was used to generate p-values to determine statistical significance for life span and paralysis assays. This test is a nonparametric comparison of medians between two groups. Each experiment was repeated at least three times with similar results.
Supplementary Material
Acknowledgments
We would like to thank the R. Morimoto and C. Link for providing the transgenic C. elegans strains used in this study. We would also like to thank J. McElwee and J. Thomas for helpful discussion and shared reagents. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. This work was conducted while MK was an AFAR Research Grant recipient. This work was also supported by a grant from The Ellison Medical Foundation to MK and BKK. KAS is supported by a post-doctoral fellowship from the Hereditary Disease Foundation. EDS was supported by NIH Training Grant P30 AG013280.
References
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bonini NM, La Spada AR. Silencing polyglutamine degeneration with RNAi. Neuron. 2005;48:715–718. doi: 10.1016/j.neuron.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Zala D, Humbert S, Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull HR, Morley JF, Morimoto RI. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol. 2007;594:167–189. doi: 10.1007/978-0-387-39975-1_15. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing Activities Protect Against Age Onset Proteotoxicity. Science. 2006 doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci U S A. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostel JL, Benner Coste L, Jacobson LA. Degradation of transgene-coded and endogenous proteins in the muscles of Caenorhabditis elegans. Biochem Biophys Res Commun. 2003;312:173–177. doi: 10.1016/j.bbrc.2003.09.248. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988a;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol. 1988b;43:B102–109. doi: 10.1093/geronj/43.4.b102. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Heydari AR, You S, Takahashi R, Gutsmann A, Sarge KD, Richardson A. Effect of caloric restriction on the expression of heat shock protein 70 and the activation of heat shock transcription factor 1. Dev Genet. 1996;18:114–124. doi: 10.1002/(SICI)1520-6408(1996)18:2<114::AID-DVG4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Khan LA, Bauer PO, Miyazaki H, Lindenberg KS, Landwehrmeyer BG, Nukina N. Expanded polyglutamines impair synaptic transmission and ubiquitin-proteasome system in Caenorhabditis elegans. J Neurochem. 2006;98:576–587. doi: 10.1111/j.1471-4159.2006.03895.x. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Koo EH, Lansbury PT, Jr, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci U S A. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD. Transgenic invertebrate models of age-associated neurodegenerative diseases. Mech Ageing Dev. 2001;122:1639–1649. doi: 10.1016/s0047-6374(01)00291-3. [DOI] [PubMed] [Google Scholar]
- Link CD, Fonte V, Hiester B, Yerg J, Ferguson J, Csontos S, Silverman MA, Stein GH. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem. 2006;281:1808–1816. doi: 10.1074/jbc.M505581200. [DOI] [PubMed] [Google Scholar]
- Martin B, Mattson MP, Maudsley S. Caloric restriction and intermittent fasting: two potential diets for successful brain aging. Ageing Res Rev. 2006;5:332–353. doi: 10.1016/j.arr.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Baillie DL. Genetic Organization in CAENORHABDITIS ELEGANS: Fine-Structure Analysis of the unc-22 Gene. Genetics. 1979;91:95–103. doi: 10.1093/genetics/91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- Parker JA, Connolly JB, Wellington C, Hayden M, Dausset J, Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci U S A. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–169. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren JR, Braeckman BP. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans. Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Weindruch RH, Walford RL. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- Youngman LD, Park JY, Ames BN. Protein oxidation associated with aging is reduced by dietary restriction of protein or calories. Proc Natl Acad Sci, USA. 1992;89:9112–9116. doi: 10.1073/pnas.89.19.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.