Summary
IscR is an Fe-S protein that functions as a transcriptional regulator of Fe-S biogenesis and other Fe-S protein encoding genes in Escherichia coli. In this study, we investigated the requirement for the ligation of the [2Fe-2S] cluster of IscR to regulate a subset of IscR target promoters (PhyaA, PydiU, P napF, and PhybO) and defined the requirements for sequence-specific binding to the IscR target site in the hyaA promoter region. In contrast to previous results with the iscR promoter, we found that the Fe-S cluster is dispensable for IscR regulation of PhyaA, PydiU, PnapF, and PhybO, since IscR mutants containing alanine substitutions of the cysteine Fe-S ligands retained IscR-dependent regulation of these promoters in vivo. In vitro assays showed that both [2Fe-2S]-IscR and an IscR mutant lacking the cluster (IscR-C92A/C98A/C104A) bound the hya site with similar affinity, explaining why the mutant protein retained its ability to repress P hyaA in vivo. Characterization of the oligomeric state of IscR showed that both apo-IscR and [2Fe-2S]-IscR were dimers in solution, and four protomers of either form bound to the hya site. Also, binding of either apo- or [2Fe-2S]-IscR to the hya site showed cooperativity, suggesting that both forms interact similarly with the target site. Analysis of mutations in the hya site using DNA competition assays showed that apo-IscR most likely recognizes an imperfect palindrome within the hya promoter. Furthermore, the strength of apo-IscR binding to PsufA, PydiU, PnapF, and PhybO IscR sites correlated with the number of matches to the hya site bases shown to be important in the competition assay. Thus, our data indicated that, unexpectedly, apo-IscR is a site-specific DNA binding protein, and the role of apo-IscR needs to be considered in developing models for how IscR globally regulates transcription.
Keywords: DNA binding, gene regulation, Fe-S biogenesis
Introduction
IscR, a [2Fe-2S] cluster-containing transcription factor, acts as a repressor or activator to control the expression of more than 40 genes in Escherichia coli1,2. IscR was first discovered for its role in regulating expression of the Isc (iron sulfur cluster) biogenesis pathway (reviewed in 3,4), which is transcribed as an operon(iscRSUA-hscBA-fdx) that includes iscR1. The finding that IscR requires a functional Isc Fe-S biogenesis pathway to repress the iscR promoter (PiscR) linked IscR [2Fe-2S] cluster occupancy to its transcription function, suggesting that Fe-S biogenesis is controlled by a homeostatic mechanism directed by [2Fe-2S]-IscR1. Recent genome-wide transcription profiling data extended these initial observations to show that IscR also controls expression of a subset of anaerobic Fe-S containing enzymes (hydrogenase-1 [Hya], hydrogenase-2 [Hyb], and the periplasmic nitrate reductase [Nap]), in addition to other proteins with Fe-S biogenesis functions (YadR [now ErpA], YhgI [now NfuA] and the Suf pathway)2,5,6. Furthermore, these results also revealed two classes of IscR binding sites2, suggesting an unconventional mode of DNA binding. In this study, we address the DNA binding properties of IscR to one class of IscR binding sites.
The two classes of IscR binding sites were inferred from DNase I footprinting of 6 IscR-regulated promoters and were further supported by phylogenetic comparisons2. A 25 bp Type 1 site (ATASYYGACTRwwwYAGTCRRSTAT) was compiled from the IscR target sites in the promoter regions of iscR, yadR, and yhgI, whereas a 26 bp Type 2 site (AWARCCCYTSnGTTTGMnGKKKTKWA) was compiled from the IscR sites upstream of the promoters for hyaA, ydiU, and sufA2. For either Type 1 or Type 2 sites, IscR protects a region of ~28–30 bp, and each promoter contained a single site except for the iscR promoter that contained three binding sites2. The length of the footprint is longer than expected for most dimeric helix-turn-helix DNA binding proteins that recognize a single palindromic sequence, and neither site revealed a common symmetrical element. Since a monomer of IscR is only 17 kDa with a single predicted helix-turn-helix motif (Fig. 1), the interaction with two apparently different DNA elements was unexpected.
Figure 1.
Diagram of IscR showing the predicted helix-turn-helix motif (http://www.uniprot.org/uniprot/P0AGK8?id=P0AGK8) and a [2Fe-2S] cluster ligation region. Inset is the amino acid sequence of the [2Fe-2S] cluster ligation region with arrows marking the putative cysteine ligand residues.
Not only do the sequences vary between the two classes of IscR binding sites, there also appear to be differences in how these sites are regulated by IscR. While PiscR contains two Type 1 sites and requires [2Fe-2S]-IscR for its repression, PsufA contains a Type 2 site and does not require the Fe-S cluster for its activation1,7. This suggests that the type of IscR site specifies whether apo- and/or [2Fe-2S] IscR binds to the site. This property of IscR was surprising since other known Fe-S transcription factors (e.g. FNR, SoxR) have only one type of site that either does (FNR) or does not (SoxR) require the Fe-S cluster for DNA binding8,9. The proposed Fe-S ligands for IscR (Cys92, Cys98, and Cys104) are distal to the predicted DNA-binding site (Fig. 1) and upon anaerobic isolation, the cluster is partially reduced ([2Fe-2S]1+-IscR), but is readily oxidized in air to ([2Fe-2S]2+-IscR)1. Because IscR appears to require its Fe-S cluster for regulation of PiscR and not for PsufA, this raises the question of whether IscR requires its [2Fe-2S] cluster for regulation of other Type 2 promoters.
In this study, we address the requirement for ligation of the [2Fe-2S] cluster for IscR regulation of promoters containing a Type 2 site. We also explore the sequence specificity of IscR binding to the hya IscR binding site as a model for describing IscR interactions with Type 2 sequences since IscR shows the strongest interaction with the hya site in footprinting assays2. To investigate regulation by IscR in vivo, we analyzed promoter fusions to lacZ along with IscR mutants lacking the ligand residues for the [2Fe-2S] cluster. Furthermore, we used fluorescence anisotropy assays to generate DNA binding isotherms and to perform equilibrium competition experiments with a library of DNA mutations to dissect the nucleotides necessary for IscR binding to DNA. Finally, size exclusion chromatography and analytical ultracentrifugation were performed along with stoichiometry experiments to determine whether multiple oligomers of IscR bound the hya site. Our studies revealed unexpected DNA binding properties of IscR.
Results
The [2Fe-2S] cluster of IscR is dispensable for regulation of promoters containing Type 2 sites
Previous studies7 showed that IscR does not require its cluster to activate PsufA, which has a Type 2 IscR binding site similar to PydiU and PhyaA2. To determine whether this lack of a requirement for the Fe-S cluster was common to other Type 2 promoters, independent alanine substitutions of the predicted Fe-S cysteine ligands of IscR (C92, C98, and C104) were tested for their ability to regulate PydiU and PhyaA, in addition to PsufA. Strains containing IscR-C92A, IscR-C98A, or IscR-C104A showed ~2-fold higher levels of activation of PsufA-lacZ than did wild-type IscR (Fig. 2A) indicating that the Fe-S cluster is not required for the transcription activation of IscR, in agreement with previous findings7. The Fe-S cluster of IscR was also dispensable for activation of PydiU since IscR-C92A, IscR-C98A, and IscR-C104A showed no defect in activation of PydiU-lacZ (Fig. 2B). The dispensability of the cluster was not specific to promoters that are activated by IscR since IscR-dependent repression of PhyaA-lacZ was even greater (~3-fold) with the IscR Cys to Ala substitutions than with wild-type IscR (Fig. 2C).
Figure 2.

Expression levels of PsufA (Panel a), PydiU (Panel b), and PhyaA (Panel c) promoters fused to lacZ were determined in strains containing either wild-type IscR (white bars), IscR- (left hash bars), IscR-C92A (right hash bars), IscR-C98A (checked bars), IscR-C104A (horizontal line bars), or IscR-C92A/C98A/C104A (vertical line bars). Strains were grown under aerobic conditions in MOPS minimal media containing 0.2% glucose and were assayed for β-galactosidase activity per O.D.600 (Miller Units, y-axis). Error bars represent data from 3 isolates on a single day. Panel d: Total IscR levels were measured by Western blots and normalized to colony forming units (CFU). Error bars represent 3 samples on various days.
In addition, the small increase in activation of PsufA and repression of PhyaA could be explained by the ~2-fold increase in IscR protein levels in strains containing IscR-C92A, IscR-C98A, or IscR-C104A, as determined by quantitative Western blots using α-IscR antibody (Fig. 2D). The increase in IscR protein levels in the mutant strains likely arises from the increase in expression of the iscR promoter, since the Fe-S form of IscR is required to repress PiscR1. A strain containing alanine substitutions of all three IscR cysteine residues (IscR-C92A/C98A/C104A) also showed increased levels of IscR protein and similar Type 2 promoter expression as any of the individual mutants (Fig. 2). Also, IscR-C92A/C98A/C104A purified under aerobic conditions did not contain an Fe-S cluster (data not shown), nor did the single mutant, IscR-C92A, when purified under anaerobic conditions (data not shown). Taken together, these data suggest that ligation of the [2Fe-2S] cluster is dispensable for IscR-mediated regulation of promoters containing a Type 2 IscR binding site.
[2Fe-2S]-IscR and IscR-C92A/C98A/C104A show no significant differences in binding affinity to the hya site
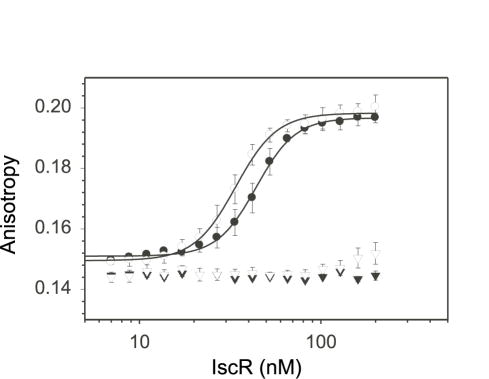
To address whether there is a preference for which form of IscR binds Type 2 sites, DNA binding by IscR-C92A/C98A/C104A and the as-purified wild-type protein was compared. Equilibrium binding assays were performed using a Texas Red labeled 30-mer dsDNA containing the 26 bp Type 2 site from the hyaA promoter as a representative Type 2 sequence2 (Table 1) and varying concentrations of either wild-type IscR (containing 60% [2Fe-2S] cluster) or IscR-C92A/C98A/C104A, which lacks the Fe-S cluster. We found that wild-type IscR and IscR-C92A/C98A/C104A bound similarly to the hya site with an apparent Kd of 34 ± 2 nM and 44 ± 3 nM, respectively, indicating that binding of IscR to the hya site is not dependent on cluster occupancy (Fig. 3). Neither form of IscR bound the DNA containing a randomized sequence (Fig. 3), which indicates that both preparations of IscR bind the hya site in a site-specific manner. While these data indicate that IscR lacking a cluster binds the hya site with high affinity, we could not precisely determine the affinity of the Fe-S only form because all of our preparations of wild-type IscR are a mixture of apo- and Fe-S-containing forms, despite attempts to remove the apo-protein. Nevertheless, experiments using wild-type IscR of varying cluster occupancies (42, 60, and 70%) indicate <1.4-fold difference in the apparent DNA binding affinity between the different occupied preparations (data not shown). In addition, no difference was observed in the apparent binding affinity of wild-type IscR assayed under anaerobic conditions, which contains some reduced [2Fe-2S]1+ cluster1, versus under aerobic conditions, which rapidly oxidizes to the [2Fe-2S]2+ cluster1. Since there was little difference in binding between preparations of the as-purified wild-type protein and the mutant lacking the cluster, these data argue that the Fe-S form has a similar affinity for the hya site as apo-IscR.
Table 1.
Identification of hya bases important for site-specific IscR binding
| Competitor | Sequence | IC50a |
|---|---|---|
| hyaA | 5′ ATAAATCCACACAGTTTGTATTGTTTTGTG | 29 |
| random | 5′ ATCATCCGTCTTATGTTAATTATAATGGTGb | >1000 |
|
| ||
| -53,-52 | 5′ ATAATCCCACACAGTTTGTATTGTTTTGTG | 33 |
| -49 | 5′ ATAAATCCTCACAGTTTGTATTGTTTTGTG | 44 |
| -46 | 5′ ATAAATCCACATAGTTTGTATTGTTTTGTG | 37 |
| -44,-43 | 5′ ATAAATCCACACATGTTGTATTGTTTTGTG | 89 |
| -42 | 5′ ATAAATCCACACAGTATGTATTGTTTTGTG | 73 |
| -41 | 5′ ATAAATCCACACAGTTAGTATTGTTTTGTG | 34 |
| -40 | 5′ ATAAATCCACACAGTTTCTATTGTTTTGTG | 39 |
| -36(T to A) | 5′ ATAAATCCACACAGTTTGTATAGTTTTGTG | 52 |
| -34/33 | 5′ ATAAATCCACACAGTTTGTATTGAATTGTG | 39 |
| -32,-30 | 5′ ATAAATCCACACAGTTTGTATTGTTATCTG | 18 |
| -54,-48,-45,-37 | 5′ ATATATCCAGACTGTTTGTAATGTTTTGTG | 80 |
| strong compiled | ATAAATCCACACAGTTTGTATTGTTTTGTGc | |
|
| ||
| -55 | 5′ ATCAATCCACACAGTTTGTATTGTTTTGTG | 394 |
| -51 | 5′ ATAAATGCACACAGTTTGTATTGTTTTGTG | 400 |
| -50(C to G) | 5′ ATAAATCGACACAGTTTGTATTGTTTTGTG | >1000 |
| -50(C to A) | 5′ ATAAATCAACACAGTTTGTATTGTTTTGTG | 268 |
| -50(C to T) | 5′ ATAAATCTACACAGTTTGTATTGTTTTGTG | >1000 |
| -47 | 5′ ATAAATCCACTCAGTTTGTATTGTTTTGTG | 190 |
| -39 | 5′ ATAAATCCACACAGTTTGAATTGTTTTGTG | 199 |
| -38 | 5′ ATAAATCCACACAGTTTGTTTTGTTTTGTG | >1000 |
| -35 | 5′ ATAAATCCACACAGTTTGTATTTTTTTGTG | 505 |
| -31 | 5′ ATAAATCCACACAGTTTGTATTGTTTGGTG | 263 |
| intermediate & weak compiled | ATAAATCCACACAGTTTGTATTGTTTTGTG | |
|
| ||
| -44,-42,-41,-40 | 5′ ATAAATCCACACATTGGTTATTGTTTTGTG | 785 |
| -42,-41 | 5′ ATAAATCCACACAGTGGGTATTGTTTTGTG | 300 |
|
| ||
| -36(T to G) | 5′ ATAAATCCACACAGTTTGTATGGTTTTGTG | 16 |
| -36(T to C) | 5′ ATAAATCCACACAGTTTGTATCGTTTTGTG | 134 |
| -48(C to T) | 5′ ATAAATCCATACAGTTTGTATTGTTTTGTG | 48 |
|
| ||
| Binding Motif | 5′ AxxxCCxxAxxxXxxxTAxGGxxxTd | |
|
| ||
| Palindrome1 | 5′ ATAAATACATACAGTTTGTATGTATTTGTG | 202 |
| Palindrome2 | 5′ ATAAATCCATCCAGTTTGGATGGATTTGTG | >1000 |
| Palindrome1–2 | 5′ ATAAATCCATACAGTTTGTATGGATTTGTG | ~10e |
IC50 values as determined through curve fitting the data using the equation for IC50 in KaleidaGraph. Representative graph in Figure 6.
Bold bases are mutated.
Italic bases are compiled from the individual competitors within that section.
Underlined bases indicate symmetry.
The IC50 for Palindrome 1–2 is an approximate value due to the limits of the curve fit.
Figure 3.
DNA binding isotherms of as-purified wild-type IscR (open symbols) and IscR-C92A/C98A/C104A (closed symbols). Protein preparations (7 to 200 nM) were incubated with either 5 nM fluorescently-labeled DNA containing the IscR hya site (circles) or 5 nM randomized site (triangles) in 40 mM Tris-Cl (pH 7.9) and 150 mM KCl. Protein binding to DNA is indicated by changes in anisotropy values (y-axis). Error bars represent assays done 3 separate days.
Surprisingly, the DNA binding isotherms yielded a sigmodial saturation curve when plotted as a linear function of IscR concentration (data not shown), indicating that binding of IscR to the 30-mer hya dsDNA sequence was cooperative. Plotting IscR concentration on a log scale (Fig. 3) and curve fitting the isotherms with the Hill equation (Equation 1) indicated that wild-type IscR and IscR-C92A/C98A/C104A had Hill coefficients of 3.8, and 4.0, respectively, establishing that both forms of IscR bind cooperatively to this sequence. Thus, taken together, these data indicate that IscR containing the [2Fe-2S] cluster interacts similarly with the hya site as the mutant lacking the cluster.
IscR is mainly a dimer in solution
Since cooperative interactions are typically exerted through subunit interactions of oligomeric proteins, we investigated whether IscR was a dimer in solution, as is often the case for prokaryotic DNA binding proteins. Analysis of either apo-IscR (generated by treatment with 2,2′-dipyridyl) or wild-type IscR (35% occupied with [2Fe-2S] cluster) by analytical size exclusion chromatography yielded an apparent molecular mass of 45 ± 2 kDa (Fig. 4A), indicating that the Fe-S cluster does not affect the oligomeric state of IscR. Consistent with this observation, there was no difference in the size exclusion elution profiles between the IscR-C92A/C98A/C104A mutant and wild-type IscR (data not shown). Also, the elution profile of the [2Fe-2S] cluster containing fractions of wild-type IscR, monitored at 420 nm, was similar to the protein fractions, monitored at 280 nm (data not shown) providing additional evidence that the [2Fe-2S] cluster does not alter the oligomeric state of IscR. However, because the apparent mass (45 kDa) was greater than that expected for a dimer (35 kDa), but less than a trimer (52 kDa), these data suggested that IscR is likely a dimer with a non-globular shape, since elution by size exclusion chromatography is dependent on both size and shape. Alternatively, it is possible that IscR is in rapid equilibrium between oligomeric states, resulting in a population of intermediate size, although we did not observe any concentration dependence of the elution profiles (data not shown).
Figure 4.
Panel a) Determination of molecular mass of IscR by size exclusion chromatography. The molecular mass (kDa) of the standards vitamin B12 (1.35 kDa), cytochrome c (12 kDa), myoglobin (17 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), gamma globin (158 kDa), and thyroglobulin (670 kDa) were plotted as a function of elution volume divided by void volume (Ve/V0). The Ve/V0 of 240 μM [2Fe-2S]- and 970 μM apo-IscR are marked with a dashed line. Panel b) Determination of the molecular mass of IscR by analytical ultracentrifugation. A global fit model (lines) with IscR in equilibrium between a dimer and tetramer was generated from sedimentation equilibrium assays. Every other data point for the 12 (circles) and 49 (squares) μM IscR-C92A/C98A/C104A at 9000 and 19200 rpm in 40 mM Tris-Cl (pH 7.4 at 25°C) and 150 mM KCl was plotted as the natural log of the absorbance as a function of radial position (cm2).
The oligomeric state of IscR-C92A/C98A/C104A was also examined with analytical ultracentrifugation sedimentation equilibrium, which allows direct measurement of the molecular mass. The analytical ultracentrifuge data showed that IscR-C92A/C98A/C104A is largely dimeric with an ill-defined larger species, which may be a tetramer (Fig. 4B). Similar results were observed with wild-type (35% occupied with [2Fe-2S] cluster) IscR (data not shown). If the larger species is modeled as a tetramer, the equilibrium constant between dimers and tetramers is approximately 35 μM, which is well-above physiological levels of IscR (~4.5 μM dimers, Figure 1D). Taken together with the size exclusion data, IscR appears to be mainly a dimer in solution.
Stoichiometry of wild-type IscR and IscR-C92A/C98A/C014A
To determine the number of IscR dimers binding to the hya site, DNA binding assays were performed using a five-fold excess of DNA over the apparent Kd of the IscR:DNA interaction. In the presence of increasing amounts of IscR, binding increased as a linear function because proteins bind DNA in a 1:1 ratio when the DNA and protein are present at concentrations well over the apparent dissociation constant10,11. At the concentration of IscR in which all of the DNA in the assay is bound (indicated by the plateau), the stoichiometry of IscR binding to the DNA sequence was calculated to be 4.1 ± 0.4 wild-type IscR protomers per hya site DNA and 4.2 ± 0.2 IscR-C92A/C98A/C104A protomers per hya site DNA (Fig. 5). Assuming that all of the protein in the preparation is active in binding, the simplest explanation to account for the solution behavior of IscR is that two IscR dimers are cooperatively binding the hya site.
Figure 5.
Stoichiometry of IscR DNA binding generated from fluorescence anisotropy assays of as-purified wild-type IscR (open symbols) and IscR-C92A/C98A/C104A (closed symbols) (269 to 1000 nM) with 250 nM Texas Red labeled-hya site DNA in 40 mM Tris-Cl (pH 7.9) and 150 mM KCl. Fraction bound (y-axis) was calculated from the anisotropy and is a function of [IscR monomers] divided by [250 nM DNA] (x-axis). Error bars represent assays done 3 separate days.
Determining the nucleotides within the hya site critical for IscR binding
Inspection of the logo for Type 2 sites does not readily reveal any single or overlapping symmetrical DNA binding elements (Fig. 8 and ref 2). However, there appears to be a partial palindrome in the hya site, which may play a role in IscR binding. Thus, to dissect the nucleotides required for IscR site-specific binding, a library of 30-mer dsDNA molecules containing one to four nucleotide mutations within the hya site was examined for their effects on IscR binding (Table 1). Any competitors with multiple mutations that showed a defect in binding (data not shown) were further analyzed with single mutations to clarify the role of each nucleotide. For this analysis, IscR-C92A/C98A/C104A was used since it bound the hya site as well as the as-purified wild-type protein but lacks the additional variable of the Fe-S occupancy. The binding of IscR-C92A/C98A/C104A to the mutated sites was measured in vitro by performing equilibrium DNA competition assays using the fluorescence-labeled wild-type hya site and varying concentrations of unlabeled mutant competitor DNA to determine of the amount of competitor DNA that decreased binding by 50% (IC50).
Figure 8.
Panel a) Comparison of IscR binding sites to the IscR binding motif with the original \ Type 2 sequence logo as a reference2. The number listed after the promoter name is the most upstream base of the IscR site relative to the transcription start site (P3 of PnapF is -121 bp relative to P1 in PnapF2). The IC50 values for each promoter are listed after the sequence. (underlined bases, symmetrical; central base, lowercase; important bases, bold; critical bases, italic) Panel b) Titration curves generated from the competition assays with 55 nM IscR-C92A/C98A/C104A in 40 mM Tris-Cl (pH 7.9), 150 mM KCl, and 5 nM labeled-hya site. The concentration of unlabeled-wild-type hya site competitor (closed circles), sufA site competitor (closed inverted triangles), ydiU site competitor (open inverted triangles), napF site competitor (closed squares), hybO site competitor (open squares), or randomized competitor (open circles) was varied from 8 to 1000 nM. Error bars represent assays done 3 separate days.
The DNA competitors were then classified into three groups: strong, intermediate, or weak competitors (Table 1, representative binding curves are shown in Fig. 6). Strong competitors showed an apparent IC50 <100 nM, which is within 3-fold of the unlabeled wild-type hya site (29 nM). Thus, the mutated bases of these competitors are the least important for IscR binding. These bases are -54A, -53A, -52T, -49A, -48C, -46C, -45A, -44G, -43T, -42T, -41T, -40G, -37T, -36T, -34T, -33T, -32T, and -30G relative to the transcription start site. Intermediate competitors reduced IscR binding between 4- and 40-fold (apparent IC50 >100 nM and <1000 nM), and include bases -55A, -51C, -47A, -39T, -35G, and -31T. Weak competitors (apparent IC50 >1000 nM) are those mutations that showed the greatest defect in DNA binding since competition was observed only at very high competitor DNA concentrations and include bases -50C and -38A. Additional substitutions of -50C (A or T) gave similar defects in binding as -50G, consistent with a critical role for this particular nucleotide (Table 1). The nucleotides that \resulted in the largest defect in DNA binding when mutated (apparent IC50 >100 nM; -55A, -51C, -50C, -47A, -39T, -38A, -35G, and -31T) were contained in a partly palindromic sequence (-55AAATCCACACAGtTTGTATTGTTTTG-30; bold, key nucleotides; underlined, palindrome; lowercase, center of symmetry). Mutation of positions that are symmetrical to one another generally gave a similar defect in binding, suggesting that IscR recognizes a palindrome. Surprisingly, the two nucleotides with the largest defect in binding (-50C and -38A; italics in sequence) are not part of the symmetrical element.
Figure 6.
Representative titration curves generated from the DNA competition assays. Fraction of 5 nM labeled-hya site DNA bound by 55 nM IscR-C92A/C98A/C104A in 40 mM Tris-Cl (pH 7.9), 150 mM KCl and in the presence of 8 to 1000 nM of either unlabeled-wild-type hya site competitor (closed circles), -55(A to C) mutant competitor (closed inverted triangles), or randomized competitor (open circles). Error bars represent assays done 3 separate days.
Mutation of -50C and -38A eliminate IscR regulation in vivo
We also examined the effect of representative mutations defective in DNA binding on PhyaA expression in vivo. Substitution of either -50(C to G) or -38(A to T) nearly eliminated repression by IscR-C92A/C98A/C104A since only ~2-fold repression was observed as compared to the typical ~70-fold repression observed for wild-type PhyaA (Fig. 7), consistent with the >25-fold defect in IscR binding to these mutant DNA sites in vitro. One of the multiple mutations from -44 to -40 (GTTTG to TTGGT) was an intermediate competitor in vitro (Table 1), and this mutation also reduced repression but not as much as either -50G or -38T, consistent with the more moderate effect on DNA binding (Table 1, Fig. 7). The defect in repression caused by these site mutations was not specific to the IscR mutant C92A/C98A/C104A since strains containing wild-type IscR also showed a similar increase in expression (Fig. 7). Neither the -50G, -38T, nor the -44 TTGGT-40 mutation had a significant effect on basal promoter function since lacZ expression in the ΔiscR strains was similar to those strains with wild-type PhyaA-lacZ (Fig. 7). Thus, for the positions examined, the in vitro competition data reflect the role of individual nucleotides in vivo.
Figure 7.
Expression levels of PhyaA containing mutations in the IscR binding site were determined in strains containing either wild-type IscR (white bars), IscR− (left hash bars), or IscR-C92A/C98A/C104A (right hash bars). Strains were grown under aerobic conditions in MOPS minimal media + 0.2% glucose. The β-galactosidase activity per O.D.600 (Miller Units, y-axis) produced from the PhyaA-lacZ was measured as previously described2. Error bars represent data from 3 isolates on a single day.
IscR binds to an imperfect palindrome within the hya promoter
Since the two most critical nucleotides, -50C and -38A, are not in the palindrome, we determined whether binding would increase by extending the symmetry to include these positions. A nucleotide symmetrical to -50C was created by substituting -36T to G, which increased IscR-C92A/C98A/C104A DNA binding ~2-fold, since less competitor DNA of the mutant (IC50 of 16 nM) was required to achieve the same amount of competition as the wild-type sequence (IC50 of 29 nM) (Table 1). The improvement in binding is dependent on the presence of the G at position -36T, since the -36A and -36C competitors had a 2- and 5-fold defect in binding, respectively (Table 1). However, changing -48C to T to improve the symmetry relative to -38A did not increase IscR-C92A/C98A/C104A binding (Table 1), suggesting that -38A is not part of the palindrome bound by IscR.
In view of the stoichiometry data of 4 subunits of IscR bound per DNA site (Fig 5), we also considered the possibility that the hya site might be composed of 2 imperfect overlapping palindromes to accommodate the binding of two IscR dimers and the large DNaseI footprint2. Two sequences were constructed to test whether symmetrical combinations of potential adjacent half-sites TCCA and TACA improved binding of IscR. Neither palindrome 1, containing two adjacent TACA half-sites, nor palindrome 2, containing two adjacent TCCA half-sites, bound IscR as well as the wild-type sequence, requiring 7 and 35 fold more DNA for 50% inhibition of binding, respectively (Table 1). In contrast, combination of both sites (TCCATACA; Palindrome1–2), which is more similar to the wild-type hya sequence, required less DNA than the wild-type hya site for 50% inhibition probably due to the -36T to G mutation. Taken together, these data suggest that, in addition to -38A, IscR can recognize key palindromic nucleotides within a larger palindrome (1AAATCCACACAGtTTGTATGGTTTT25; bold, key nucleotides; underlined, palindrome; lowercase, center of symmetry), but leaves open the question of how four subunits of IscR bind this site since this sequence has only one axis of symmetry. Nevertheless, we suggest that these nucleotides define a 9 bp IscR binding motif (1AxxxCCxxAxxxXxxxTAxGGxxxT25; Table 1).
Binding and regulation of additional IscR Type 2 promoters by clusterless IscR
Comparison of the Type 2 IscR sites from the sufA and ydiU promoter regions2 to the IscR binding motif (1AxxxCCxxAxxxXxxxTAxGGxxxT25; Table 1) shows that both the sufA and ydiU sites contain 5 of the 9 bases in this binding motif(Fig. 8A). Using these sequences as competitors in the binding assays showed that neither the sufA or the ydiU site competed as well as the hya site (Fig. 8B), consistent with the contribution of nucleotides within the motif in high affinity DNA binding.
Of the promoters shown to be directly regulated by IscR, only PnapF and PhybO were not classified as either a Type 1 or a Type 2 promoter2. Comparison of the IscR footprint at PnapF from Giel et al.2 with the IscR binding motif showed that the region contains 5 of the 9 nucleotides (Fig. 8A). Consistent with the low number of matches, the napF site has a similar competition curve as the sufA site (Fig. 8B). Inspection of the hybO regulatory region revealed a site from +2 to +27 matching 7 of the 9 nucleotides within the recognition motif (Fig. 8A), but this site was not observed in the IscR footprints12, which only visualized from −5 to −197 relative to the transcription start site. Competition of this putative hybO IscR site showed that the hybO site competed as well as the hya site (Fig. 8B), although the hybO site has 1 less nucleotide of the IscR binding motif than the hya site. β-galactosidase assays using the IscR mutant lacking the [2Fe-2S] cluster, IscR-C92A, showed that both PnapF and PhybO were repressed by the mutant 20- and 2.1-fold, respectively (Fig. 9). These data showed that the [2Fe-2S] cluster of IscR was dispensable for repression of PhybO and PnapF in vivo as found with the other Type 2 sites. Thus, a general feature of Type 2 IscR binding sites is the ability of either apo- or [2Fe-2S]-IscR to bind and regulate transcription.
Figure 9.
Expression levels of PnapF (Panel a) or PhybO (Panel b) promoters fused to lacZ were determined in strains containing either wild-type IscR (white bars), IscR- (left hash bars), or IscR-C92A (right hash bars). These strains were grown under aerobic conditions in minimal media, and β-galactosidase activity per O.D.600 (Miller Units, y-axis) was determined. Error bars represent data from 3 isolates on a single day.
Discussion
Characterization of IscR has expanded from its function in feedback regulation of its own promoter1, to that of a global regulator with two different binding sites2, and now to the surprising finding that both [2Fe-2S]- and apo-IscR can bind Type 2 sites in a sequence-specific manner. We also present a model in which IscR has the novel property of differentially regulating Type 1 and Type 2 promoters based on the amounts of [2Fe-2S]-IscR and apo-IscR, which we propose reflects the Fe-S cluster status of cells.
Characteristics of a Type 2 site
Previous transcription profiling and DNase I footprinting results2 indicated that IscR binding sites could be subdivided into 2 types based on sequence similarity. In the present study, detailed analysis of a representative Type 2 site from Phya, which showed the strongest binding, revealed important features of Type 2 sites and allowed us to propose a binding motif. First, the key nucleotides in the Type 2 IscR binding motif were only a subset of the improved palindrome (1AAATCCACACAGtTTGTATGGTTTT25, bold, binding motif; underline, palindrome; Table 1). Second, only one of the most critical bases (italic), -50C, was part of the palindrome. The other critical base, -38A, does not appear to be part of the palindrome, but it may still play a role in site-specific binding.
Although not all the bases in the IscR binding motif (1AxxxCCxxAxxxXxxxTAxGGxxxT25, Table 1) are conserved among the Type 2 promoters in E. coli, there appears to be a correlation in the relative strength of IscR binding and the number of matches to the motif. The sufA, ydiU, and napF sites, which contain the least number of matches to the IscR binding motif (5 of 9), did not bind as well as the hya site, which contains 8 of the 9 motif bases (Fig. 8). In comparison, the hybO site, which contains 7 of the 9 motif bases, bound as well as the hya site (Fig. 8). All of these Type 2 sites contain the critical -50C of the hya site, but only the hya site is missing the G symmetrical to -50C, which in the hya site is position -36. Thus, the correlation between binding strength and the number of matches to the motif provides additional evidence for the function of this sequence motif in IscR DNA binding. Identifying additional IscR sites using genomic analysis should provide further support for the role of this sequence element.
Comparison of the Type 2 binding motif to the Type 1 binding sites reveals that the iscR1 site has 3 of the 9 bases, the iscR2 and yhgI sites have 4 of the 9 bases, and only the yadR site has 5 of the 9 bases2. Although the yadR site has 5 matches to the Type 2 binding motif, it is missing the C and A that in the hya site (-50C and -38A) were critical for binding. Furthermore, this same A is absent in all other Type 1 sites. Further analysis shows that the Type 1 sites either contain the CC pair (X4CCX19) or the symmetrical GG pair (X19GGX4) of the Type 2 motif (X4CCX13GGX4). Thus, the presence of the CC and GG dinucleotides in addition to the A of position 18 in the motif may play a role in distinguishing the two types of sites and will be the subject of future work.
Comparison of the Type 2 binding motif to the Type 2 logo (Fig. 8a) proposed from the phylogenetic sequence conservation of three footprinted sites (hyaA, sufA, ydiU)2 shows some notable differences. For example, the overrepresentation of a string of T’s in the logo (Fig. 8a; positions 22–24) suggested a possible role in IscR binding. However, no DNA binding function of these T’s was indicated from our mutational analysis. Rather, it seems more likely that the T’s are overrepresentated in the logo because of the overlap with the -35 hexamer (TTGACA) of the sigma 70 binding site, which is located in a similar position relative to the IscR site in all three promoters used to generate the logo. In fact, PhyaA has two possible -35 hexamers, TTGTTT or TTGTGC, which may explain why two transcription start sites were observed in vitro. While the first T of the downstream predicted -35 hexamer corresponds to the T-rich region, the second T of the predicted upstream -35 hexamer (TTGTTT) replaces the first G of the GG dinucleotide. In comparison, both the sufA and the ydiU sites contain a G in that position because PsufA and PydiU predicted -35 elements are 3 and 2 bp, respectively, further downstream with respect to the IscR site. In contrast, for PnapF and PhybO, the location of the IscR binding site overlaps the transcription start site and thus lacks the stretch of T’s, perhaps explaining why PnapF and PhybO were not identified as containing a Type 2 site using phylogenetic footprinting2. Thus, genomic analysis of promoter regions for IscR binding sites should take into consideration the position of promoter elements.
Another surprising finding is that the IscR binding site in PhyaA, PsufA, and PydiU is located in a similar position (within 2–3 bp) with respect to the -35 hexamer of each promoter even though PhyaA is repressed by IscR, and PsufA and PydiU are activated by IscR. The ability of IscR to repress and activate from a similar location within the promoter may be due to the slight differences in spacing to the predicted -35 elements or the weak binding of IscR to the sufA and ydiU sites compared to the strong binding of IscR to the hya site. If the latter is the case, then IscR function may be reminiscent of OhrR13, which converts from a repressor to an activator by a decrease in binding affinity.
IscR is a member of the helix-turn-helix Rrf2 family of proteins
The Rrf2 family of transcription factors are widely distributed in bacteria, but overall have not been well studied. Rrf2 from Desulfovibrio vulgaris, the founding member of this family, was discovered for its role in expression of thehigh-molecular-mass cytochrome redox protein complex 14. Other members include RirA, an iron responsive regulator found in the symbiotic bacterium Rhizobium leguminosarum15, and NsrR, thenitric oxide sensor discovered in Nitrosomonas europaea16. Most Rrf2 family members contain three Cys residues in the C-terminal region (http://ca.expasy.org/cgi-bin/prosite-search-ac?PDOC01035), similar to IscR, suggesting that they may ligate an Fe-S cluster. Although the fourth ligand to the IscR Fe-S cluster is not known, variations in this ligand could explain the ability of different Rrf2 proteins to respond to specific signals (e.g. Fe levels, NO, or Fe-S cluster status).
Every member of the Rrf2 family contains a predicted N-terminal helix-turn-helix DNA-binding motif (http://ca.expasy.org/cgi-bin/prosite-search-ac?PDOC01035), but members have apparently evolved distinct DNA binding specificities reflecting differences in function of some family members. For example, NsrR is also found in E. coli along with IscR17, but NsrR only matches three amino acids (underlined) in the predicted helix-turn-helix of IscR: 28-LADISER-QGIS-LSYLEQLFSRLRK-51 (http://www.uniprot.org/uniprot/P0AGK8?id=P0AGK8). As expected from the low sequence similarity between the IscR and NsrR helix-turn-helix residues, the DNA binding motif for NsrR in γ-proteobacteria (gATGyATTTxAAATrCAtc)18 does not share obvious similarity with the Type 2 IscR binding motif (1AxxxCCxxAxxxXxxxTAxGGxxxT25; Table 1), and only one promoter, PnapF, is common to both the IscR and NsrR regulons 2,19. Thus, while protein structure prediction algorithms suggest that Rrf2 family members share a common structural organization, divergence at the protein sequence level may explain their ability to recognize distinct DNA target sites. However, no data is available to address whether any other Rrf2 family members have two types of target sites as found with IscR, and it is unclear how the single predicted helix-turn-helix in IscR is able to distinguish between Type 1 and Type 2 sites.
How do four subunits of IscR bind to the hya site DNA?
Since IscR is mainly a dimer in solution (Fig. 4), we were surprised to find that four protomers bound to the 30 mer hya DNA site (Fig. 5). This result suggested two possible oligomeric arrangements. First, one tetramer of IscR bound to a single operator DNA site, which would be similar to Lac repressor binding (reviewed in 20). However, the fact that Lac repressor is a tetramer in the absence of DNA21, Lac repressor binding to its operator is non-sigmoidal21, and its operator is shorter (~20 bp)22 with only 2 of the 4 protomers making specific contacts23, suggest that the Lac repressor may not be the best model for understanding IscR:DNA stoichiometry.
Rather, the properties of IscR observed in this study are more consistent with those of QacR, IdeR, and DtxR in which two dimers have been shown by X-ray crystallography and DNaseI footprinting to contact longer operator sites (~30 bp) that are composed of two overlapping binding sites24,25,26,27,28. As observed with IscR, all of these proteins bind their sites cooperatively24,26,29,30. Surprisingly, the cooperativity of DtxR, IdeR, and QacR binding to their DNA target does not arise from dimer-dimer interactions since the dimers bind on different faces of the DNA helix25,26,28. In addition, while QacR, DtxR, and IdeR bind symmetrical elements, each helix-turn-helix motif of QacR contacts different bases within each half site, resulting in asymmetric binding. Our analysis of hya site mutants indicate that IscR may bind DNA asymmetrically, similar to QacR. Further, because QacR DNA binding widens the major groove and undertwists the DNA, a mechanism was suggested in which binding of one dimer causes a conformational change in the DNA, which favors binding of a second dimer30. In summary, given the similar DNA binding properties of IscR to QacR, the simplest explanation to account for the data reported here is that two IscR dimers bind cooperatively to the hya DNA sequence. Additional experiments will be required to determine if cooperative binding of IscR results from a DNA induced conformational change or from dimer-dimer interactions.
Apo-IscR plays a role in regulating Type 2 promoters
While previous data have shown that apo-IscR can activate the Type 2 promoter, PsufA7, our data showed that other promoters containing a Type 2 IscR binding motif, PhyaA, PydiU, PnapF, and PhybO, were also regulated by apo-IscR (Fig. 2, 9). In addition, both [2Fe-2S]-IscR and apo-IscR bound the hya site with similar affinity in vitro, suggesting that both forms of IscR can regulate PhyaA and other Type 2 promoters in vivo. FNR, another Fe-S transcription factor in E. coli, only forms dimers and binds DNA in the presence of its oxygen-labile [4Fe-4S] cluster8,31. SoxR, a [2Fe-2S]-containing transcription activator in the MerR family, can bind its single target DNA site in the presence or absence of its Fe-S cluster. Because SoxR binds independently of its [2Fe-2S] cluster, it also represses transcription of its own promoter independently of its [2Fe-2S] cluster32. However, SoxR can only activate transcription from the divergent PsoxS when its [2Fe-2S] cluster is present and oxidized9. Although SoxR and IscR may be similar in their ability to function as transcription factors in the absence of their Fe-S clusters, SoxR only binds a single DNA site, whereas IscR has multiple targets containing either the Type 1 and Type 2 sites. Thus, IscR is unusual in its ability differentially bind its target sites depending on the presence or absence of its cluster.
Regulation of [2Fe-2S]-IscR and its impact on expression of Type 2 promoters
Although the [2Fe-2S] cluster of IscR is not required for DNA binding of Type 2 promoters, the amount of the Fe-S form of IscR may indirectly control expression of Type 2 promoters by determining the levels of total IscR protein produced from the iscR promoter. Because IscR requires its Fe-S cluster to repress the iscR promoter1, any condition that leads to a decrease in [2Fe-2S]-IscR should increase the amount of total IscR. Thus, stress conditions that increase expression from the iscR promoter could also change expression of Type 2 promoters, if the Type 2 binding site is not completely occupied by IscR under normal growth conditions, such as seen by Yeo et al.7 for the sufA promoter. This indeed appears to be the case for PsufA, PhyaA, and PnapF, since changes in expression in strains containing IscR mutants lacking the cluster were correlated with increased IscR levels, indicating that none of these IscR binding sites are completely occupied under our standard growth condition (Fig. 2, 9). Also, stress conditions may affect the oxidation state of the [2Fe-2S] cluster, but similar in vitro binding of IscR under anaerobic and aerobic conditions to the hya DNA site suggests that O2-mediated oxidation of [2Fe-2S]1+-IscR to [2Fe-2S]2+-IscR1 does not affect IscR binding to Type 2 sites (data not shown). Furthermore, the previously proposed homeostatic mechanism for controlling iscR expression1 that renders expression of the isc operon sensitive to the demand for Fe-S biogenesis would not only alter expression of promoters that require [2Fe-2S]-IscR but also the Type 2 promoters that are not saturated for binding. While this makes expression of the iscR promoter the key factor in determining the overall global regulatory impact of IscR, it also reveals the mechanistic complexity of this system and underscores the need to determine the requirement for [2Fe-2S]-IscR for each promoter. In summary, our data showed that IscR does not require its cluster for regulation of Type 2 promoters, and future work will focus on the role of the [2Fe-2S] cluster for direct regulation of Type 1 promoters or indirect regulation of the Type 2 promoters.
Materials and Methods
Strain Construction
Chromosomally-encoded mutants of IscR in which cysteine residues were substituted with alanine were constructed in several steps. First, the kan cassette from pKD1333, which has flanking FRT (FLP recognition target) sites, was placed 6 bases after the iscR termination codon on pPK59601 with the primers EcoRI-FRT1 and EcoRI-FRT2 (Supp. Table 1). The iscR gene on the resulting plasmid, pPK7312, was then mutated using QuikChange (Stratagene) site-directed mutatgenesis to create the single and multiple cysteine to alanine substitutions. The iscR variants were PCR-amplified using primers iscR-83-51 and iscS100 (Supp. Table 1), and the DNA was electroporated into BW25993 overexpressing λ Red recombinase from pKD4633. The correct recombinants were selected for kan and confirmed with DNA sequencing. The insertion of the kan cassette six bp after the iscR translational stop codon was given the allele number zfh-3600::kan. The mutations were then transduced into MG1655 or its derivatives (see Table 2) using P1vir and selecting for the adjacent kan cassette. Finally, the kan cassette was removed via transformation with pCP2033, which encodes FLP recombinase, and the resulting chromosomal region was sequenced to ensure the presence of the mutation and the loss of the kan cassette.
Table 2.
Strains and plasmids used in this work
| Strain/plasmid | Relevant genotype | Source |
|---|---|---|
| Bacterial strains | ||
| MG1655 | λ-F- rph-1 | Laboratory stock |
| PK4854 | MG1655 ΔiscR | 1 |
| BW25993/pKD46 | lacIqhsdR 514 ΔaraBADAH33ΔrhaBADLD78 pKD46 | 23 |
| PK7318 | BW25993 zfh-3600::kan | This study |
| PK7813 | BW25993 iscR-C92A zfh-3600::kan | This study |
| PK7826 | BW25993 iscR-C98A zfh-3600::kan | This study |
| PK7394 | BW25993 iscR-C104A zfh-3600::kan | This study |
| PK7887 | BW25993 iscR-C92A/C98A/C104A zfh-3600::kan | This study |
| PK7359 | MG1655 zfh-3601::FRT | This study |
| PK7823 | MG1655 iscR-C92A zfh-3601::FRT | This study |
| PK7833 | MG1655 iscR-C98A zfh-3601::FRT | This study |
| PK7807 | MG1655 iscR-C104 zfh-3601::FRT | This study |
| PK7896 | MG1655 iscR-C92A/C98A/C104A zfh-3601::FRT | This study |
| PK6879 | MG1655 PsufA-lacZ | 2 |
| PK7362 | PK7359 PsufA-lacZ | This study |
| PK6880 | PK4854 PsufA-lacZ | 2 |
| PK7825 | PK7823 PsufA-lacZ | This study |
| PK7837 | PK7833 PsufA-lacZ | This study |
| PK7809 | PK7807 PsufA-lacZ | This study |
| PK7899 | PK7896 PsufA-lacZ | This study |
| PK8004 | MG1655 PydiU-lacZ | 2 |
| PK8130 | PK7359 PydiU-lacZ | This study |
| PK8005 | PK4854 PydiU-lacZ | 2 |
| PK8131 | PK7823 PydiU-lacZ | This study |
| PK8132 | PK7833 PydiU-lacZ | This study |
| PK8133 | PK7807 PydiU-lacZ | This study |
| PK8134 | PK7896 PydiU-lacZ | This study |
| PK7573 | MG1655 PhyaA-lacZ | 2 |
| PK7386 | PK7359 PhyaA-lacZ | This study |
| PK6886 | PK4854 PhyaA-lacZ | 2 |
| PK8006 | PK7823 PhyaA-lacZ | This study |
| PK8009 | PK7833 PhyaA-lacZ | This study |
| PK7797 | PK7807 PhyaA-lacZ | This study |
| PK7900 | PK7896 PhyaA-lacZ | This study |
| PK8182 | MG1655 PhyaA(-44TTGGT-40)-lacZ | This study |
| PK8188 | PK4854 PhyaA(-44TTGGT-40)-lacZ | This study |
| PK8194 | PK7896 PhyaA(-44TTGGT-40)-lacZ | This study |
| PK8758 | MG1655 PhyaA(-50G)-lacZ | This study |
| PK8759 | PK4854 PhyaA(-50G)-lacZ | This study |
| PK8760 | PK7896 PhyaA(-50G)-lacZ | This study |
| PK8788 | MG1655 PhyaA(-38T)-lacZ | This study |
| PK8789 | PK4854 PhyaA(-38T)-lacZ | This study |
| PK8790 | PK7896 PhyaA(-38T)-lacZ | This study |
| PK7538 | MG1655 PnapF-lacZ | 2 |
| PK8000 | PK7359 PnapF-lacZ | This study |
| PK7539 | PK4854 PnapF-lacZ | 2 |
| PK8008 | PK7823 PnapF-lacZ | This study |
| PK7574 | MG1655 PhybO-lacZ | 2 |
| PK7800 | PK7359 PhybO-lacZ | This study |
| PK6888 | PK4854 PhybO-lacZ | 2 |
| PK8007 | PK7823 PhybO-lacZ | This study |
| PK7599 | BL21(DE3) ΔhimA::tet + p6161 | 2 |
| PK7879 | BL21(DE3) ΔhimA::tet ΔiscR::kan + pPK7820 | This study |
| PK7881 | BL21(DE3) ΔhimA::tetΔiscR::kan + pPK7862 | This study |
| Plasmids | ||
| pKD46 | Phage λgam -bet-exo genes under ParaB control | 23 |
| pPK5960 | StuI-EcoRV 985-bp iscR in pACYC184 | 1 |
| pKD13 | FRT-kan-FRT | 23 |
| pPK7312 | EcoRI 1303-bp FRT-kan-FRT in pPK5960 | This study |
| pPK6161 | iscR in pET-11a | 1 |
| pPK7820 | iscR-C92A in pET-11a | This study |
| pPK7862 | iscR-C92A/C98A/C104A in pET-11a | This study |
Removal of the kan cassette leaves an 82 bp FRT sequence between iscR and iscS (allele number zfh-3601::FRT), which is non-polar on iscS and iscU as determined by Western blot analysis using α-IscS and α-IscU-specific antibodies (data not shown). The extra 82 bp also does not appear to affect IscR activity because IscR-mediated repression of PhyaA, PsufA, PydiU, PnapF, and PhybO was similar in wild-type strains and strains with the 82 bp sequence between iscR and iscS (data not shown).
Mutations within the IscR binding site of the hyaA promoter were created by QuikChange site-directed mutatgenesis (Stratagene) using pPK68342, which contains PhyaA fused to α-lacZ sequence adjacent to a kan cassette. The mutated plasmids were PCR-amplified, and the DNA was electroporated into BW25993/pKD46, creating chromosomal promoter-lacZ fusions at the lac locus as previously described2,34. The promoter-lacZ fusions were moved into MG1655, PK4854, and PK7896 using P1vir transduction by virtue of the adjacent kan cassette.
β-galactosidase assays and Western blot analysis
Three independent isolates of each strain were grown in MOPS minimal media with 0.2% glucose35 under aerobic conditions to an O.D.600 of ~0.2 and assayed for β-galactosidase activity36. Data from the average of three isolates on a single day are shown, and replication on three independent days gave similar results.
To correlate β-galactosidase activity with IscR protein levels, parallel samples were removed and analyzed for IscR protein levels by Western blotting. The cell pellets, along with purified IscR standards, were solubilized by heat (95°C) and electrophoresed on a 12% SDS-PAGE gel. After transfer to a nitrocellulose membrane, the membrane was blocked with 5% milk. IscR was detected using α-IscR rabbit primary antibodies and fluorescein isothiocyanate-labeled goat anti-rabbit antibodies (BD Pharmingen). The fluorescence was measured with a FMBIO II fluorescence scanner (Hitachi) and quantified using ImageQuant v. 1.2 (Molecular Dynamics). All samples were analyzed in triplicate. To determine the number of IscR molecules per cell, the number of colony forming units (CFU) was measured by viability plating from aerobic cultures grown to varying O.D.600.
Purification of IscR
Wild-type IscR was purified from PK7599 as described previously2. Apo-IscR was generated from wild-type IscR by treatment with 1.76 mg/mL 2,2′-dipyridyl for 4 hours under anaerobic conditions at room temperature and subsequent passage through a Sephadex-G25 spin column. IscR-C92A was purified from PK7879 under anaerobic conditions following the same protocol as for wild-type IscR, and IscR-C92A/C98A/C104A was purified from PK7881 similar to wild-type but under aerobic conditions at 4°C. Protein concentration and iron and sulfide content were determined colorimetrically as described previously1, and protein concentration was standardized by the protein molarity determined from the amino acid composition analysis (Molecular Analysis Facility, University of Iowa).
Size Exclusion Chromatography
A Superose 12 size exclusion column (25 ml) (Amersham Pharmacia) attached to a high-pressure liquid chromatography system (Beckman) was calibrated using vitamin B12 (1.35 kDa), cytochrome c (12 kDa), myoglobin (17 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), gamma globin (158 kDa), and thyroglobulin (670 kDa). Wild-type IscR (2,400 and 240 μM, 35% occupied with cluster) and apo-IscR (970 μM) were analyzed separately under anaerobic conditions in 50 mM Tris-Cl pH 7.4, 200 mM KCl, and 10% glycerol. The data for the size standards were fitted using a log plot of molecular mass as a function of elution volume divided by void volume (Ve/V0).
Analytical Ultracentrifugation
IscR-C92A/C98A/C104A at 12, 31, and 49 μM in 40 mM Tris-Cl pH 7.4 at 25°C and 150 mM KCl was spun at 4°C at 3000 initial, 9000, 12000, 16400, 19200, and 24000 rpm in a 4 cell rotor in a Beckman Coulter XL-A analytical ultracentrifuge until reaching equilibrium (~12 h). After the 24000 rpm spin, the sample was allowed to re-equilibrate to 16200 rpm, and superposition with the earlier 16400 rpm data indicated that no large irreversible aggregates formed during the experiment (data not shown). IscR absorbance values were recorded at A280 and A230. Although a small amount of IscR degraded from 17.2 kDa to 14.6 kDa (as determined by ESI-MS) during centrifugation, this was taken into consideration during the modeling. The models used global fitting of the data and were similar to the Laue approach37,38 with programs written for Igor Pro (Wavemetrics).
DNA binding fluorescence assays
DNA binding isotherms were generated by measuring changes in fluorescence polarization when IscR bound a 30-mer dsDNA linked to a Texas Red fluorophore (IDT) at the 5′ end of the top strand (Table 1). The 30-mer dsDNA contained either the 26 bp hya site or a 26 bp randomized hya site obtained by using the algorithm by Stothard at http://bioinformatics.org/sms/. Complementary strands of DNA were annealed by heating at 95°C for 5 min in 40 mM Tris-Cl (pH 7.9), 30 mM KCl before slow cooling to room temperature. IscR concentrations from 7 to 200 nM were incubated with 5 nM labeled-DNA, 40 mM Tris-Cl (pH 7.9), and 150 mM KCl for 10 min at room temperature. 150 mM KCl was chosen because parallel experiments with IscR-C92A/C98A/C104A and the randomized hya site using 30 mM or 70 mM KCl showed some non-specific binding (data not shown). Experiments shown were performed under aerobic conditions, although anaerobic binding assays were also performed and gave similar results (data not shown). Measurements of fluorescence polarization were taken using a Beacon 2000 Fluorescence Polarization system (Invitrogen) and Texas Red filters (Andover Corp.) with a 10 s delay to exclude ambient light. Data for the wild-type hya site were fitted using a 4-parameter Hill equation39,40 (SigmaPlot ver. 6.00):
| [1] |
where ‘A’ is the anisotropy of the sample, ‘Afree’ is the anisotropy with no IscR, ‘Abound’ is the anisotropy with 200 nM IscR, and ‘n’ is the Hill coefficient.
Stoichiometry assays were performed using the same solution conditions except with 250 nM labeled-DNA and IscR concentrations ranging from 268 nM to 1600 nM. For wild-type IscR, fraction bound was determined using the equation:
| [2] |
where ‘A’ is the anisotropy of the sample, ‘Afree’ is the anisotropy with no IscR, ‘Abound’ is the anisotropy with 1600 nM IscR, and ‘q’ is the quenching factor, which was between 0.87 and 0.91 for wild-type IscR41. ‘q’ was calculated by dividing the total fluorescence of the 1600 nM IscR sample by the total fluorescence of the 0 nM IscR sample. Because IscR-C92A/C98A/C104A did not quench the Texas Red fluorophore, the fraction bound was calculated using:
| [3] |
DNA equilibrium competition assays were used to measure the binding of a series of mutations within the hya site. These assays included 5 nM fluorophore-labeled wild-type hya site DNA in 40 mM Tris-Cl (pH 7.9) and 150 mM KCl, and varying concentrations of 30-mer HPLC-purified unlabeled dsDNA competitor (IDT, Table 1) ranging from 8 to 1000 nM. 55 nM IscR-C92A/C98A/C104A was added last, and DNA binding was measured as above. The fraction of fluorescent probe bound in the presence of competitors was calculated as above except that Abound is the anisotropy at 55 nM IscR in the absence of competitors. The data were fitted using the IC50 equation (KaleidaGraph ver.4.0):
| [4] |
where ‘FBmax’ is the reaction with 55 nM IscR and no competitor. The assays to generate binding isotherms, stoichiometry values, and the competition data were repeated on three independent occasions.
Supplementary Material
Acknowledgments
We acknowledge Helmut Beinert for analyzing iron and sulfide content of purified protein; Larry Vickery for providing the α-IscS and α-IscU-specific antibodies; Brendan Wanta for assistance with β-galactosidase assays; Kevin Myers for strain construction; Hwan Youn and Timothy Cordes for advice about fluorescence anisotropy assays; Dr. James Keck for the use of his Beacon 2000 fluorescence polarization fluorometer; and the Kiley lab for comments on the manuscript. Analytical ultracentrifugation data were obtained with assistance from Darrell McCaslin at the University of Wisconsin-Madison Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin-Madison and grants BIR-9512577 (NSF) and S10 RR13790 (NIH). DNA sequencing and mass spectrometry were performed by the University of Wisconsin-Madison Biotechnology Center. A.D.N. is a trainee of the NIH Molecular Biosciences Predoctoral Training Grant GM07215, and J.L.G. was a trainee of the NIH Biotechnology Predoctoral Training Grant GM08349, both to the University of Wisconsin-Madison. This work was supported by the NIH grant GM45844 to P.J.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–75. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–81. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 4.Fontecave M, Ollagnier-de-Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch Biochem Biophys. 2008;474:226–37. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Loiseau L, Gerez C, Bekker M, Ollagnier-de-Choudens S, Py B, Sanakis Y, de Mattos JT, Fontecave M, Barras F. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–31. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelini S, Gerez C, Ollagnier-de-Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–91. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 7.Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol. 2006;61:206–18. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 8.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: 4Fe-4S to 2Fe-2S conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–92. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–46. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtuszewski K, Hawkins ME, Cole JL, Mukerji I. HU binding to DNA: Evidence for multiple complex formation and DNA bending. Biochem. 2001;40:2588–98. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–59. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 12.Giel JL. Escherichia coli PhD. University of Wisconsin-Madison; 2007. Role of IscR in regulation of iron-sulfur biogenesis. [Google Scholar]
- 13.Oh SY, Shin JH, Roe JH. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J Bact. 2007;189:6284–92. doi: 10.1128/JB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keon RG, Fu R, Voordouw G. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp vulgaris Hildenborough. Arch Microbiol. 1997;167:376–83. doi: 10.1007/s002030050458. [DOI] [PubMed] [Google Scholar]
- 15.Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AW. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiol. 2002;148:4059–71. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- 16.Beaumont HJ, Lens SI, Reijnders WN, Westerhoff HV, van Spanning RJ. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol Microbiol. 2004;54:148–58. doi: 10.1111/j.1365-2958.2004.04248.x. [DOI] [PubMed] [Google Scholar]
- 17.Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bact. 2006;188:874–81. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: Comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:415–31. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filenko N, Spiro S, Browning DF, Squire D, Overton TW, Cole J, Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bact. 2007;189:4410–7. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis M. The Lac repressor. C R Biol. 2005;328:521–48. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Riggs AD, Suzuki H, Bourgeoi S. Lac repressor-operator interaction. 1 Equilibrium studies. J Mol Biol. 1970;48:67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- 22.Bahl CP, Wu R, Stawinsky J, Narang SA. Minimal length of the lactose operator sequence for the specific recognition by the lactose repressor. Proc Natl Acad Sci USA. 1977;74:966–70. doi: 10.1073/pnas.74.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell CE, Lewis M. Crystallographic analysis of Lac repressor bound to natural operator O1. J Mol Biol. 2001;312:921–6. doi: 10.1006/jmbi.2001.5024. [DOI] [PubMed] [Google Scholar]
- 24.Wisedchaisri G, Chou CJ, Wu M, Roach C, Rice AE, Holmes RK, Beeson C, Hol WG. Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis. Biochem. 2007;46:436–47. doi: 10.1021/bi0609826. [DOI] [PubMed] [Google Scholar]
- 25.Wisedchaisri G, Holmes RK, Hol WG. Crystal structure of an IdeR-DNA complex reveals a conformational change in activated IdeR for base-specific interactions. J Mol Biol. 2004;342:1155–69. doi: 10.1016/j.jmb.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 26.Chen CS, White A, Love J, Murphy JR, Ringe D. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochem. 2000;39:10397–407. doi: 10.1021/bi0009284. [DOI] [PubMed] [Google Scholar]
- 27.White A, Ding X, vanderSpek JC, Murphy JR, Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998;394:502–6. doi: 10.1038/28893. [DOI] [PubMed] [Google Scholar]
- 28.Grkovic S, Brown MH, Schumacher MA, Brennan RG, Skurray RA. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J Bact. 2001;183:7102–9. doi: 10.1128/JB.183.24.7102-7109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou CJ, Wisedchaisri G, Monfeli RR, Oram DM, Holmes RK, Hol WG, Beeson C. Functional studies of the Mycobacterium tuberculosis iron-dependent regulator. J Biol Chem. 2004;279:53554–61. doi: 10.1074/jbc.M407385200. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002;21:1210–8. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiley PJ, Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1999;22:341–52. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo E, Leautaud V, Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17:2629–36. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bact. 2005;187:1135–60. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bact. 1974;119:736–47. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1972. [Google Scholar]
- 37.Brunzelle JS, Jordan DB, McCaslin DR, Olczak A, Wawrzak Z. Structure of the two-subsite beta-d-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane. Arch Biochem Biophys. 2008;474:157–66. doi: 10.1016/j.abb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Laue TM. Sedimentation equilibrium as thermodynamic tool. Methods Enzymol. 1995;259:427–52. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- 39.Morgan PH, Mercer LP, Flodin NW. General model for nutritional responses of higher organisms. Proc Natl Acad Sci USA. 1975;72:4327–31. doi: 10.1073/pnas.72.11.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjorve E. Shapes and functions of species-area curves: a review of possible models. J Biogeography. 2003;30:827–35. [Google Scholar]
- 41.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–12. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.