Abstract
Ten E. coli aminoacyl-tRNAs (aa-tRNAs) were assessed for their ability to decode cognate codons on E. coli ribosomes by using three assays that evaluate the key steps in the decoding pathway. Despite a wide variety of structural features, each aa-tRNA exhibited similar kinetic and thermodynamic properties in each assay. This surprising kinetic and thermodynamic uniformity is likely to reflect the importance of ribosome conformational changes in defining the rates and affinities of the decoding process as well as the evolutionary “tuning” of each aa-tRNA sequence to modify their individual interactions with the ribosome at each step.
Introduction
Protein synthesis in E. coli is a complex system in which 45 elongator aminoacyltRNA (aa-tRNA) substrates (Sprinzl et al., 1998) quickly and accurately decode the 61 different sense codons on the ribosome. Each aa-tRNA is chemically distinct, consisting of different combinations of sequences, lengths, post-transcriptional modifications, and esterified amino acids. For decoding to occur, these very different molecules must bind the ribosome at the same discrete site and undergo several intermediate steps before entering the ribosomal A site where peptide bond formation takes place. It is generally accepted that all elongator aa-tRNAs go through the same pathway and thus encounter the same succession of defined ribosomal environments. However, it is unclear whether the individual aa-tRNAs transit the ribosome with identical thermodynamic and kinetic properties. For example, an aa-tRNA with a GC rich anticodon may initially bind tighter to its codon than an aa-tRNA with an AU rich anticodon. Similarly, a tRNA esterified to a small non-polar amino acid may pass from the entry site into the A site more rapidly than a tRNA esterified to a bulky aromatic amino acid.
Available data sets do not agree as to whether individual aa-tRNAs act similarly or differently during decoding. Several in vivo experiments suggest that different aa-tRNAs and even synonymous codons are decoded at different rates. In one such experiment, an insertion of three identical test codons was placed in the leader sequence of a gene regulated by the pyrE attenuator, thereby coupling translation and transcription and revealing a six fold difference in transcriptional attenuation for 12 different codons (Bonekamp et al., 1989). Another study used pulse chase experiments to measure expression of β-galactosidase which contained an insert of eight identical test codons late in the lacZ gene and found a five fold difference in the rate of protein production for four different codons (Sørensen and Pedersen, 1991). Finally, a study which measured how effectively 29 different codons could compete with a common frameshift site, which served as a kinetic standard, found a 60 fold range of incorporation efficiency (Curran and Yarus, 1989). While all of these experiments suggest that different aa-tRNAs are incorporated into the ribosome differently, they all measure the decoding rate indirectly so the observed differences may not necessarily reflect intrinsic kinetic or thermodynamic differences in how individual aa-tRNAs interact with the ribosome. In contrast, a limited number of biochemical experiments using purified E. coli ribosomes suggest that aa-tRNAs may function similarly to one another. Similar dissociation rates of eight different aa-tRNAs from the ribosomal A and P sites suggest that they bind to the sites in a thermodynamically equivalent manner (Fahlman et al., 2004). However, the very slow non-enzymatic release of aa-tRNAs from ribosomes may not provide insight into how tRNAs behave in the very fast decoding pathway. The only experiments that directly and quantitatively compared the performance of aa-tRNAs in kinetically relevant steps in the elongation cycle used only a few aa-tRNAs that were tested in different laboratories under slightly different reaction conditions (Thomas et al., 1988; Pape et al., 1998; Cochella and Green, 2005; Kothe and Rodnina, 2007; Ling et al., 2007). However, these few experiments do suggest that different aa-tRNAs may be selected into the ribosome similarly.
The goal of this work was to directly compare the decoding properties of a group of chemically diverse aa-tRNAs under identical conditions by testing the initial binding to the entry site as well as the rates of EF-Tu GTPase activation and aa-tRNA accommodation into the A site. This should establish whether or not the ribosome is sensitive to the chemical and structural differences among aa-tRNAs during decoding.
Results
Assessing Different aa-tRNAs in the Key Steps of the Decoding Pathway
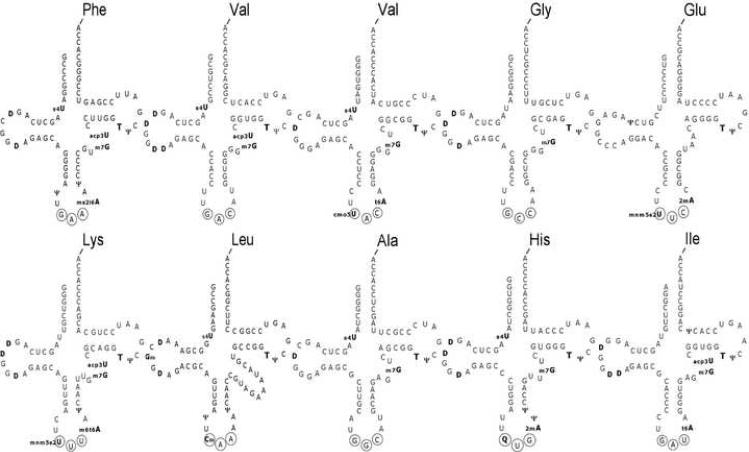
Ten different E. coli aa-tRNAs were chosen to represent the diverse physical and chemical properties of this class of molecules (Figure 1). Their esterified amino acids include side chains that are both positively and negatively charged and vary in size from glycine to phenylalanine. Their anticodon sequences vary from three G-C pairs to three A-U pairs. Seven of the tRNAs are members of the most common D4V5 structural class (Kim et al., 1974), while tRNAGlu has a different tertiary architecture, tRNAHis has an additional residue in the acceptor stem, and tRNALeu has an extended variable loop. Each tRNA also has a set of post-transcriptional modifications, some of which are common and others unique. All these structural differences could potentially effect how each aa-tRNA could fit into the ribosome and thereby modify their affinity or reactivity in the translation process. Finally, the ten tRNAs are present in log phase E. coli in quite different concentrations, ranging from 0.95% of the total tRNA for tRNAAla to 7.3% for tRNAGlu (Dong et al., 1996).
Figure 1.
Secondary structures of ten E. coli aa-tRNAs used for this study. Anticodon nucleotides are circled and post-transcriptionally modified nucleotides are in bold and abbreviated according to Sprinzl et al. (1998).
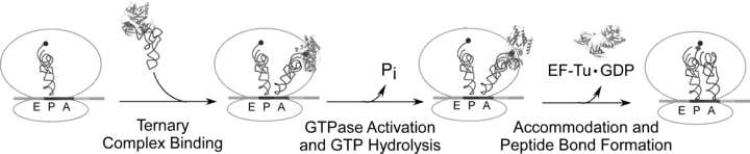
The ability of each of the aa-tRNAs to decode their respective fully complementary cognate codons on E. coli ribosomes was evaluated using three different quantitative assays which monitored different stages of the decoding process (Figure 2). First, a filter retention assay was used to determine the equilibrium dissociation constant for the binding of aa-tRNA·EF-Tu·GTP ternary complexes to the ribosomal entry site. Second, the subsequent rate of GTP hydrolysis by EF-Tu on the encoded ribosomes was determined using a rapid quench assay. Finally, a second rapid quench assay was used to determine the rate of peptide bond formation after the aa-tRNA had reached the peptidyl transferase center at the end of the decoding process. All three assays used identically encoded E. coli 70S ribosomal substrates, containing either deacyl tRNAfMet or fMet-tRNAfMet in the P site and an mRNA with a codon in the A site that was fully complementary to the anticodon of each aa-tRNA tested. Therefore, each aa-tRNA was evaluated in an identical sequence context. Since future experiments will examine the decoding properties of mutant tRNA transcripts, to ensure proper folding of both modified and unmodified tRNAs the classic translation buffer containing 10 mM MgCl2 was used. Although buffers which contain lower concentrations of magnesium ions and have added polyamines show an increase in the accuracy of aa-tRNA selection on near-cognate codons their decoding parameters on cognate codons are quite similar to the classic buffer (Pape et al., 1998; Gromadski and Rodnina, 2004; Gromadski et al., 2006).
Figure 2.
Key steps in the decoding pathway.
Binding of Ternary Complex to the Ribosomal Entry Site
The reversible binding of the preformed ternary complex of aa-tRNA, EF-Tu, and GTP to the ribosome can be measured by using the H84A mutation of E. coli EF-Tu which blocks the irreversible GTP hydrolysis step and thereby prevents completion of the decoding reaction (Scarano et al., 1995). Experiments using the fluorescent nucleotide mant-GTP have established that the H84A mutation blocks decoding after a conformational change in the active site of EF-Tu, known as GTPase activation, that is rate limiting for GTP hydrolysis (Daviter et al., 2003). It is thought that this histidine residue is necessary to stabilize the water molecule that attacks the terminal phosphate of GTP during hydrolysis (Daviter et al., 2003). Thus, the equilibrium binding constant determined using EF-Tu(H84A) ternary complexes equals the product of the initial binding, codon recognition, and GTPase activation equilibria defined by previous kinetic experiments (Pape et al., 1998).
Modifications of the nitrocellulose filter binding assay employed by Daviter et al. (2003) were made in order to improve sensitivity and accuracy. First, each tRNA was [3'-32P] labeled using tRNA nucleotidyltransferase and aminoacylated with a non-radioactive amino acid (Wolfson and Uhlenbeck, 2002), thereby yielding aa-tRNAs with higher specific activities than can be achieved via acylation with 14C labeled amino acids. Second, since the E. coli ternary complex does not bind to nitrocellulose filters, a dual membrane filter system was employed to permit more accurate data at the extremes of the binding curve (Fahlman and Uhlenbeck, 2004). A typical binding experiment involved mixing a constant, low concentration (100 pM) of ternary complex containing a [32P] labeled aa-tRNA to a series of concentrations of 70S ribosomes containing saturating concentrations of mRNA and tRNAfMet bound to the P site. After a two minute incubation, all reactions are simultaneously filtered on a 96 well filtration block and washed with buffer. Trial experiments showed that the short incubation time was sufficient to permit complete binding at all ribosome concentrations but was not long enough for the much slower non-enzymatic binding of any aa-tRNA or deacyl tRNA that had not bound EF-Tu (Figure 3) (Fahlman and Uhlenbeck, 2004). This made it unnecessary to purify the ternary complex before the assay.
Figure 3.
Binding of aa-tRNA to the entry site of E. coli 70S ribosomes. Representative equilibrium binding curve for Val-tRNAGACVal·EF-Tu(H84A)·GTP ternary complex to programmed ribosomes in RB buffer at 20°C. The curve corresponds to a single binding isotherm with Kd = 1.5 nM and a fraction bound at saturation of 68%. Ternary complex containing Val-tRNAGACVal(●), Val-tRNAGACVal with no EF-Tu·GTP present (∎), ternary complex containing Val-tRNAGACVal with a mRNA containing a non-cognate AAA codon (◆) and with no mRNA present (▴).
A typical data set using Val-tRNA(GAC)Val is shown in Figure 3. Since the ternary complex was not purified away from deacyl tRNA prior to the reaction, a P1 nuclease digestion of the [32P] labeled aa-tRNA was performed to establish the fraction of aminoacylated tRNA present (Wolfson and Uhlenbeck, 2002). For the example shown in Figure 3, 68% of the input [3'-32P] tRNA(GAC)Val was aminoacylated (data not shown). Therefore, the 32% of input radioactivity which did not bind ribosomes reflects the deacylated tRNA(GAC)Val present in the assay that does not bind EF-Tu(H84A) and could not bind ribosomes non-enzymatically during the short incubation time. Consequently, virtually all available aa-tRNA was bound to EF-Tu(H84A) and the resulting ternary complex was fully bound to ribosomes. The equilibrium binding curve shown in Figure 3 formed within two minutes and remained stable over time. The complex was in kinetic equilibrium since an excess of non-radiolabeled ternary complex was able to compete the labeled ternary complex off ribosomes (data not shown). Since the ribosomes were not in excess of the ternary complex throughout the experiment, the complete binding equation for a simple binding equilibrium was used to fit the data to a Kd = 1.5 nM. Control experiments without mRNA or with an mRNA containing a non-cognate (AAA) codon showed very little binding (Figure 3). In addition, a control reaction without added EF-Tu·GTP showed that non-enzymatic binding does not occur during the two minute incubation (Figure 3).
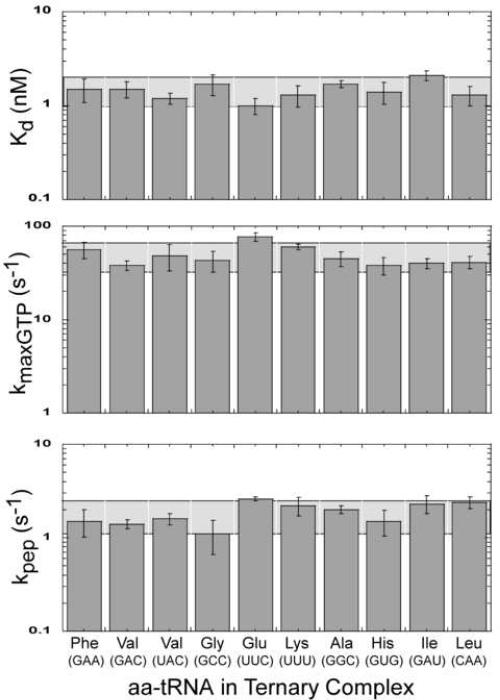
Similar binding curves were obtained for the nine other aa-tRNAs with their complementary codons with mean Kd values ranging from 1.0 nM to 2.1 nM which only slightly exceeds the experimental error of the assay (Table I). Thus, despite different anticodon sequences, the ten ternary complexes bind ribosomes with very similar affinities. The Kd values agree well with the previously determined Kd = 3 nM for Phe-tRNAPhe ternary complexes in a similar binding assay (Daviter et al., 2003). In addition, Kd values of 1.2 and 0.9 nM for ternary complexes containing Trp-tRNATrp and Ala-tRNA(UGC)Ala have been reported using similar assays (Cochella and Green, 2005; Kothe and Rodnina, 2007). Therefore, it appears likely that ternary complexes of all elongator aa-tRNAs bind to ribosomes with similar affinities.
Table I.
Binding and kinetic data for different ternary complexes in the decoding cycle
| aa-tRNA (anticodon) | Kd (nM) | K(ribosome)1/2 (μM | kmaxGTP (s-1) | kpep (s-1) |
|---|---|---|---|---|
| Phe (GAA) | 1.5 (0.42) | 1.4 (0.68) | 56 (11) | 1.5 (0.49) |
| Val (GAC) | 1.5 (0.29) | 0.99 (0.12) | 38 (4.6) | 1.4 (0.16) |
| Val (UAC) | 1.2 (0.16) | 1.5 (0.72) | 48 (15) | 1.6 (0.22) |
| Gly (GCC) | 1.7 (0.42) | 1.2 (0.80) | 43 (11) | 1.1 (0.44) |
| Glu (UUC) | 1.0 (0.20) | 1.7 (0.37) | 77 (8.3) | 2.6 (0.13) |
| Lys (UUU) | 1.3 (0.33) | 1.3 (0.25) | 60 (4.2) | 2.2 (0.49) |
| Ala (GGC) | 1.7 (0.14) | 1.9 (0.25) | 45 (7.8) | 2.0 (0.20) |
| His (GUG) | 1.4 (0.36) | 1.3 (0.62) | 38 (8.0) | 1.5 (0.46) |
| Ile (GAU) | 2.1 (0.25) | 1.1 (0.38) | 40 (5.1) | 2.3 (0.50) |
| Leu (CAA) | 1.3 (0.30) | 1.0 (0.38) | 41 (6.2) | 2.4 (0.34) |
Numbers in parentheses are standard error of the mean. Kd and kpep values are based on at least 3 independent replicates and K(ribosome)1/2 and kmaxGTP are determined from curves fit to at least 6 apparent kGTP values at different ribosome concentrations.
Rate of GTP Hydrolysis
Following ternary complex binding to a cognate codon in the ribosomal entry site, the active site of EF-Tu stimulates hydrolysis of GTP to GDP and Pi. This fast chemical step appears to be rate limited by a conformational change in the structure of EF-Tu. This change can be detected by the fluorescent GTP analogue, mant-GTP (Rodnina et al., 1995).
The rate of GTP hydrolysis was measured by preparing ternary complex with wild type EF-Tu, non-radioactive aa-tRNA, and [γ-32P] GTP (Pape et al., 1998). After removing excess unbound GTP by gel filtration, the resulting ternary complex of approximately 600 nM is mixed with an excess of ribosomes bound to mRNA and deacyl tRNAfMet in a stop quench apparatus. The fraction of GTP hydrolyzed at each time point was subsequently determined by thin layer chromatography (TLC) (Cochella and Green, 2005). A representative time course can be described by a single exponential curve as shown in Figure 4A. The ~60% of GTP hydrolyzed at completion does not change with ribosome concentration and therefore is likely representative of the fraction of GTP initially bound as ternary complex at the beginning of each reaction. Since the rate of GTP hydrolysis is a function of ribosome concentration, it was necessary to perform the experiment at several ribosome concentrations to obtain the rate at saturation. A fit to a Michaelis Menten curve with a kmaxGTP = 38 s-1 and K(ribosome)1/2 = 0.99 μM was obtained for the ternary complex containing Val-tRNA(GAC)Val (Figure 4B). The tight affinity of the ternary complex for the ribosome suggests this high K1/2 is due to fast decoding and slow dissociation rates, as has been observed for Phe-tRNAPhe (Pape et al., 1998).
Figure 4.
Rate of ribosome stimulated GTP hydrolysis. (a) Representative timecourse for GTP hydrolysis with 300 nM ternary complex containing Val-tRNAGACVal, 3.5 μM encoded ribosomes in RB buffer at 20°C. The data is described by a single exponential fit with kGTP apparent = 30 s-1. (b) Ribosome saturation curve for Val-tRNAGACVal fit to a single Michaelis Menten expression with kGTP at saturation = 38 ± 4.6 s-1.
Similar experiments determining kGTP as a function of ribosome concentration were performed for ternary complexes containing nine other aa-tRNAs (Table I). The values of the extrapolated kmaxGTP vary from 38 s-1 to 77 s-1 among the ten aa-tRNAs which is only slightly outside the range of experimental error. The kmaxGTP values determined here are similar to the rates reported for Phe-tRNAPhe (~60 s-1 (Pape et al., 1998) and 110 s-1 (Gromadski and Rodnina, 2004)) and Ala-tRNA(UGC)Ala (40 s-1 (Kothe and Rodnina, 2007)). Values of K(ribosome)1/2 were also comparable to the K(ribosome)1/2 of 2 μM determined for the ternary complex containing Trp-tRNATrp (Cochella et al., 2007). Thus, ternary complexes containing chemically different aa-tRNAs are able to activate the hydrolysis of GTP by EF-Tu similarly. All together, the uniform Kd's of different ternary complexes in the entry site, the uniform K(ribosome)1/2 values, and similar kmaxGTP rates for different aa-tRNAs suggest that the individual rates for the substeps of EF-Tu binding, codon-anticodon association and dissociation are also similar.
Rate of Peptide Bond Formation
After GTP hydrolysis, aa-tRNA is released from EF-Tu and the 3' end repositions into the peptidyl transferase center in the large ribosomal subunit while still maintaining the codon-anticodon interactions in the small subunit (Valle et al., 2003). This conformational change, termed accommodation, can be detected kinetically by a change in fluorescence intensity of tRNAs containing a fluorophore at one of several locations, and occurs at the same rate as kpep, the observed rate of peptide bond formation (Pape et al., 1998; Blanchard et al., 2004). The accommodation step is much slower than the rate of peptide bond formation, estimated to be faster than 300 s-1 (Bieling et al., 2006), and is therefore rate limiting. Based on single molecule FRET experiments (Blanchard et al., 2004) and molecular dynamics simulations (Sanbonmatsu et al., 2005) it is likely that several conformational changes are associated with accommodation and it is not known which one actually limits the rate of peptide bond formation. While kpep is the sum of the accommodation rate and the rate of rejection during the proofreading steps, cognate aa-tRNA complexes are expected to have negligible rejection rates (Pape et al., 1998). Therefore, kpep reports on the rate of aa-tRNA accommodation into the active site of the ribosome.
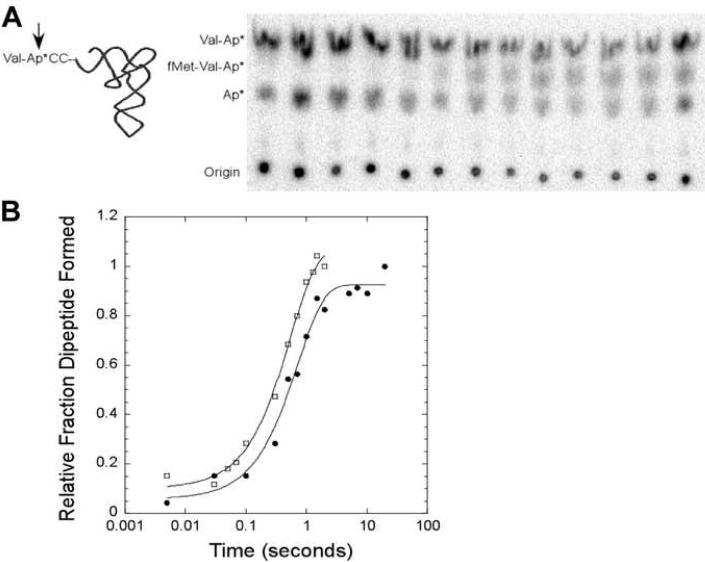
The standard assay to measure kpep involves preparing ribosomes with [35S] radiolabeled fMet-tRNAfMet in the P site and then mixing with a non-radiolabeled ternary complex in a rapid quench apparatus (Pape et al., 1998; Cochella and Green, 2005). Unreacted [35S] fMet and the [35S] fMet-aa product are hydrolyzed from the tRNA by treatment with base and the radiolabeled products of the reaction are separated by HPLC or paper electrophoresis (Pape et al., 1998; Cochella and Green, 2005). We have developed a variant of this assay that is more sensitive and also uses 20 fold less of the A site tRNA than the standard assay. It involves mixing ribosomes with saturating non-radiolabeled fMet-tRNAfMet in the P site with a very low concentration of ternary complex containing a [32P] aa-tRNA in a rapid mixing device. After quenching the reaction an aliquot is treated with S1 nuclease which converts the dipeptidyl-tRNA product to [32P] fMet-aa-AMP and unreacted aa-tRNA to [32P] aa-AMP. In addition, since any deacyl tRNA present in the reaction which can not form ternary complex is converted to [32P] AMP, it is not necessary to purify the ternary complex prior to the reaction. The three S1 digestion products can be separated by TLC or paper electrophoresis (Figure 5A). The fraction of dipeptide formed at each time point is calculated using the sum of all three S1 nuclease products. However, the amount of [32P] AMP does not vary over time and serves as an internal control that tRNAs are not deacylating during the experiment or subsequent analysis. The rate of peptide bond formation for Val-tRNA(GAC)Val determined by this new method (1.4 s-1) matches the rate (1.8 s-1, Figure 5B) determined in a parallel reaction using [35S] fMet-tRNAfMet and analyzing the hydrolyzed products by paper electrophoresis (Cochella and Green, 2005).
Figure 5.
Observed rate of formation of the peptide bond. (a) Assay employs an aa-tRNA that is 32P labeled at the terminal adenosine and loaded into the A site by EF-Tu·GTP. After Val-tRNAGACVal reaction with fMet-tRNAfMet in the P site, products are cleaved by S1 nuclease (arrow) to release valyl-AMP, fMet-valyl-AMP, and free AMP which are separated by TLC. (b) Representative data for accommodation assay performed with a [3' 32P] labeled Val-tRNAGACVal (●, kpep = 1.4 s-1, raw data shown in (a)) or a 35S labeled P site fMet-tRNAfMet (⧠, kpep = 1.8 s-1) in RB buffer at 20°C fit to a single exponential curve. Data is normalized to 100% dipeptide formation at the final timepoint of each reaction. .
Values for kpep were obtained for nine additional aa-tRNAs using this new assay. As with the previous decoding steps, all aa-tRNAs tested had similar rates (Table I). Even though the aa-tRNA set chosen had a range of anticodon strengths and chemically and structurally different esterified amino acids, a narrow range of kpep values (1.1 to 2.6 s-1) was observed for all ten aa-tRNAs. The previously calculated rates of accommodation for Trp-tRNATrp (3 s-1 (Cochella et al., 2007)) and Phe-tRNAPhe (2 s-1 (Gromadski et al., 2006) and 1.4 s-1 (Ling et al., 2007)) determined using the standard assay agree well with the rates obtained in Table I. This suggests that all aa-tRNAs accommodate from the entry site of the ribosome to the A site at similar rates.
Discussion
Different aa-tRNAs Have Uniform Decoding Properties
The ribosome is an enzyme that must change its substrate specificity at each step in the polymerization reaction. In order to compare how a set of aa-tRNA substrates functions on ribosomes, three quantitative assays were chosen that monitor different substeps in the elongation cycle. Although all the aa-tRNAs were expected to function in translation, it was possible that their extensive physical and chemical differences would lead to measurable differences in how well decoding occurred at one or more steps in the pathway. An overall uniform aa-tRNA selection rate could theoretically be achieved by having a single rate limiting step while still allowing variation in the affinities or rates of individual aa-tRNAs at other steps. However, no large differences were observed among any of the elongator aa-tRNAs tested in each step of the pathway. Figure 6 shows that for each step in the decoding process most aa-tRNAs have values within a two-fold error range of the overall mean value. Thus, Gly-tRNAGly with its small amino acid and GC rich anticodon binds and accommodates into the A site similarly to Phe-tRNAPhe with its bulky amino acid and AU rich anticodon.
Figure 6.
Summary of decoding properties of ten aa-tRNAs. Two-fold range of the overall mean value for each step is shown as a horizontal shaded bar. Anticodons for each aa-tRNA are shown in parentheses.
It is remarkable that the kinetic and thermodynamic equivalence of aa-tRNAs extends to all of the assays used since each assay measures a distinctly different substep in the decoding pathway and therefore reflects the behavior of aa-tRNAs in spatially different regions of the ribosome. Also, while aa-tRNAs have a similar overall architecture, they are very different in structural detail. Each tRNA has its own characteristic nucleotide sequence and anticodon with different GC content and a unique set of post-transcriptional modifications that permits its chemically unique amino acid to be incorporated according to its corresponding codon. Since different aa-tRNAs also bind equivalently to the ribosomal A and P sites (Fahlman et al., 2004), it appears that the ribosome can ignore the substantial physical and chemical differences among its aa-tRNA substrates throughout the decoding process. This is surprising since there is structural data suggesting that extensive contacts form between aa-tRNAs and the ribosome throughout translation which would presumably be unique for each aa-tRNA (Ogle et al., 2001; Valle et al., 2003; Selmer et al., 2006).
It is important to point out that the results presented here were obtained using ribosomes in the process of forming the first peptide bond with initiator tRNA in the P site. These conditions were chosen to permit comparison with the X-ray and Cryo-EM structures and with similar biochemical data from other laboratories. However, it is possible that Kd, kGTP, and kpep will differ for ribosomes undergoing elongation at a later point in the sequence once Shine Dalgarno interactions are broken and the exit channel is filled. Significant differences in the structure of the ribosome are observed when this occurs (Yusupova et al., 2006). In addition, since “codon context” effects are known to exist (Buckingham, 1990; Irwin et al., 1995), the kinetic and thermodynamic parameters of the A site tRNA may also depend on the identity of the P site tRNA. Although likely, it remains to be shown whether the uniform decoding properties determined here will extend to elongating ribosomes with different P site tRNAs.
Conformational Changes Dominate Kinetic and Thermodynamic Properties
How can these very different aa-tRNA substrates undergo each step in the decoding pathway in such an equivalent manner? One possibility is that the multiple ribosomal conformational changes that occur during decoding dominate the kinetic and thermodynamic measurements. Indeed, there is substantial structural and biochemical data indicating that ribosomal conformational changes accompany each substep in decoding and contribute to the corresponding rate and equilibrium constants. The Kd value describing initial binding of ternary complex to ribosomes partially reflects structural changes in the flexible L7/L12 protein (Oleinikov et al., 1993; Diaconu et al., 2005) as well as a rearrangement of the GTPase activation center in the large subunit (Valle et al., 2003). In addition, Kd reflects a rearrangement of the small subunit that occurs when a cognate codon-anticodon helix forms (Ogle et al., 2001; Ogle et al., 2002). The observed rate of GTP hydrolysis, kGTP, has been shown to be rate limited by a “GTPase activation” conformational change in EF-Tu that can be detected by a fluorescent GTP analogue (Rodnina et al., 1995; Pape et al., 1998). Since kGTP is much slower when a near cognate codon is used (Pape et al., 1999; Gromadski and Rodnina, 2004; Gromadski et al., 2006), it appears that this conformational change involves the entire 70S ribosome, perhaps by communication between the intersubunit bridges (O'Connor and Dahlberg, 1995; Huggins et al., 2005; Liiv and O'Connor, 2006). Finally, kpep, the observed rate of peptide bond formation, is rate limited by a conformational change termed accommodation which involves the large scale movement of the acceptor arm of tRNA from where it was bound to EF-Tu to the A site (Valle et al., 2003). The accommodation step can be independently detected by a change in fluorescent intensity of fluorophores attached to the A site tRNA (Pape et al., 1998) or by FRET between fluorescently labeled A and P site tRNAs (Blanchard et al., 2004). Thus, the observed uniformity among aa-tRNAs at every step in the pathway may simply reflect the fact that the aa-tRNAs are passively fed through a large molecular machine that operates at a fixed rate for each step.
aa-tRNA Tuning is Also Necessary for Uniformity
There is abundant evidence that the translation process is not entirely governed by the ribosomal movements. Instead, tRNAs are active participants in the translation mechanism since each tRNA possesses a unique combination of structural elements that modulate its performance in translation. This phenomenon, termed “tRNA tuning”, has most extensively been documented using suppression of amber codons by suppressor tRNAs in E. coli. Mutating sequence elements in the anticodon stem-loop of suppressor tRNAs dramatically modulates their suppression efficiency (Yarus et al., 1986a; Yarus et al., 1986b; Hou and Schimmel, 1988; Kleina et al., 1990; Schultz and Yarus, 1994a; Schultz and Yarus, 1994b; McClain et al., 1998). In addition, several of the post-transcriptional modifications present in the anticodon loops of certain tRNAs have been found to be essential for efficient and accurate decoding (Takai et al., 1996; Krüger et al., 1998; Konevega et al., 2004; Murphy et al., 2004; Yamada et al., 2005). In a few cases there is also evidence that sequence elements in the D stem of certain tRNAs can promote codon misreading (Raftery et al., 1986; Smith and Yarus, 1989; Cochella and Green, 2005). Biochemical experiments exploring the mechanistic rationale of how tRNAs are tuned are more limited, although it is clear that structural elements present in tRNAs can modify the initial binding of ternary complex as well as kGTP and kpep (Cochella and Green, 2005). Since it has been shown here that different aa-tRNAs function equivalently in translation, it appears that aa-tRNAs are not tuned to maximize translational efficiency, but rather to optimize it in a way that ensures that each aa-tRNA undergoes translation in a uniform manner.
The idea that uniform thermodynamic properties results from tRNA tuning is already well established for the binding of aa-tRNAs to EF-Tu. Elongator tRNAs bind EF-Tu with similar affinities when they are esterified to their cognate amino acids (Louie et al., 1984; Louie and Jurnak, 1985), but can bind several thousand fold weaker or tighter when misacylated (LaRiviere et al., 2001; Asahara and Uhlenbeck, 2002; Dale et al., 2004). Recent protein and tRNA mutagenesis experiments have shown that sequences in the T stem of tRNAs are primarily responsible for tuning affinities for EFTu (Sanderson and Uhlenbeck, 2007a; Sanderson and Uhlenbeck, 2007b). It has also been shown that uniform A and P site binding of different aa-tRNAs is due to the presence of post-transcriptional modifications and a cognate esterified amino acid (Fahlman et al., 2004) Dale and Uhlenbeck unpublished). Tuning has similarly been demonstrated in pre-tRNA binding to RNase P between the 5' leader sequence in pre-tRNA and the tRNA body (Sun et al., 2006).
Role of Uniformity in Protein Synthesis
The uniform performance of different aa-tRNAs at different steps in the decoding pathway raises the question, why have aa-tRNAs evolved to be so similar? Biochemical experiments measuring the misincorporation of aa-tRNAs on codons containing single nucleotide mismatches may provide insight into the purpose of uniform cognate decoding. The rate of misincorporation of Phe-tRNAPhe is similar on codons with a single mismatch at each position (Gromadski et al., 2006). This suggests that the complex network of interactions in the decoding center (Ogle et al., 2001) evolved to give a similar threshold of accuracy, independent of the position of the mismatched nucleotides. In addition, limited data suggests that misincorporation rates are also similar for the near-cognate codons of different aa-tRNAs. The slow forward rates of incorporation and rapid rates of rejection are very similar for Phe-tRNAPhe, Trp-tRNATrp, and Ala-tRNAUGCAla on codons mismatched at the first position despite their quite different codon composition (Cochella and Green, 2005; Gromadski et al., 2006; Kothe and Rodnina, 2007). Since proofreading is determined by the competition between the forward and reverse rate constants of aa-tRNA incorporation, it is possible that the uniform cognate decoding rates observed in this paper at both selection steps may reflect the selective pressure which maintains similar translational accuracy at all codons.
Although the aa-tRNAs tested have very similar intrinsic decoding properties, this does not necessarily mean that all codons are translated at the same rate in vivo. It is well established that aa-tRNA concentrations in cells can vary considerably and are used to modify the expression of proteins from different genes based on the use of synonymous codons (Folley and Yarus, 1989; Dong et al., 1996; Ermolaeva, 2001). The relative expression of different tRNA genes in bacteria is regulated by growth conditions (Dong et al., 1996), making it possible to alter gene expression at a global level. Similarly, upon amino acid starvation aa-tRNA concentrations vary among different isoacceptor tRNAs (Elf et al., 2003; Dittmar et al., 2005; Sorensen et al., 2005), permitting differential protein expression. The uniform decoding properties of aa-tRNAs observed here suggest that these global regulatory mechanisms primarily depend upon the concentration of active ternary complex to control the rate of protein synthesis.
Experimental Procedures
Materials
Tight coupled 70S ribosomes from E. coli MRE600 cells were prepared as described (Powers and Noller, 1991). Final ribosome pellets were resuspended in ribosome binding buffer (RB buffer-- 50 mM HEPES [pH 7.0], 30 mM KCl, 70 mM NH4Cl, 10 mM MgCl2, and 1 mM DTT) and were stored and activated as previously described (Fahlman et al., 2004). The mRNAs used were derivatives of the initiation region of the T4 gp32 mRNA with the following sequence: 5'-GGCAAGGAGGUAAAAAUGXXXGCACGU-3', where XXX indicates the codon complementary to the anticodon of the A site tRNA.
The EF-Tu(H84A) plasmid containing a C-terminal histidine tag and TEV protease sequence was a generous gift of Rachel Green. It was over-expressed in BL21-Gold (DE3) cells and purified by Ni-NTA chromatography following the Qiagen procedure. The histidine tag was subsequently removed by a His-tagged TEV protease and EF-Tu(H84A) was purified from the cleaved tag with Ni-NTA agarose (Lucast et al., 2001). The purified protein was then dialyzed into RB buffer with 10% glycerol, aliquoted, flash frozen and stored at -80°C.
Purified E. coli tRNAs were purchased from Sigma or Subriden RNA. [3'-32P]labeling of tRNA and aminoacylation with purified cognate aminoacyl tRNA synthetases was performed as previously described with typical aminoacylation yields of 70% as determined by the P1 or S1 nuclease assay (Wolfson and Uhlenbeck, 2002).
Ternary Complex Binding Assay
Equilibrium binding of ternary complexes to the entry site of the ribosome was determined as previously described (Daviter et al., 2003) with several modifications: [3'-32P] labeled aa-tRNA was used with a final ternary complex concentration <1 nM. In 36 μL reactions, ternary complexes were incubated for two minutes at 20°C with 12 different ribosome concentrations ranging from 0.05 to 100 nM containing 3 fold excess mRNA and 10 fold excess tRNAfMet in the P site. 12 μL of each sample was filtered over nitrocellulose (Whatman 0.45 μm) and nylon (Amersham 0.45 μm) in duplicate and washed with 10 fold excess RB buffer (Fahlman and Uhlenbeck, 2004). At least three independent replicates were performed for each aa-tRNA. Data was quantified using a phosphoimager (Molecular Dynamics) and binding constants were determined by fitting the data to a single Michaelis Menten binding isotherm using KaleidaGraph software (Synergy Software). Experimental error included the variation between different serial dilutions and ribosome preparations.
Kinetic Experiments
The rate of GTP hydrolysis was determined essentially as previously described (Cochella and Green, 2005) with the exception that the ternary complex mix was not purified, but was passed through two P30 spin columns to remove excess [γ-32P] GTP. Six to twelve different apparent kGTP values were used to fit each kmaxGTP curve. Since each point on the kmaxGTP curve represents an independent GTP hydrolysis experiment composed of at least 12 time points, any error in the apparent kGTP rate would affect the value of kmaxGTP.
The rate of peptide bond formation was determined by first converting EF-Tu·GDP to EF-Tu·GTP by heating 0.55 μM EF-Tu, 55 μM GTP, 3 mM phosphoenolpyruvate, and 12 U/mL pyruvate kinase in RB buffer at 37°C for 20 minutes. Then ternary complex is formed by adding 0.5 mM EF-Tu·GTP to 50 nM [3'-32P] labeled aa-tRNA in RB buffer and incubating on ice for 20 minutes. 500 nM ribosomes are programmed with 1.5 μM mRNA and 1 μM fMet-tRNAfMet in the P site at 20°C. Equal volumes of ternary complex and ribosomes were mixed in a quench flow apparatus at 20°C (Kintek). Reactions were quenched in 5 mM NaOAc (pH 4.5), 100 μM EDTA. Samples were analyzed by digesting 1 μL of the quenched reaction with 4 μL of 100 U/μL nuclease S1 for 10 minutes at room temperature followed by development on PEI cellulose TLC plates or cellulose paper electrophoresis plates. TLC solvents were glacial acetic acid/1 M NH4Cl/H2O (5:10:85) for reactions with valine, alanine, glycine, and isoleucine or isopropanol/HCl/H2O (70:15:15) for phenylalanine and glutamate. Paper electrophoresis was performed at 1000V for 20 minutes in PyrAc buffer (50 mM pyridine, 3.5 M acetate, pH 3) for reactions involving histidine, lysine, and arginine. At least three independent experiments were performed for each aa-tRNA with at least 12 different time points represented on each curve. Rates were determined by fitting each curve with a single exponential equation.
Control experiments using Val-tRNAGACVal in the standard kpep assay were performed by simultaneously aminoacylating the tRNA and forming ternary complex by incubating 1 μM tRNAGACVal, 250 μM valine, 4 mM ATP, 10 U/mL pyrophosphatase, 0.5 μM valyl synthetase, 5 μM EF-Tu, 50 μM GTP, 3 μM phosphoenol pyruvate, and 12 U/mL pyruvate kinase in RB at 37°C for one hour. 2 μM heat activated ribosomes were programmed with 6 μM mRNA and 0.4 μM [35S]fMet-tRNAfMet. Reactions were performed similarly to the 32P assay and were quenched with 0.5 M KOH. Product analysis was performed by heating the samples at 37°C for ten minutes to hydrolyze the [35S]fMet or [35S]fMet-Val from the tRNA followed by cellulose paper electrophoresis at 1000V for 20 minutes in PyrAc buffer.
Acknowledgements
The project described above was supported by R01-GM37552 from the National Institutes of Health, and 1C06RR018850-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci U S A. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Beringer M, Adio S, Rodnina MV. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat Struct Mol Biol. 2006;13:423–428. doi: 10.1038/nsmb1091. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Bonekamp F, Dalbøge H, Christensen T, Jensen KF. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J Bacteriol. 1989;171:5812–5816. doi: 10.1128/jb.171.11.5812-5816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham RH. Codon context. Experientia. 1990;46:1126–1133. doi: 10.1007/BF01936922. [DOI] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Dale T, Sanderson LE, Uhlenbeck OC. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004;43:6159–6166. doi: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Ermolaeva MD. Synonymous codon usage in bacteria. Curr Issues Mol Biol. 2001;3:91–97. [PubMed] [Google Scholar]
- Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Folley LS, Yarus M. Codon contexts from weakly expressed genes reduce expression in vivo. J Mol Biol. 1989;209:359–378. doi: 10.1016/0022-2836(89)90003-x. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Huggins W, Ghosh SK, Nanda K, Wollenzien P. Internucleotide movements during formation of 16 S rRNA-rRNA photocrosslinks and their connection to the 30 S subunit conformational dynamics. J Mol Biol. 2005;354:358–374. doi: 10.1016/j.jmb.2005.09.060. [DOI] [PubMed] [Google Scholar]
- Irwin B, Heck JD, Hatfield GW. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270:22801–22806. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- Kim SH, Sussman JH, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, Rich A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974;71:4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina LG, Masson JM, Normanly J, Abelson J, Miller JH. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol. 1990;213:705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. Rna. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U, Rodnina MV. Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell. 2007;25:167–174. doi: 10.1016/j.molcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Krüger MK, Pedersen S, Hagervall TG, Sørensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol. 1998;284:621–631. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Liiv A, O'Connor M. Mutations in the intersubunit bridge regions of 23 S rRNA. J Biol Chem. 2006;281:29850–29862. doi: 10.1074/jbc.M603013200. [DOI] [PubMed] [Google Scholar]
- Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. Rna. 2007;13:1881–1886. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A, Jurnak F. Kinetic studies of Escherichia coli elongation factor Tuguanosine 5'-triphosphate-aminoacyl-tRNA complexes. Biochemistry. 1985;24:6433–6439. doi: 10.1021/bi00344a019. [DOI] [PubMed] [Google Scholar]
- Louie A, Ribeiro NS, Reid BR, Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- Lucast LJ, Batey RT, Doudna JA. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques. 2001;30:544–546. 548, 550. doi: 10.2144/01303st06. passim. [DOI] [PubMed] [Google Scholar]
- McClain WH, Schneider J, Bhattacharya S, Gabriel K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc Natl Acad Sci U S A. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F. V. t., Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- O'Connor M, Dahlberg AE. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Oleinikov AV, Perroud B, Wang B, Traut RR. Structural and functional domains of Escherichia coli ribosomal protein L7/L12. The hinge region is required for activity. J Biol Chem. 1993;268:917–922. [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. Embo J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. Embo J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery LA, Bermingham JR, Jr., Yarus M. Mutation in the D arm enables a suppressor with a CUA anticodon to read both amber and ochre codons in Escherichia coli. J Mol Biol. 1986;190:513–517. doi: 10.1016/0022-2836(86)90020-3. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. Embo J. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson LE, Uhlenbeck OC. The 51-63 base pair of tRNA confers specificity for binding by EF-Tu. Rna. 2007a doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson LE, Uhlenbeck OC. Exploring the specificity of bacterial elongation factor Tu for different tRNAs. Biochemistry. 2007b;46:6194–6200. doi: 10.1021/bi602548v. [DOI] [PubMed] [Google Scholar]
- Scarano G, Krab IM, Bocchini V, Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Lett. 1995;365:214–218. doi: 10.1016/0014-5793(95)00469-p. [DOI] [PubMed] [Google Scholar]
- Schultz DW, Yarus M. tRNA structure and ribosomal function. I. tRNA nucleotide 27-43 mutations enhance first position wobble. J Mol Biol. 1994a;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- Schultz DW, Yarus M. tRNA structure and ribosomal function. II. Interaction between anticodon helix and other tRNA mutations. J Mol Biol. 1994b;235:1395–1405. doi: 10.1006/jmbi.1994.1096. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy F. V. t., Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Smith D, Yarus M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. J Mol Biol. 1989;206:503–511. doi: 10.1016/0022-2836(89)90497-x. [DOI] [PubMed] [Google Scholar]
- Sorensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Bjork GR, Ehrenberg M. Over expression of a tRNA(Leu) isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol. 2005;354:16–24. doi: 10.1016/j.jmb.2005.08.076. [DOI] [PubMed] [Google Scholar]
- Sørensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. Embo J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Takaku H, Yokoyama S. Codon-reading specificity of an unmodified form of Escherichia coli tRNA1Ser in cell-free protein synthesis. Nucleic Acids Res. 1996;24:2894–2899. doi: 10.1093/nar/24.15.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LK, Dix DB, Thompson RC. Codon choice and gene expression: synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc Natl Acad Sci U S A. 1988;85:4242–4246. doi: 10.1073/pnas.85.12.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc Natl Acad Sci U S A. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Matsugi J, Ishikura H, Murao K. Bacillus subtilis tRNA(Pro) with the anticodon mo5UGG can recognize the codon CCC. Biochim Biophys Acta. 2005;1728:143–149. doi: 10.1016/j.bbaexp.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Yarus M, Cline S, Raftery L, Wier P, Bradley D. The translational efficiency of tRNA is a property of the anticodon arm. J Biol Chem. 1986a;261:10496–10505. [PubMed] [Google Scholar]
- Yarus M, Cline SW, Wier P, Breeden L, Thompson RC. Actions of the anticodon arm in translation on the phenotypes of RNA mutants. J Mol Biol. 1986b;192:235–255. doi: 10.1016/0022-2836(86)90362-1. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]