Summary
trkB is a tyrosine protein kinase gene highly related to trk, a proto-oncogene that encodes a receptor for nerve growth factor (NGF) and neurotrophin-3 (NT-3). trkB expression is confined to structures of the central and peripheral nervous systems, suggesting it also encodes a receptor for neurotrophic factors. Here we show that brain-derived neurotrophic factor (BDNF) and NT-3, but not NGF, can induce rapid phosphorylation on tyrosine of gp145trkB, one of the receptors encoded by trkB. BDNF and NT-3 can induce DNA synthesis in quiescent NIH 3T3 cells that express gp145trkB. Cotransfection of plasmids encoding gp145trkB and BDNF or NT-3 leads to transformation of recipient NIH 3T3 cells. In these assays, BDNF elicits a response at least two orders of magnitude higher than NT-3. Finally, 125I-NT-3 binds to NIH 3T3 cells expressing gp145trkB; binding can be competed by NT-3 and BDNF but not by NGF. These findings indicate that gp145trkB may function as a neurotrophic receptor for BDNF and NT-3.
Introduction
The trk proto-oncogene encodes a tyrosine protein kinase, gp140trk, that serves as a receptor for certain neurotrophic factors including nerve growth factor (NGF) (Hempstead et al., 1991; Kaplan et al., 1991; Klein et al., 1991) and the recently isolated neurotrophin-3 (NT-3) (Cordon-Cardo et al., 1991). At least two other mammalian genes highly related to the trk proto-oncogene have been identified (Barbacid et al., 1991). These genes, designated trkB and trkC, are expressed in multiple structures of the nervous system, suggesting that they may also encode neurogenic receptors (Klein et al., 1989, 1990a, 1990b; Middlemas et al., 1991; Lamballe et al., submitted).
The mammalian trkB locus has an intricate transcription pattern. At least six different trkB transcripts, ranging in size from 2.0 kb to 9.0 kb, have been identified in mouse brain (Klein et al., 1989, 1990a). Even more numerous transcripts have been detected in rat tissues (Middlemas et al., 1991). Preliminary sequence analysis of cDNA clones corresponding to some of these transcripts predicts the synthesis of multiple proteins. To date, two trkB gene products, designated gp145trkB and gp95trkB, have been identified (Klein et al., 1990a; Middlemas et al., 1991). The deduced amino acid sequence of gp145trkB reveals the basic features characteristic of tyrosine protein kinase receptors. Comparison of the mouse gp145trkB sequence with that of the human gp140trk receptor reveals a high degree of homology in their respective extracellular (51%) and catalytic kinase (88%) domains.
The trkB gene product gp95trkB represents a new class of noncatalytic cell surface receptors (Klein et al., 1990a; Middlemas et al., 1991). cDNA sequence analysis predicts that amino acid residues 1 to 465 of gp95trkB are identical to those of gpl45trkB. These residues encompass the signal peptide, extracellular domain, transmembrane region, and the first 12 cytoplasmic residues. However, gp95trkB has a very short cytoplasmic domain of 23 residues, of which the last 11 have no resemblance to any of the gpl45trkB sequences. Analysis of rat cDNA clones predicts the existence of a second noncatalytic isoform of almost identical sequence to gp95trkB except for its 9 carboxy-terminal residues (Middlemas et al., 1991).
In situ hybridization studies have indicated that trkB is expressed in numerous structures of the central and peripheral nervous systems (Klein et al., 1989, 1990b). During development, trkB expression can be first detected in 9.5-day-old embryos, a time when the central and peripheral nervous systems are in the process of differentiating into anatomically recognizable structures. In later-stage embryos, the pattern of trkB expression parallels the developing structures of the central and peripheral nervous systems (Klein et al., 1990b). In the adult brain, trkB mRNAs can be found in most structures, including the cortical layers, the thalamus, and the hippocampus (particularly the pyramidal cell layer), the lateral choroid plexus, and the ependymal lining of certain ventricles. A more restricted pattern of expression can be observed in the cerebellum, where only the Purkinje cell layer and the caudal peduncle have detectable levels of trkB expression (Klein et al., 1990b).
Subsequent studies utilizing probes specific for the various trkB transcripts have indicated that the catalytic (gp145trkB) and noncatalytic (gp95trkB) receptors are expressed in different structures (Klein et al., 1990a). Analysis of coronal sections of the adult mouse brain revealed gp145trkB-coding transcripts in the cortical layers, the thalamus, and the hippocampus, therefore suggesting a primary role for gp145trkB in neurons. In contrast, the noncatalytic gp95trkB receptor may not be expressed in neurons since transcripts encoding gp95trkB have been found in the choroid plexus and the ependymal cell layer of certain ventricles (Klein et al., 1990a). This pattern of expression raises the possibility that gp95trkB may play a role in the transport and/or clearance of the putative trkB ligand. The localization of transcripts encoding the various trkB receptors in the peripheral nervous system remains to be determined.
The recent identification of gp140trk as a functional receptor for NGF and NT-3 (Cordon-Cardo et al., 1991; Hempstead et al., 1991; Kaplan et al., 1991; Klein et al., 1991; Nebreda et al., 1991) has raised the possibility that at least some of the trkB products may serve as receptors for neurotrophic factors. NGF and NT-3 are members of a highly related family of peptides that also includes brain-derived neurotrophic factor (BDNF) (Barde et al., 1982; Leibrock et al., 1989) and the recently isolated neurotrophin-4 (NT-4) (Hallböök et al., 1991). As mature factors, these proteins share close to 60% amino acid identity, including their six cysteine residues that presumably confer on them related tertiary structures (Hallböök et al., 1991; Hohn et al., 1990; Jones and Reichardt, 1990; Maisonpierre et al., 1990a; Rosenthal et al., 1990). These factors, particularly BDNF and NT-3, are expressed in a wide variety of neural structures, many of which also contain trkB transcripts (Ernfors et al., 1990b; Hofer et al., 1990; Maisonpierre et al., 1990a, 1990b; Phillips et al., 1990). We undertook the present studies to examine whether any of these neurotrophic factors may serve as ligands for gp145trkB, a tyrosine protein kinase receptor encoded by the mammalian trkB gene.
Results
Coexpression of trkB with BDNF or NT-3 Induces Cell Transformation
We have recently shown that NGF, and to a lesser extent NT-3, is a mitogen for NIH 3T3 cells expressing gp140trk receptors (Cordon-Cardo et al., 1991). Moreover, NIH 3T3 cells become morphologically transformed when they express the trk proto-oncogene in the presence of exogenous NGF. Cotransfection of expression plasmids encoding the trk proto-oncogene and NGF or NT-3 also led to the morphological transformation of NIH 3T3 cells in a dose-dependent manner (Cordon-Cardo et al., 1991). These results suggest that transformation of NIH 3T3 cells can be used as an experimental assay to investigate the ligand–receptor relationships between other trk-related genes and the NGF family of neurotrophic molecules.
To test this hypothesis, we cotransfected NIH 3T3 cells with Moloney murine sarcoma virus long terminal repeat-driven mammalian expression plasmids (see Experimental Procedures) encoding gp145trkB (pFRK44) and each of the three neurotrophic factors, NGF (pLTRSNGF), BDNF (pLL42), and NT-3 (pLL43) (Table 1). As illustrated in Figure 1, cotransfection of saturating amounts of pFRK44 DNA (2 μg) with serial dilutions of pLL42 DNA resulted in the efficient transformation of NIH 3T3 cells. Quantitative analysis of these cotransfection assays indicated that BDNF can transform NIH 3T3 cells with an efficiency of 4 × 104 ffu per μg of pLL42 DNA, a transforming activity about one order of magnitude lower than that observed with trk oncogenes isolated from human tumors (Barbacid et al., 1991). The transforming activity of BDNF is entirely dependent on the expression of gp145trkB receptors, since pLL42 DNA failed to induce any detectable morphological transformation when transfected alone into NIH 3T3 cells (Table 1).
Table 1.
Transformation of NIH 3T3 cells by Cotransfection of pFRK44, an Expression Plasmid Encoding gp145trkB, with Expression Plasmids Encoding BDNF, NT-3, and NGF
| Cotransfection of pFRK44 DNAa with |
Transforming Activity (Foci per 1.5 × 105 Cells) | ||
|---|---|---|---|
| Neurotrophic Factor | Plasmidb | DNA (ng) | |
| – | – | – | 0 |
| BDNF | pLL42 | 2000 | TMTCc |
| 200 | TMTC | ||
| 20 | >500 | ||
| 2 | 80 | ||
| NT-3 | pLL43 | 2000 | 200 |
| 200 | 44 | ||
| 20 | 6 | ||
| 2 | 0 | ||
| NGF | pLTRSNGF | 2000 | 0 |
| 200 | 0 | ||
Two micrograms of pFRK44 DNA was used in each case.
Transfection of 2 μg of either pLTRSNGF, pLL42, or pLL43 alone did not induce morphological transformation of NIH 3T3 cells.
TMTC, too many to count.
Figure 1. Transformation of NIH 3T3 Cells by Cotransfection of trkB with BDNF or NT-3.
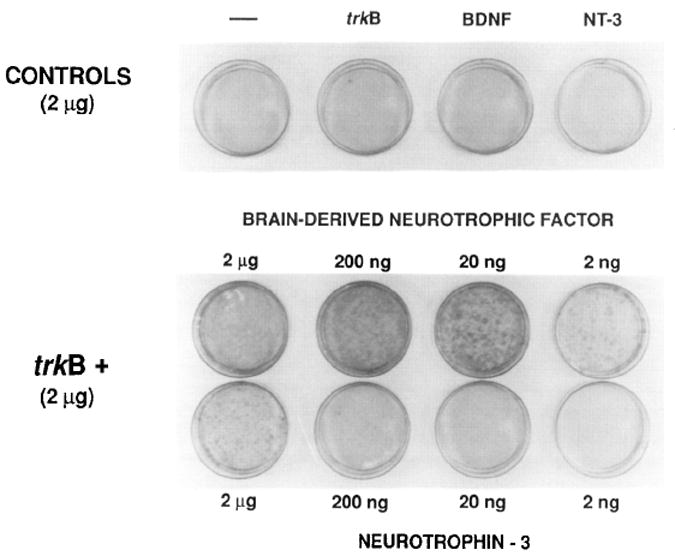
NIH 3T3 cells were transfected with saturating amounts (2 μg) of the trkB kinase expression plasmid pFRK44 in either the absence or presence of various amounts (2 μg to 2 ng) of the pLL42 (BDNF) or pLL43 (NT-3) expression plasmids. As controls, pLL42 and pLL43 DNAs (2 μg) were transfected in the absence of pFRK44 DNA. Foci of transformed cells were counted under the microscope 12 days after transfection (Table 2) and the plates subsequently stained with Giemsa.
Cotransfection of pFRK44 (trkB) and pLL43 (NT-3) DNAs also resulted in morphological transformation of the recipient NIH 3T3 cells, albeit with a 200-fold lower transformation efficiency (Table 1). Coexpression of pFRK44 and pLTRSNGF DNAs did not lead to detectable morphological transformation (Table 1). This result is in agreement with our previous report that 125I-labeled NGF does not bind to NIH 3T3 cells expressing the gp145trkB kinase (Klein et al., 1991). Next, we performed similar experiments in which NIH 3T3 cells were cotransfected with saturating amounts of each of the plasmids encoding the neurotrophic factors in the presence of various amounts of gp145trkB-coding pFRK44 DNA. As illustrated in Table 2, pFRK44 DNA was able to transform the recipient NIH 3T3 cells only in the presence of the BDNF- and NT-3-coding plasmids, pLL42 and pLL43, respectively. Quantitative analysis revealed higher levels (50- to 100 fold) of transforming activity when pFRK44 was cotransfected in the presence of BDNF (pLL42) than when cotransfected with NT-3 (pLL43) (Table 2).
Table 2.
Transformation of NIH 3T3 Cells by Cotransfection of Expression Plasmids Encoding BDNF, NT-3, and NGF with Varying Amounts of pFRK44 DNA
| Cotransfected DNA |
Transforming Activity (Foci per 1.5 × 105 Cells) | |
|---|---|---|
| Neutrophic Factor DNAa | pFRK44 DNA (ng) | |
| pLL42 (BDNF) | 100 | TMTCb |
| 10 | >350 | |
| 1 | 120 | |
| 0.1 | 9 | |
| pLL43 (NT-3) | 100 | 189 |
| 10 | 16 | |
| 1 | 3 | |
| pLTRSNGF (NGF) | 1000 | 0 |
| 100 | 0 | |
One microgram of either pLTRSNGF, pLL42, or pLL43 DNA was used in each case.
TMTC, too many to count.
Phosphorylation of gp145trkB Receptors by BDNF and NT-3
Recent studies have shown that NGF and NT-3, but not BDNF, induce the rapid phosphorylation of gpl40trk on tyrosine residues (Cordon-Cardo et al., 1991; Kaplan et al., 1991; Klein et al., 1991). To examine whether BDNF and/or NT-3 could induce a similar response with the gp145trkB kinase, we incubated Z52-17 cells, a NIH 3T3–derived cell line transfected with pFRK44 DNA, with each of the neurotrophic factors NGF, BDNF, and NT-3. BDNF and NT-3 were derived from Sf9 insect cells infected with recombinant baculoviruses or from COS cells transfected with cytomegalovirus/SV40-derived BDNF and NT-3 expression vectors (see Experimental Procedures). As shown in Figure 2, BDNF and NT-3 stimulated the rapid phosphorylation of tyrosine residues in gp145trkB receptors regardless of whether they were synthesized in mammalian (Figure 2B) or insect (Figure 2C) cells. In contrast, neither NGF nor control supernatants from COS or Sf9 cells induced detectable phosphorylating activity.
Figure 2. BDNF and NT-3 Stimulate Phosphorylation of Tyrosine Residues in the trkB Tyrosine Kinase Receptor gpl45trkB.
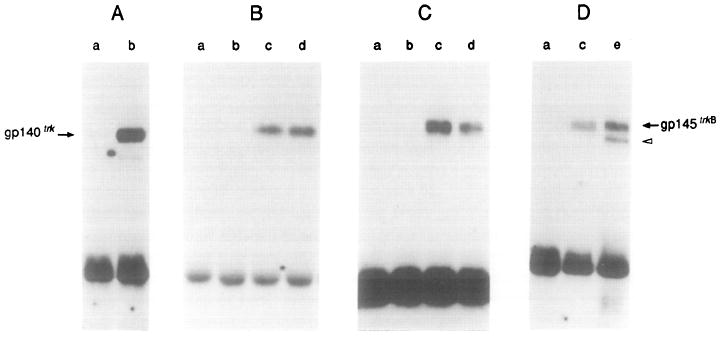
(A) gp140trk-expressing E25-48 cells incubated in the absence (lane a) or presence (lane b) of 50 ng/ml NGF.
(B) gp145trkB-expressing Z52-17 cells incubated with a 1:10 dilution of supernatant from mock-transfected COS cells (lane a), 50 ng/ml NGF (lane b), a 1:10 dilution of supernatant from COS cells transfected with pVN1 (BDNF) (lane c), or a 1:10 dilution of supernatant derived from COS cells transfected with pVN2 (NT-3) (lane d).
(C) gp145trkB-expressing Z52-17 cells incubated with supernatant from Sf9 cells infected with wild-type baculovirus (lane a) or 50 ng/ml NGF (lane b), 50 ng/ml baculovirus-derived BDNF (lane c), or 50 ng/ml baculovirus-derived NT-3 (lane d).
(D) Lane a, Z52-17 cells incubated with supernatant from Sf9 cells infected with wild-type baculovirus. Lane c, Z52-17 cells incubated with 50 ng/ml baculovirus-derived BDNF. Lane e, Z82-41 cells transformed by cotransfection of pFRK44 (trkB) and pLL42 (BDNF) DNAs in the absence of ligand.
All incubations were performed for 10 min at 37°C. Cells were lysed and incubated with rabbit polyclonal antibodies elicited against gp140trk (Martin-Zanca et al., 1989) (A) or gp145trkB (Klein et al., 1990a) (B–D). Immunoprecipitates were analyzed by 8% SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose filters, and blotted with the anti-phosphotyrosine monoclonal antibody 4G10 (UBI) as described in Experimental Procedures. Filters were exposed to Kodak X-Omat film for 24 hr at −70°C with the help of an intensifying screen. The migration of gp140trk and gp145trkB receptors is indicated by arrows. The open arrowhead indicates the migration of a phosphorylated precursor of gp145trkB.
Next, we investigated the extent of phosphorylation of gp145trkB receptors expressed in Z82-41 cells, a transformed cell line generated by cotransfection of trkB and BDNF DNAs. As shown in Figure 2D, these cells expressed constitutively phosphorylated gp145trkB receptors. The extent of phosphorylation of these receptors in the absence of ligand was at least comparable to that of gp145trkB expressed in Z52-17 cells in the presence of saturating amounts (50 ng/ml) of BDNF. These results indicate that NIH 3T3 cells transformed by cotransfection of pFRK44 and pLL42 DNAs may owe their phenotype to the constitutive activation of the gp145trkB receptor kinase by an autocrine loop. In addition to gp145trkB, Z82-41 cells contain a second molecular species constitutively phosphorylated on tyrosine residues, and this species is likely to correspond to partially glycosylated gp145trkB precursors (Figure 2D). These precursors do not become phosphorylated as a response to exogenously added ligands, suggesting that they are not located in the cell membrane (Figure 2D). Therefore, their constitutive phosphorylation in Z82-41 cells raises the possibility that BDNF can recognize trkB receptors intracellularly before they are completely processed and properly anchored in the plasma membrane. Whether this interaction elicits signal transduction remains to be determined.
Mitogenic Activity of BDNF and NT-3
We have previously shown that gp140trk receptors mediate the mitogenic activity of their cognate NGF and NT-3 ligands on NIH 3T3 cells (Cordon-Cardo et al., 1991). To examine whether BDNF and NT-3 have similar activity on cells expressing gp145trkB receptors, we incubated quiescent Z52-17 cells with partially purified BDNF and NT-3 factors derived from Sf9 insect cells infected with the corresponding recombinant baculoviruses. As controls, we utilized E25-48 cells, a NIH 3T3 cell line that expresses gp140trk receptors (Klein et al., 1991). As shown in Figure 3A, NGF, and to a lesser extent NT-3, stimulated the incorporation of 3H-labeled thymidine in these E25-48 cells. Neither BDNF nor control supernatants obtained from Sf9 cells infected with wild-type baculoviruses had any significant effect. When the same experiment was performed on Z52-l 7 cells, both BDNF and NT-3 induced DNA synthesis in a dose-dependent manner (Figure 3B). Quiescent Z52-17 cells could not be stimulated either by NGF or by control supernatants (Figure 3B). Similar results were obtained when we replaced these partially purified baculovirus-expressed factors with crude preparations derived from BDNF-and NT-3-transfected COS cells. Finally, the parental NIH 3T3 cells did not respond to any of the neurotrophic factor preparations used in these experiments (data not shown).
Figure 3. Stimulation of 3H-Labeled Thymidine Incorporation by BDNF and NT-3.
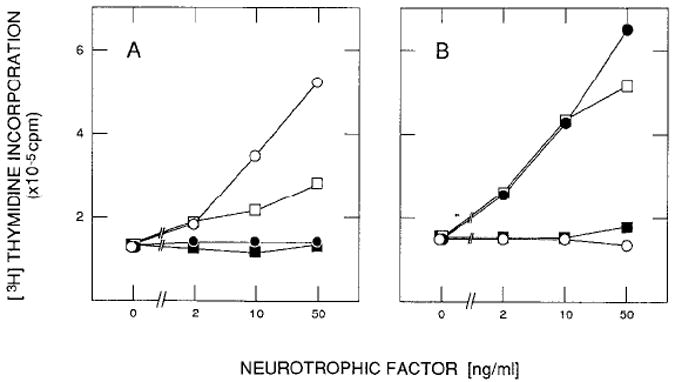
Quiescent gp140trk-expressing E25-48 cells (A) and gp145trkB-expressing Z52-17 cells (B) were incubated in DMEM containing 5 μg/ml insulin and the indicated amounts of NGF (open circles), baculovirus-derived BDNF(filled circles), baculovirus-derived NT-3 (open squares), or supernatant from wild-type baculovirus-infected Sf9 insect cells (filled squares). Incorporation of 3H-labeled thymidine into DNA was measured in duplicate samples 22 hr after stimulation as described in Experimental Procedures. Values are not corrected for DNA synthesis in untreated resting cultures.
To determine the percentage of Z52-17 cells responsive to BDNF and NT-3, we conducted DNA synthesis assays in the presence of 5-bromodeoxyuridine (BrdUrd) (Table 3). As illustrated in Figures 4A and 4B, stimulation of quiescent NIH 3T3 and Z52-17 cells with 20% calf serum induced more than 80% of these cells to enter S phase within 24 hr. Neither insulin nor NGF elicited significant DNA synthesis (Figures 4C–4F). In contrast, supernatants derived from COS cells transfected with BDNF and NT-3 expression plasmids induced 64% and 47% of Z52-17 cells to enter S phase, respectively (Figures 4H and 4J; Table 3). As negative controls, neither BDNF- nor NT-3-containing COS cell–derived supernatants had significant mitogenic activity on the parental NIH 3T3 cells (Figures 4G and 4I). Immunocytochemistry studies revealed that about 80% of Z52-17 cells expressed detectable levels of gp145trkB receptors (data not shown). Therefore, the percentage of Z52-17 cells responding to BDNF is similar to that responding to 20% calf serum.
Table 3.
Mitogenic Activity of BDNF and NT-3 on gp145trkB-Expressing Z52-17 Cells
| BrdUrd Incorporation |
||||
|---|---|---|---|---|
| NIH 3T3 Cells |
Z52-17 Cells |
|||
| Additionsa | Positive/Total | Percentage Positive | Positive/Total | Percentage Positive |
| None | 16/421 | 3.8% | 41/372 | 10.9% |
| 20% Calf serum | 525/630 | 83.3% | 532/602 | 88.3% |
| BDNF (COS cells) | 30/254 | 11.8% | 186/289 | 64.3% |
| BDNF (Sf9 cells) | 20/396 | 5.0% | 151/447 | 33.7% |
| NT-3 (COS cells) | 31/349 | 8.8% | 97/206 | 47.0% |
| NT-3 (Sf9 cells) | 19/393 | 4.8% | 186/576 | 32.2% |
| NGF | 13/226 | 5.7% | 39/280 | 13.9% |
All samples contained 5 μg/ml insulin. BDNF and NT-3 derived from COS cells were used as 1:10 dilutions of cell free supernatants. BDNF and NT-3 purified from Sf9 cells were used at final concentrations of 50 ng/ml. NGF (UBI) was also used at a final concentration of 50 ng/ml.
Figure 4. Induction of DNA Synthesis by BDNF and NT-3.
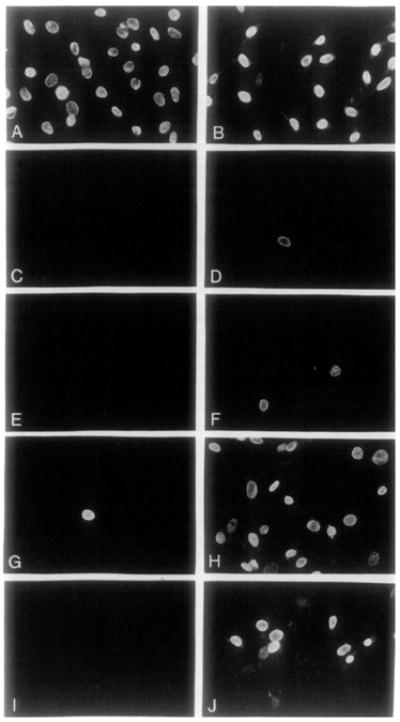
Quiescent NIH 3T3 (left-hand panels) and gpl45trkB-expressing Z52-17 cells (right-hand panels) were incubated in the presence of 100 μM BrdUrd plus 5 μg/ml insulin and the following additions: 20% calf serum (A, B), no additions(C, D), 50 ng/ml NGF(E, F), a1:10 dilution of supernatants derived from COS cells expressing BDNF (G, H), or a 1:10 dilution of supernatants derived from COS cells expressing NT-3 (I, J). Cells were fixed after 24 hr and analyzed for immunofluorescence using a mouse anti-BrdUrd monoclonal antibody and donkey anti-mouse IgG conjugated with Texas red. Final magnification 290 x.
The more limited mitogenic effects of NT-3 on NIH 3T3 cells expressing gp145trkB receptors (Table 3) is in agreement with the results obtained in transformation assays. Purified preparations (50 ng/ml) of baculovirus-derived BDNF and NT-3 also had mitogenic effects, although they appeared to be somewhat less efficient. Whether this difference is due to their processing in Sf9 insect cells or to loss of activity during purification remains to be determined.
Binding of 125I-Labeled NT-3 to gp145trkB
The above results suggest that gpl45trkB is a functional receptor for BDNF and NT-3. However, formal proof of a direct interaction between these molecules and the gp145trkB kinase requires binding studies. To this end, we iodinated purified BDNF and NT-3 proteins isolated from baculovirus-infected Sf9 cells as indicated in Experimental Procedures. NT-3 was efficiently iodinated using the Bolton-Hunter reagent, which primarily labels lysine residues. Other iodination methods that lead to the specific labeling of tyrosine residues yielded less efficient results. NGF became equally well labeled with either method. Unfortunately, parallel efforts to iodinate the BDNF protein have been unsuccessful thus far. Therefore, we had to limit our direct binding studies to the NT-3 protein.
As shown in Figure 5, incubation of NIH 3T3 cells expressing gp140trk (E25-48 cells) or gp145trkB (B35-41 and Z52-17 cells) receptors with 1 nM 125I-labeled NT-3 at 4 °C resulted in its efficient binding. Moreover, a significant fraction of the observed 125I-labeled NT-3 binding could be readily competed by a 75-fold excess of unlabeled NT-3. In contrast, the parental NIH 3T3 cells exhibited very limited binding (less than 20% of that bound to E25-48, B35-41, or Z52-17 cells), none of which could be competed with a similar excess of unlabeled neurotrophic factor (Figure 5).
Figure 5. Binding of NT-3 to Cells Expressing trk and trkB Tyrosine Protein Kinases.
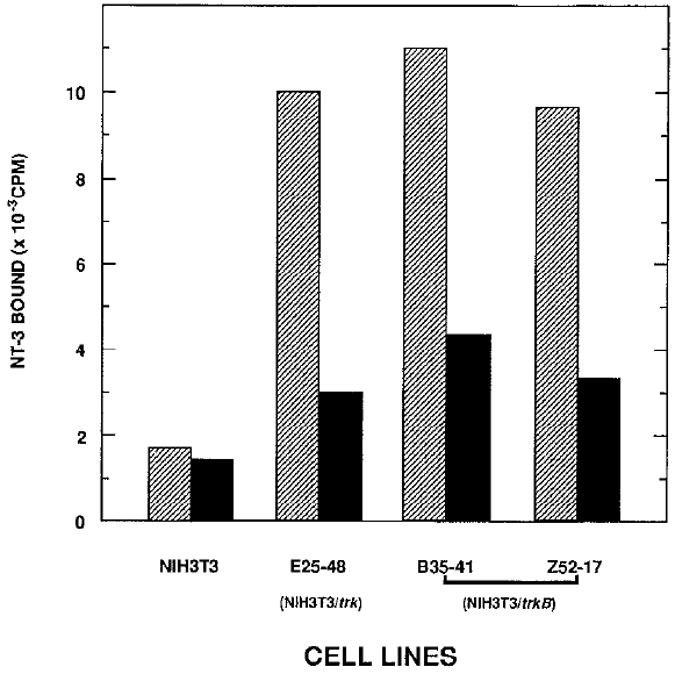
Cells, including parental NIH 3T3 cells and NIH 3T3 cells expressing the human trk (E25-48) and mouse trkB (B35-41, Z52-17) proto-oncogenes, were incubated with 1 nM 125I-labeled NT-3 at 4°C for 2 hr in the absence (hatched bars) or presence (filled bars) of 75 nM purified unlabeled NT-3. Unbound NT-3 was removed at the end of the incubation, and the radioactivity associated with the cells was measured as described in Experimental Procedures. Results are expressed as the mean of duplicate samples.
To overcome the unavailability of 125I-labeled BDNF, we utilized competitive binding assays to determine whether BDNF may also bind to gpl40trk and/or gpl45trkB receptors. In agreement with our previous studies, NGF but not BDNF could displace binding of 125I-labeled NT-3 to gp140trk-expressing E25-48 cells (data not shown). These results add further support to the concept that BDNF does not interact with gp140trk receptors. In contrast, BDNF readily displaced 125I-labeled NT-3 from binding to gp145trkB-expressing B35-41 cells (Figure 6). Moreover, BDNF was more effective in inhibiting 125I-labeled NT-3 binding than unlabeled NT-3. Similar results were obtained with Z52-17 cells (data not shown). These observations indicate that BDNF may bind to gp145trkB receptors with higher affinity than NT-3. Finally, NGF failed to displace 125I-labeled NT-3 from binding to B35-41 cells (Figure 6), a result in agreement with our previous observations that 125I-labeled NGF does not bind to cells expressing the gp145trkB kinase (Klein et al., 1991).
Figure 6. BDNF Competes with 125I-Labeled NT-3 for Binding to gp145trkB Receptors.
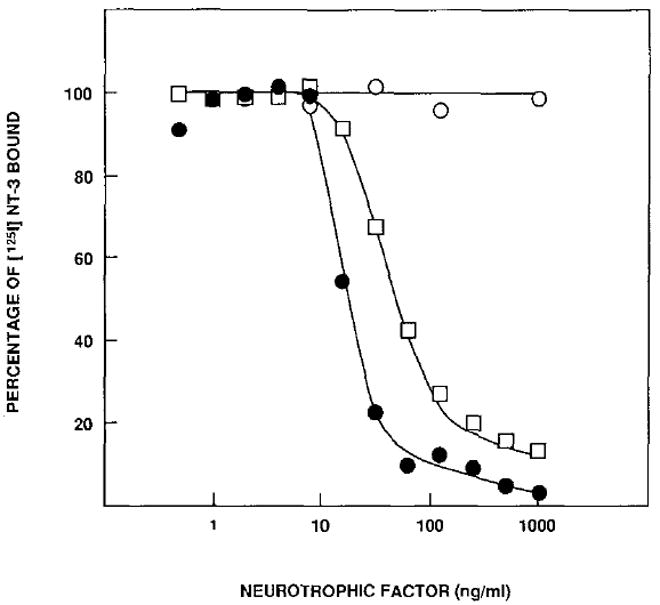
B35-41 cells were preincubated with unlabeled NGF (open circles), purified NT-3 (open squares), or purified BDNF (filled circles) at 4°C for 1 hr. 125I-labeled NT-3 was added to a final concentration of 5 ng/ml, and the cells were incubated for an additional 2 hr. The percentage of 125I-labeled NT-3 bound to the cells was determined as described in Experimental Procedures. Nonspecific binding to control NIH 3T3 cells (≤ 10%) was subtracted from the results presented. The data shown here represent the average of two separate experiments with duplicate samples.
Discussion
Transfection of NIH 3T3 cells with expression plasmids encoding gpl40trk and its cognate ligands NGF and NT-3 results in transformation of the recipient cells (Cordon-Cardo et al., 1991). In the present studies, we have taken advantage of the mitogenic properties of the trk family of receptors to establish a functional ligand-receptor relationship between gp145trkB and the neurotrophic factors BDNF and NT-3. We have demonstrated that constitutive expression of either BDNF or NT-3 in NIH 3T3 cells expressing the gp145trkB kinase induces their morphological transformation. These transformation assays may prove useful to identify receptors for novel neurotrophic factors such as the recently isolated neurotrophin-4 (Hallböök et al., 1991). Moreover, they may help to identify the ligands of orphan neurogenic receptors such as the members of the elkleck family of tyrosine kinases (Letwin et al., 1988; Lindberg and Hunter, 1990; Lai and Lemke, 1991).
The identification of the gp145trkB tyrosine kinase as a receptor for BDNF is supported by additional experimental evidence including the following: first, the induction of DNA synthesis by BDNF in quiescent NIH 3T3 cells expressing gp145trkB receptors but not in their parental NIH 3T3 cells; second, the rapid phosphorylation on tyrosine residues of gp145trkB molecules in cells exposed to BDNF; third, the constitutive tyrosine phosphorylation of gp145trkB in cells coexpressing BDNF; and fourth, the efficient displacement of 125I-labeled NT-3 from binding to cells expressing gp145trkB receptors. These results, taken together, firmly establish gp145trkB as a functional receptor for BDNF.
In addition, gpl45trkB is a receptor for NT-3. 125I-labeled NT-3 binds efficiently to cells expressing gp145trkB and induces its rapid phosphorylation on tyrosine residues. Moreover, the interaction of NT-3 with gpl45trkB receptors has functional relevance since NT-3 can induce quiescent cells expressing gp145trkB to enter S phase. However, the mitogenic activity of NT-3 on these cells appears to be more limited than that of BDNF since NT-3 exhibits 100- to 200-fold lower transforming activity when coexpressed with gpl45trkB receptors. It could be argued that some of these differences may reflect less efficient expression of NT-3 in NIH 3T3 cells. However, we have recently shown that coexpression of NT-3 with trkC receptors induces the transformation of NIH 3T3 cells with an efficiency comparable to that of BDNF when cotransfected with gp145trkB (Lamballe et al., submitted). Therefore, NT-3 is likely to mediate its biological responses through trkB as well as trkC receptors.
Each of the members of the NGF family of neurotrophins binds to the previously identified low affinity NGF receptor, gp80LNGFR (Rodriguez-Tébar and Barde, 1988; Ernfors et al., 1990a; Hallböök et al., 1991). However, we have shown that NIH 3T3 cells do not express these receptors (Klein et al., 1991). Therefore, the mitogenic properties of BDNF and NT-3 reported in this study are likely to be mediated solely by gp145trkB receptors. Our results, however, do not exclude the possibility that gp145trkB may form complexes eitherwith gp80LNGFR (Hempstead et al., 1991) or with other molecules to mediate the neurotrophic effects of BDNF and NT-3. Whether such multireceptor complexes exist and whether they modulate the neurotrophic activities of BDNF and NT-3 are interesting possibilities that should be explored.
The trkB gene is expressed in most structures of the peripheral and central nervous systems of the developing mouse embryo (Klein et al., 1989, 1990b). In the adult, available information regarding trkB expression is limited to the dorsal region of the cerebrum and certain areas of the cerebellum (Klein et al., 1990a, 1990b). In situ hybridization analysis conducted with probes specific for the various trkB transcripts has indicated that gp145trkB receptors are primarily expressed in the cerebral cortex, hippocampus, and thalamus. Other structures such as the choroid plexus and the ependymal layer of the ventricles contain transcripts encoding the noncatalytic gp95trkB protein (Klein et al., 1990a). In addition, trkB transcripts have been identified in a subset of sensory neurons that are known to respond to BDNF and NT-3. BDNF transcripts are also widely distributed in the adult brain (Ernfors et al., 1990b; Hofer et al., 1990; Maisonpierre et al., 1990a; Phillips et al., 1990). The highest levels of expression have been observed in the hippocampus (both pyramidal and granular neurons), cortex, amygdala, and the thalamic and hypothalamic nuclei. In the cerebellum, BDNF appears to be expressed in the internal granular layer (Hofer et al., 1990), whereas trkB transcripts are most prominent in the Purkinje cell layer (Klein et al., 1990b). On the other hand, NT-3 is expressed in a wide variety of tissues (Hohn et al., 1990; Maisonpierre et al., 1990a; Rosenthal et al., 1990). Whether in some of these tissues NT-3 interacts with receptors other than those encoded by trkB and trkC (Lamballe et al., submitted) remains to be determined.
The biological properties of BDNF appear to be as diverse as its pattern of expression. For instance, BDNF promotes the survival and differentiation of developing neurons of the dorsal root ganglion (Lindsay et al., 1985; Kalcheim et al., 1987). In addition, BDNF stimulates the survival as well as the differentiation of septal cholinergic neurons, which are known to degenerate in Alzheimer’s disease (Alderson et al., 1990; Knüsel et al., 1991). BDNF also promotes the survival of dopaminergic neurons of the substantia nigra and appears to protect them against selective destruction by 1-methyl-4-phenylpyridinium, a chemical known to cause Parkinson’s disease in humans (Hyman et al., 1991; Knüsel et al., 1991). In contrast, NT-3 appears to have a more limited range of action since it supports the survival of sensory and sympathetic but not of cholinergic or dopaminergic neurons (Hohn et al., 1990; Maisonpierre et al., 1990a; Rosenthal et al., 1990). The identification of the gp145trkB tyrosine kinase as a functional receptor for BDNF and NT-3 should make it possible to identify the primary cellular targets of this neurotrophic factor and help to elucidate its role in the normal brain as well as its potential involvement in neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases.
Experimental Procedures
Cells and Expression Plasmids
Cells, including NIH 3T3 (Jainchill et al., 1969) B35-41 and E25-48 (Klein et al., 1991), Z52-17, and Z82-41 cells, were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% calf serum. Z52-17 cells were obtained by transfecting NIH 3T3 cells with pFRK44, a pMEXneo-derived expression plasmid (Martin-Zanca et al., 1989) containing the mouse trkB cDNA encoding the catalytic gp145trkB protein (Klein et al., 1989). Z82-41 cells were generated by cotransfection of NIH 3T3 cells with pFRK44 and pLL42 DNAs. pLL42 is a pMEX-derived expression plasmid (Martin-Zanca et al., 1989) that contains an 850 bp insert encoding the human prepro-BDNF(Jones and Reichardt, 1990). pLL43 is a similar pMEX-derived plasmid that contains a 930 bp insert corresponding to the human prepro-NT-3 (Jones and Reichardt, 1990). pLTRSNGF is a pBR322-based expression plasmid (Izant and Weintraub, 1984) containing a 960 bp insert encoding the mouse prepro-NGF flanked by the Moloney murine sarcoma virus long terminal repeat and the SV40 polyadenylation signal (Clegg and Reichardt, unpublished data). pVN1 and pVN2 expression plasmids were generated by amplifying the corresponding inserts of pLL42 and pLL43 and subcloning them into pCM7B, a COS cell expression vector that contains the cytomegalovirus enhancer/promoter element and the SV40 polyadenylation signal (Seed, 1987). Gene transfer assays in NIH 3T3 cells were performed by the calcium phosphate precipitation technique, as previously described (Graham and van der Eb, 1973).
Neurotrophic Factors
Murine 2.5S NGF was purchased from Upstate Biotechnology, Inc. BDNF and NT-3 were purified from supernatants of Sf9 insect cells infected with recombinant baculoviruses pAcS27 (BDNF) and pAcS28 (NT-3), as recently described (Cordon-Cardo et al., 1991). Purified BDNF and NT-3 were approximately 95% pure as judged by Coomassie blue–stained SDS–polyacrylamide gels, and amino acid sequencing showed that NT-3 was properly processed at the amino terminus (Cordon-Cardo et al., 1991). Transient expression of BDNF and NT-3 using a COS cell expression system was performed essentially as described (Seed and Aruffo, 1987). Approximately 5 × 106 COS cells were transfected with 10 μg of pVNl (BDNF) and pVN2 (NT-3) DNAs and 24 hr later split at a 1:2 dilution. After an additional 24 hr incubation in DMEM containing 10% calf serum, cells were placed in serum-free DMEM containing 5 μg/ml insulin and transferrin (Sigma). Cell-free supernatants were harvested 4 days later.
Immunoblotting
Detection of phosphorylated tyrosine residues in gp140trk and gp145trkB receptors was carried out by Western blot analysis as previously described (Klein et al., 1991). Cell extracts were immunoprecipitated with a rabbit polyclonal antiserum raised against a peptide corresponding to the 14 carboxy-terminal amino acid residues of the human trk proto-oncogene product (Martin-Zanca et al., 1989) or against the mouse trkB tyrosine kinase domain expressed in bacteria (Klein et al., 1990a). Mouse anti-phosphotyrosine monoclonal antibody 4G10 (UBI) was then used as primary antibody after transfer of the immunoprecipitated proteins onto nitrocellulose filters. Filters were subsequently incubated with rabbit anti-mouse IgG (Dako) before probing with 125I-labeled protein A.
DNA Synthesis
Induction of DNA synthesis in quiescent cells following stimulation by neurotrophic factors was measured by incorporation of 3H-labeled thymidine into newly synthesized DNA as previously described (Cordon-Cardo et al., 1991). The percentage of cells entering S phase was determined by immunofluorescence assays on cells incubated in the presence of BrdUrd (Cordon-Cardo et al., 1991). Fixed cells were incubated with a 1:50 dilution of a mouse monoclonal anti-BrdUrd antibody (Becton Dickinson), washed with PBS, and subsequently incubated with a 1:50 dilution of a donkey polyclonal anti-mouse IgG antiserum conjugated with Texas red (Amersham).
Binding Assays
Purified baculovirus-expressed NT-3 was radioiodinated using 125I-labeled Bolton-Hunter reagent (2 mCi per reaction; 4000 Ci/mmol; Du Pont) as previously described (Rodriguez-Tébar and Barde, 1988). The labeled protein was separated by gel filtration through a G-25 column (medium) preequilibrated with 50 mM sodium acetate (pH 4.0) containing 150 mM NaCl and 0.5% (wt/vol) BSA and was eluted with the same buffer at a flow rate of 0.2–0.25 ml/min. Fractions of ~0.5 ml were collected, and the radioactivity in each fraction was determined in an Atomlab 100 gamma counter. The fractions containing the protein (3–4 fractions) were pooled. Analysis of the purity of 125I-labeled NT-3 by 20% SDS–polyacrylamide gel electrophoresis did not reveal other significant protein species present in the NT-3 preparation.
Competition binding assays were done essentially as previously described (Jing et al., 1990; Klein et al., 1991). Cells (2 × 105 per sample) were preincubated with various concentrations (0.5 ng/ml to 1 μg/ml) of unlabeled factors at 4°C for 1 hr. The same volume of 125I-labeled NT-3 was added to each sample to reach a final concentration of 5 ng/ml. Cells were further incubated at 4 °C for 2 hr, washed four times with ice-cold DMEM containing 0.1% BSA, and lysed in 100 μl of 1 N NaOH, and radioactivity was counted in a G5500 gamma counter (Beckman).
Acknowledgments
We are indebted to Drs. J. I. Tu and H. M. Tsay for allowing us to use their facilities to iodinate the neurotrophic factors. We also wish to thank Mr. Ed O’Rourke, Ms. Linda K. Long, and Ms. Caroline Zarou for excellent technical assistance and Dr. A. Aruffo for providing the COS expression vector and his advice with the COS cell expression system. K. R. J. is supported by an American Cancer Society postdoctoral fellowship. L. F. R. is an Investigator of the Howard Hughes Medical Institute.
References
- Alderson RF, Alterman AL, Barde Y-A, Lindsay RM. Brain-derived neurotrophic factor increases survival and differentiated functions of rat septal cholinergic neurons in culture. Neuron. 1990;5:297–306. doi: 10.1016/0896-6273(90)90166-d. [DOI] [PubMed] [Google Scholar]
- Barbacid M, Lamballe F, Pulido D, Klein R. The trk family of tyrosine protein kinase receptors. Biochim Biophys Acta Rev Cancer. 1991 doi: 10.1016/0304-419x(91)90010-i. in press. [DOI] [PubMed] [Google Scholar]
- Barde Y-A, Edgar D, Thoenen H. Purification of a new neurotrophic factor from mammalian brain. EMBO J. 1982;1:549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Cardo C, Tapley P, Jing S, Nanduri V, O’Rourke E, Lamballe F, Kovary K, Klein R, Jones KR, Reichardt LF, Barbacid M. The trk tyrosine protein kinase mediates the mitogenic properties of nerve growth factor and neurotrophin-3. Cell. 1991;66:173–l83. doi: 10.1016/0092-8674(91)90149-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Ibáñez CF, Ebenndal T, Olson L, Persson H. Molecular cloning and neurotrophic activities of a protein with structural similarities to nerve growth factor: developmental and topographical expression in the brain. Proc Natl Acad Sci USA. 1990a;87:5454–5458. doi: 10.1073/pnas.87.14.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Wetmore C, Olson L, Persson H. Identification of cells in rat brain and peripheral tissues expressing mRNA for members of the nerve growth factor family. Neuron. 1990b;5:511–526. doi: 10.1016/0896-6273(90)90090-3. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hallböök F, Ibáñez CF, Persson H. Evolutionary studies of the nerve growth factor family reveal a novel member abundantly expressed in Xenopus ovary. Neuron. 1991;6:845–858. doi: 10.1016/0896-6273(91)90180-8. [DOI] [PubMed] [Google Scholar]
- Hempstead BL, Martin-Zanca D, Kaplan DR, Parada LF, Chao MV. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature. 1991;350:678–683. doi: 10.1038/350678a0. [DOI] [PubMed] [Google Scholar]
- Hofer M, Paglisui SR, Hohn A, Leibrock J, Barde Y-A. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9:2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn A, Leibrock J, Bailey K, Barde Y-A. Identification and characterization of a novel member of the nerve growth factor/brain-derived neurotrophic factor family. Nature. 1990;344:339–341. doi: 10.1038/344339a0. [DOI] [PubMed] [Google Scholar]
- Hyman C, Hofer M, Barde Y-A, Juhasz M, Yancopoulos GD, Squinto SP, Lindsay RM. BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature. 1991;350:230–232. doi: 10.1038/350230a0. [DOI] [PubMed] [Google Scholar]
- Izant JG, Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984;36:1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- Jainchill JL, Aaronson SA, Todaro GJ. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing S, Spencer T, Miller K, Hopkins C, Trowbridge IS. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J Cell Biol. 1990;110:283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KR, Reichardt LF. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci USA. 1990;87:8060–8064. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalcheim C, Barde Y-A, Thoenen H, Le Douarin NM. In vivo effect of brain-derived neurotrophic factor on the survival of developing dorsal root ganglion cells. EMBO J. 1987;6:2871–2873. doi: 10.1002/j.1460-2075.1987.tb02589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Martin-Zanca D, Parada LF. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature. 1991;350:158–160. doi: 10.1038/350158a0. [DOI] [PubMed] [Google Scholar]
- Klein R, Parada LF, Coulier F, Barbacid M. trkB, a novel tyrosine protein kinase receptor expressed during mouse neural development. EMBO J. 1989;8:3701–3709. doi: 10.1002/j.1460-2075.1989.tb08545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R, Conway D, Parada LF, Barbacid M. The trkB tyrosine protein kinase gene codes for a second neurogenic receptor that lacks the catalytic kinase domain. Cell. 1990a;61:647–656. doi: 10.1016/0092-8674(90)90476-u. [DOI] [PubMed] [Google Scholar]
- Klein R, Martin-Zanca D, Barbacid M, Parada LF. Expression of the tyrosine kinase receptor gene trkB is confined to the murine embryonic and adult nervous system. Development. 1990b;109:845–850. doi: 10.1242/dev.109.4.845. [DOI] [PubMed] [Google Scholar]
- Klein R, Jing S, Nanduri V, O’Rourke E, Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- Knüsel B, Winslow JW, Rosenthal A, Burton LE, Seid DP, Nikolics K, Hefti F. Promotion of central cholinergic and dopaminergic neuron differentiation by brain-derived neurotrophic factor but not neurotrophin-3. Proc Natl Acad Sci USA. 1991;88:961–965. doi: 10.1073/pnas.88.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Lemke G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron. 1991;6:691–704. doi: 10.1016/0896-6273(91)90167-x. [DOI] [PubMed] [Google Scholar]
- Leibrock J, Lottspeich F, Hohn A, Hofer M, Hengerer B, Masiakowski P, Thoenen H, Barde Y-A. Molecular cloning and expression of brain-derived neurotrophic factor. Nature. 1989;341:149–152. doi: 10.1038/341149a0. [DOI] [PubMed] [Google Scholar]
- Letwin K, Yee S-P, Pawson T. Novel protein-tyrosine kinase cDNAs related to fpslfes and eph cloned using anti-phosphotyrosine antibody. Oncogene. 1988;3:621–678. [PubMed] [Google Scholar]
- Lindberg RA, Hunter T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the ephlelk family of protein kinases. Mol Cell Biol. 1990;10:6316–6324. doi: 10.1128/mcb.10.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay RM, Thoenen H, Barde Y-A. Placode and neural crest-derived sensory neurons are responsive at early developmental stages to brain-derived neurotrophic factor. Dev Biol. 1985;112:319–328. doi: 10.1016/0012-1606(85)90402-6. [DOI] [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Squinto S, Ip NY, Furth ME, Lindsay RM, Yancopoulos GD. Neurotrophin-3: a neurotrophic factor related to NGF and BDNF. Science. 1990a;247:1446–1451. doi: 10.1126/science.247.4949.1446. [DOI] [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Friedman B, Adlerson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990b;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol. 1989;9:24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middlemas DS, Lindberg RA, Hunter T. trkB, a neural receptor protein-tyrosine kinase: evidence for a full-length and two truncated receptors. Mol Cell Biol. 1991;11:143–153. doi: 10.1128/mcb.11.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebreda AR, Martin-Zanca D, Kaplan DR, Parada LF, Santos E. Induction by NGF of meiotic maturation of Xenopus oocytes expressing the trk protooncogene product. Science. 1991;252:558–561. doi: 10.1126/science.1850550. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Hains JM, Laramee GR, Rosenthal A, Winslow JW. Widespread expression of BDNF but not NT-3 by target areas of basal forebrain cholinergic neurons. Science. 1990;250:290–294. doi: 10.1126/science.1688328. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tébar A, Barde Y-A. Binding characteristics of brain-derived neurotrophic factor to its receptors on neurons from the chick embryo. J Neurosci. 1988;8:3337–3342. doi: 10.1523/JNEUROSCI.08-09-03337.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal A, Goeddel DV, Nguyen T, Lewis M, Shih A, Laramee GR, Nikolics K, Winslow JW. Primary structure and biological activity of a novel human neurotrophic factor. Neuron. 1990;4:767–773. doi: 10.1016/0896-6273(90)90203-r. [DOI] [PubMed] [Google Scholar]
- Seed B. An LFA-3 cDNA encodes a phospholipid-linked membrane protein homologous to its receptor CD2. Nature. 1987;329:840–842. doi: 10.1038/329840a0. [DOI] [PubMed] [Google Scholar]
- Seed B, Aruffo A. Molecular cloning of the CD2 antigen, the T cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]


