Abstract
The alanine helix provides a model system for studying the energetics of interaction between water and the helical peptide group, a possible major factor in the energetics of protein folding. Helix formation is enthalpy-driven (−1.0 kcal/mol per residue). Experimental transfer data (vapor phase to aqueous) for amides give the enthalpy of interaction with water of the amide group as ≈−11.5 kcal/mol. The enthalpy of the helical peptide hydrogen bond, computed for the gas phase by quantum mechanics, is −4.9 kcal/mol. These numbers give an enthalpy deficit for helix formation of −7.6 kcal/mol. To study this problem, we calculate the electrostatic solvation free energy (ESF) of the peptide groups in the helical and β-strand conformations, by using the delphi program and parse parameter set. Experimental data show that the ESF values of amides are almost entirely enthalpic. Two key results are: in the β-strand conformation, the ESF value of an interior alanine peptide group is −7.9 kcal/mol, substantially less than that of N-methylacetamide (−12.2 kcal/mol), and the helical peptide group is solvated with an ESF of −2.5 kcal/mol. These results reduce the enthalpy deficit to −1.5 kcal/mol, and desolvation of peptide groups through partial burial in the random coil may account for the remainder. Mutant peptides in the helical conformation show ESF differences among nonpolar amino acids that are comparable to observed helix propensity differences, but the ESF differences in the random coil conformation still must be subtracted.
Baker and Hubbard (1) found that water molecules cluster around peptide C⩵O groups in protein x-ray structures. Ben-Naim (2) and Honig and coworkers (3, 4) used model compound data to argue that interaction of water with peptide C⩵O groups in protein secondary structures should be an important factor in the energetics of protein folding. If the free energy of the interaction is as small as −0.5 kcal/mol, and if half the peptide groups in a 100-residue protein are stripped of water molecules when folding is complete, then breaking the interactions between water and peptide C⩵O groups should cost 25 kcal. If some of these water-peptide interactions are not broken in forming a molten globule folding intermediate, they would represent a very important source of stabilizing free energy.
The alanine peptide helix provides a suitable model system for determining the strength of the interaction between water and the helical peptide group and for examining the effects of side chains on this interaction. Modeling studies indicate that side chains can block the access of water to backbone C⩵O groups in a helix (5, 6) in a side chain-specific and rotamer-specific (6) manner. Formation of the alanine helix is known to be enthalpy-driven with an enthalpy change of −1.0 kcal/mol per residue (7, 8). A high-resolution x-ray structure of an alanine peptide helix is available (9). We show below that if amides such as N-methylacetamide are used as models for the free peptide group in an unfolded alanine peptide, then the enthalpy of the peptide hydrogen bond is not sufficient to drive helix formation, and some additional source of enthalpy is needed. The enthalpy of interaction between amides and water is large (≈−11.5 kcal/mol, Table 1), considerably larger than the enthalpy computed by quantum mechanics for the helical peptide hydrogen bond in the gas phase (−4.9 kcal/mol, ref. 10). Thus, the missing enthalpy needed for helix formation is −11.5–1.0 + 4.9 = −7.6 kcal/mol. An important component of the missing enthalpy is probably the enthalpy of interaction between water and the peptide groups in the helix.
Table 1.
Gas → water transfer energetics for amides: Nonpolar and electrostatic contributions to the enthalpy and standard free energy
| Solvation energies†, kcal/mol | Acetamide | N-methyl acetamide | N,N-dimethyl acetamide | Propionamide |
|---|---|---|---|---|
| ΔGobs‡ | −9.7 | −10.07 | −8.54 | −9.38 |
| ΔGvdw+cavity‡ | 1.95 | 2.16 | 2.32 | 2.12 |
| ΔGpol‡ | −11.65 | −12.23 | −10.86 | −11.50 |
| ΔG*es‡ | −11.75 | −12.20 | ND | −11.57 |
| ΔH*obs‡ | −16.32 | −17.07 | −16.56 | −17.45 |
| ΔHvdw+cavity‡ | −4.67 | −5.58 | −6.25 | −5.41 |
| ΔHpol‡ | −11.65 | −11.49 | −10.31 | −12.04 |
| Polar ASA (Å2)§ | 95 | 51 | 32 | 88 |
| Nonpolar ASA (Å2) | 97 | 173 | 217 | 130 |
ND, not determined. ASA, accessible surface area.
The units are kcal/mol, and the temperature is 25°C.
ΔG is the standard Gibbs free energy of transfer from the gas phase to the dilute aqueous solution (or the solvation free energy), and ΔH is the corresponding enthalpy of transfer, or solvation enthalpy; both are referred to the standard state of Ben-Naim (17), (see also refs. 15 and 16). ΔGpol and ΔHpol are the polar contributions to the solvation free energy and enthalpy, respectively. They are found by subtracting the sum of the cavity and van der Waals terms from the solvation free energy of enthalpy. ΔG*es is the calculated value, based on delphi and the parse parameter set, of the ESF. The transfer free energy data from amides are from Wolfenden (17) and Wolfenden et al. (18). The enthalpy data for amides are from Della Gatta et al. (19). The alkane transfer data, which are used to give the sum of the cavity and van der Waals terms, are from Cabani et al. (42) and Ben-Naim and Marcus (43). The equations used to fit the alkane data are ΔH = 0.101 − 0.0278x and ΔG = 0.727 + 0.0064x, where x is ASA in Ȧ2; the data point for methane was not used in fitting in both cases.
Values of polar and nonpolar ASA were calculated with naccess 2.1 (S. Hubbard and J. Thornton, University College, London) by using a probe radius of 1.4 A° and van der Waals radii from Chothia (44).
To study this problem, we calculate values of the electrostatic solvation free energy (ESF), by using the delphi program and the parse parameter set of partial charges and Born radii (11), of the peptide groups in alanine peptides in helical, β-strand, and random coil conformations, and also in mutant peptides. The delphi program treats the solvent as a continuum of high dielectric constant and the protein as a low dielectric cavity with fixed real and partial charges and uses a finite difference method to solve the Poisson-Boltzmann equation. This approach to determining peptide solvation has the attractive feature that there are no adjustable parameters except for peptide geometry, once the decision is made to use the parse parameters. Comparing delphi and a second, independent method based on Langevin dipoles (12) gives ESF values for tripeptides that agree within 1–2 kcal/mol when the same partial charges are used in both methods (13). The partial charges and radii in the parse parameter set reproduce closely the solvation free energies of the set of 67 model compounds on which they are based (11); they may be regarded as empirical parameters, useful for predicting solvation free energies for new compounds of the same type (here, peptide groups) but not to be used for other purposes. The set of model compounds used to formulate the parse charge set does not yet include peptides, although it does include amides. The procedure used to subtract the cavity and van der Waals contributions from the experimental solvation free energy affects the values assigned to the parse charges. Before presenting electrostatic calculations of solvation for peptide groups in alanine peptides, we first consider simple amides for which both experimental and theoretical data for solvation free energies are available.
Amides as Models for the Interaction with Water of the Free Peptide Group
Transfer data (vapor phase to aqueous) are available for both the enthalpy and free energy of interaction of the amide group with water. The experimentally observed transfer quantities are called solvation free energy and enthalpy after they have been corrected to the standard state introduced by Ben-Naim (14), which takes account of the different volumes available to a solute molecule in the gas and liquid phases. The standard state correction is large in the case of free energy but modest in the case of enthalpy (see the discussion by Lee, refs. 15 and 16). Amides have long been used as models for the interaction of water with the free peptide group (17), and earlier studies reveal that amides interact very strongly with water. Wolfenden and coworkers (17, 18) succeeded in measuring directly the free energy of transfer of amides from the vapor phase to aqueous solution by a procedure based on isotopic labeling and detection. The experimental solvation enthalpies of amides (19) are determined by combining results from separate experiments, yielding the heat of solution and the heat of vaporization.
The enthalpy of interaction between the amide polar groups and water is the largest single factor when either the solvation free energy or enthalpy is measured in an amide transfer experiment (vapor phase to aqueous solution), but other terms also enter in. For amides, the enthalpy of interaction with water is approximately equal to the ESF, which may be calculated theoretically by a model with specified partial charges and atomic radii. To determine the enthalpy of interaction with water from an amide transfer experiment (which gives the solvation enthalpy), it is necessary to estimate the sum of two other terms and subtract them. The cavity term, which arises from forming a cavity in water for the solute, is chiefly entropic and makes a large unfavorable contribution to the solvation free energy. Solvent reorganization at the cavity surface introduces an enthalpic contribution into the cavity term (see discussion for nonpolar solutes, refs. 15 and 16). The van der Waals term arises from interactions between solute and solvent after insertion of the solute in the cavity.
We follow the procedure suggested by Sitkoff et al. (11) of approximating the sum of these two terms by experimental transfer data for nonpolar alkanes, evaluated at corresponding values of water-accessible surface area. There are no adjustable parameters in this procedure. When used together with the delphi program and parse parameter set, this approximation gives predicted values of solvation free energy that reproduce the experimental values closely (11). By using alkane data to represent the sum of the cavity and van der Waals terms, the van der Waals interactions between water and the polar atoms of an amide are assumed to be the same as those between water and an alkane. The uncertainty caused by this approximation is difficult to evaluate (see discussion, refs. 11 and 20). Makhatadze and Privalov (21) used a similar procedure for evaluating the sum of the cavity and van der Waals terms, but their analysis of transfer enthalpies is based on assigning group enthalpies and assuming that they are additive; their value for the enthalpy of interaction between water and the peptide group (−14.2 kcal/mol) differs from the average value given here in Table 1 (−11.5 kcal/mol). They find that the interaction enthalpy is almost independent of temperature near 25°C.
The data in Table 1 show that the enthalpy and standard free energy of interaction of the amide polar groups with water are nearly equal for all four amides. This equality should not be surprising, because the polar interaction with water may be considered as H-bonding for amides in water, and the enthalpy of an amide-water H-bond is substantial whereas the loss in entropy from binding one water molecule in liquid water is small (compare ref. 21). The enthalpy of a water-amide H-bond lies between −5 and −6 kcal/mol, as computed by quantum mechanics for the gas phase (22), whereas the entropy loss from binding a water molecule is only 0–2 kcal/mol in terms of free energy, according to thermodynamic data for crystalline hydrate salts (23). Table 1 also shows that the calculated ESF values of these amides are equal to the standard free energies of the polar interaction with water, as expected (11). Different amides yield similar ESF values (Table 1) as well as similar values of the enthalpy of interaction with water.
There is no close correlation between polar accessible surface area and these values of polar free energy or enthalpy. The polar accessible surface area decreases 3-fold from acetamide to N,N-dimethylacetamide whereas the ESF value decreases by less than 10%. Because the ESF value is close to the enthalpy of interaction with water for amide polar groups, we use predicted values of ESF to estimate the enthalpy of interaction with water of peptide groups in alanine peptides.
Calculated ESF Values for Peptide Groups in Alanine Peptides
Because the peptide C⩵O and NH groups have large dipoles, main-chain dipole-dipole interactions have a major influence on the electrostatic potential of the peptide backbone and on conformational transitions such as the α→β transition (24, 25). Brant and Flory (24) found that they were unable to account for the measured dimensions of polypeptide chains without taking main-chain dipole-dipole interactions into account. Neighboring C⩵O and NH dipoles are antiparallel in the extended (β) conformation but parallel in the α conformation, so that main-chain dipole-dipole interactions strongly favor β over α. Peptide dipoles in the main chain also interact strongly with water dipoles, and side-chain perturbation of the electrostatic interactions between water and main chain dipoles is predicted to be a main factor determining helix propensities (5, 13, 25, 26).
The predictions of ESF values below follow the same procedure as those used to fit solvation free energies of amides. The geometry of the peptide is first assigned by using insight ii, whereas the delphi program and the parse parameter set are used to calculate the ESF values (ΔG*es in Table 1) of peptide groups at various positions in the peptide. The results depend on the assigned geometry as well as on the use of the parse parameter set, and it should be kept in mind that experimental data for the solvation free energies of peptides are not yet available. The partial charges and radii assigned to the peptide NH and C⩵O groups may be changed when such information becomes available.
Fig. 1 plots ESF values of peptide groups in a blocked alanine peptide containing eight alanine residues in the β-strand conformation (no peptide H-bonds). In the numbering system used here, the amide group formed by joining the acetyl and N-methyl blocking groups (to make N-methylacetamide) is not counted as a peptide group and the number of peptide groups equals the number of alanine residues. In β-strand peptides with from two to five alanine residues, the exterior (N terminal and C terminal) peptide groups have uniformly high ESF values (−8.5 to −8.3 and −8.8 to −8.7 kcal/mol, respectively) and interior peptide groups have lower and nearly constant ESF values near −7.9 kcal/mol, as in Fig. 1 with eight residues.
Figure 1.
The calculated ESF of the peptide groups in an eight-residue alanine peptide, β-strand conformation are plotted against residue number. With the numbering system used here, the number of peptide groups equals the number of alanine residues because, in a series of peptides with varying numbers of alanine residues, the amide bond created by joining the N-terminal acetyl and C-terminal N-methyl blocking groups (to make N-methylacetamide) is not counted as a peptide bond. The ESFs have been calculated as outlined (13) by using delphi and the parse parameter set (11); the internal dielectric constant is 2, and the external dielectric constant is 80. The peptide geometry is generated by using insight ii with the φ,ψ backbone angles set to −120° and 120°, respectively, for the β-strand.
These results indicate that amides are not close models for the interaction of the peptide group with water. The ESF value for N-methylacetamide differs by −4 kcal/mol from the value for an interior alanine peptide group. An analysis by quantum mechanics of the charge distribution of the peptide group compared with amides (27) predicts that at least a blocked dipeptide is needed to provide a satisfactory model for the peptide bond. Regarding solvation of peptide groups in β-strand peptides, it is necessary to distinguish exterior from interior peptide groups, and the solvation of the central peptide group of a tripeptide differs substantially from the dipeptide value. These results agree with the conclusion (13) that solvent accessibility is a major factor determining the ESF.
An earlier study by Yang and Honig (28) used delphi calculations, among other approaches, to examine the determinants of stability in alanine peptide helices. They found that the electrostatic solvation free energy of peptide groups in the unfolded peptide is comparable in size to the enthalpy of the peptide hydrogen bond, and they concluded that hydrogen bonds do not drive helix formation, but rather the burial of nonpolar surface area is the main determinant.
ESF Values for Alanine Peptides in the Helical Conformation
Results are shown in Fig. 2 for two blocked alanine peptides in the helical conformation: one peptide with five Ala residues (5-mer) and one with 15 residues (15-mer). The results are more complex than the ones for the β-strand conformation, both because the accessibility of water to peptide groups changes in successive residues at each end of the helix and because hydrogen bonding has a major affect on the electrostatic potential (24, 25). The C⩵O of Ala-1 is H-bonded to the NH of Ala-5 and the NH groups of Ala residues 1–4 do not make peptide H-bonds, although in the helical geometry specified here the C⩵O of the N-terminal acetyl group H-bonds to the NH of Ala-4. At the C-terminal end of the 15-mer helix, the NH of Ala-15 is H-bonded to the C⩵O of Ala-11 and the C⩵O groups of Ala-12–15 do not make peptide H-bonds, although the C-terminal NHCH3 group H-bonds to the C⩵O of Ala-12.
Figure 2.
The calculated ESFs, in kcal/mol, of the peptide groups in two helical alanine peptides are plotted against residue number. The peptides contain either five or 15 alanine residues, plus N-terminal acetyl and C-terminal N-methylamide blocking groups. The backbone φ,ψ angles are −65° and −40° respectively. For other information, see legend to Fig. 1.
A large drop in the absolute value of the peptide ESF occurs in going from Ala-1 to Ala-5 in the 15-mer helix, accompanying the formation of the first peptide group in which both the C⩵O and NH groups make peptide H-bonds. A striking feature of the 15-mer helix results is that the ESF values of peptide groups keep on changing after Ala-5, in contrast to the step function behavior predicted by standard H-bond models of helix formation; the changing values result from the long-range nature of the dipole-dipole interactions within the helix (5, 13, 24). A much smaller drop in absolute ESF occurs in the 5-mer between Ala-1 and Ala-5; the 5-mer forms only a single peptide H-bond plus two H-bonds provided by the acetyl and amide blocking groups. Likewise, a large increase in absolute ESF value occurs at the C-terminal end of the 15-mer helix in going from Ala-11 to Ala-15, as the C⩵O groups cease to be H-bonded. In the interior of the helix, where both the NH and C⩵O moieties of the peptide groups are H-bonded, the ESF value of a peptide group lies between −2.4 and −2.7 kcal/mol for alanine residues 6–10. Approximately −0.5 kcal/mol partitions with the peptide NH and −2.0 kcal/mol with the peptide C⩵O (data not shown).
The results in Fig. 2 indicate that interior peptide groups in a helix are significantly solvated, and the free energy is large enough to be highly important in the energetics of protein folding. The ESF of an interior helical peptide group is only about one-half the gas phase energy of the H-bond formed between water and an amide C⩵O group (22), but there are many peptide groups in a protein, and their solvation introduces a major factor in the energetics of folding. The existence of significant solvation in helical peptide groups is consistent with a molecular dynamics study of the solvation of the hydrogen-bonded formamide dimer (29), which found that the dimer has more affinity for water than for CCl4 by −2.2 kcal/mol. A high-resolution x-ray structure of a 37-residue peptide containing chiefly alanine residues (9) shows water molecules clustering around peptide C⩵O groups but no fixed H-bonds between water and C⩵O groups. Clustering of water molecules near C⩵O groups is consistent with a molecular dynamics simulation (30) of water molecules surrounding a peptide helix and also with an NMR study (31) of the solvation of a peptide helix. An infrared study of the amide I′ and amide II′ bands shown by a dimeric coiled–coil helix indicates that solvent-exposed peptide groups are solvated (45).
ESF Values of Alanine Peptides in the Random Coil Conformation
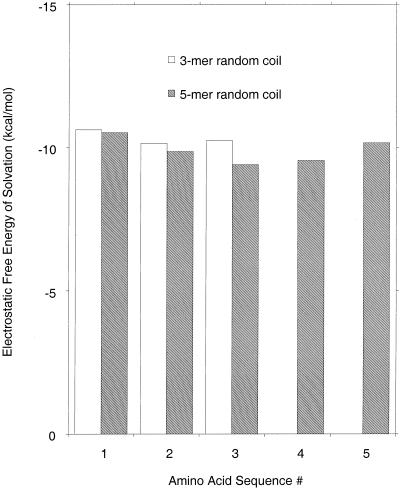
Results for two blocked alanine peptides in the random coil conformation, containing three or five residues, are shown in Fig. 3. The random coil has been defined previously (13). Structures in the Protein Data Bank provide a library of sequences with specified torsional angles, and residue conformations are chosen at random from this library. There are several differences between these random coil results and the ones for β-strand peptides (Fig. 1). In the β-strand peptides, there is a major difference in ESF between exterior and interior peptide groups, but all interior groups have the same ESF value and this remains true as the number of alanine residues is varied. In contrast, the ESF values of the peptide groups in the random coil change steadily with residue position and are different in the three- and five-residue peptides. The decrease in solvation with increasing peptide length is explained by reduced solvent accessibility of the peptide groups in longer peptides (13).
Figure 3.
The calculated ESF values, in kcal/mol, are plotted against residue number for two blocked alanine peptides in the random coil conformation. The peptides contain either three or five alanine residues. The definition of the random coil conformation is given in the text and in ref. 13, and the method of calculating the results follows ref. 13.
Because the ESF value changes both with residue position and the number of residues in the peptide, it is not possible to give a single value for the ESF of a peptide group in the random coil. The ESF values for the three- and five-residue peptides in the random coil (Fig. 3) are larger in absolute value than that of interior peptide groups in the β-strand conformation (−7.9 kcal/mol) because the random coil contains a mixture of α, β, and “other conformations,” and α has a larger absolute value ESF than β (5, 13, 26, 28, 32, 33), as illustrated here in Figs. 1 and 2. Data for the proportions of α, β, and other conformations are given by Smith et al. (34) for the various amino acid residues and for two models of the random coil based on structures in the Protein Data Bank. [One model (all) includes residues contained within secondary structure, whereas the second model (coil) excludes them.] In the coil model, the proportions of β, α, and other conformations for the alanine residue are 0.51, 0.32, and 0.17, respectively (34). Thus, the fact that the peptide absolute ESF value is higher in the α than in the β conformation explains why alanine peptide absolute ESF values are higher in the short random coil peptides shown here than in the β-strand conformation, but the dependence of ESF on peptide length in the random coil, versus the invariance observed in the β-strand, adds an additional twist. The importance of understanding the ESF behavior of the random coil was pointed out by Tobias and Brooks (36) in an early simulation of alanine and valine peptide helix formation.
Origin of the Favorable Enthalpy Change for Alanine Helix Formation
The problem posed in the Introduction is the existence of a major deficit (−7.6 kcal/mol) in accounting for the enthalpy change (−1.0 kcal/mol) that drives alanine helix formation, when experimental data for amides (−11.5 kcal/mol) are used to model the enthalpy of interaction between water and the peptide group in the random coil and a quantum mechanics value (−4.9 kcal/mol) is used for the enthalpy of the peptide H-bond in a helix, and other sources of enthalpy are ignored. The deficit is reduced to −1.5 kcal/mol when ESF calculations are used to model the peptide-water interaction in the random coil by the value (−7.9 kcal/mol) for interior peptide groups in β-strand peptides, and a value (−2.5 kcal/mol) is given to the ESF of interior peptide groups in a helix, but calculations for some random coil peptides indicate that ESF values in the random coil are a complex, not yet well understood, subject.
In considering the enthalpy needed to drive helix formation, one term has not yet been discussed: the electrostatic free energy difference between the α and β conformations arising from main-chain dipole-dipole interactions (5, 13–15). Quantum mechanics calculations indicate that the α conformation (without peptide H-bonds) is less favorable than β in the gas phase by about 2.5 kcal/mol (32, 33, 35) whereas, in solution, increased solvation of peptide groups in the α conformation increases the stability of the α conformation relative to β. The ESF value of an interior peptide group in the β conformation is −7.9 kcal/mol (Fig. 1) whereas in the α conformation it is −9.6 kcal/mol for the first interior peptide group in a 5-mer (Fig. 2). Because the overall free energy difference between α and β in solution (without peptide H-bonds) is unfavorable for helix formation, this factor adds to the need for additional enthalpy to drive helix formation.
ESF Values of Mutant Peptides in Relation to Helix Propensities
Both theoretical analysis (5, 13, 26) and experimental data (6) have been used to argue that side-chain perturbation of the interaction between water and main-chain polar groups should be a main determinant of helix propensities. Calculation of ESF values for mutant peptides provides a direct test of the size of this perturbation without introducing adjustable parameters. The geometry of the peptide is held fixed in the α or β conformation, and the size of the perturbation is calculated from the difference between the ESF values of the mutant and an all-Ala peptide.
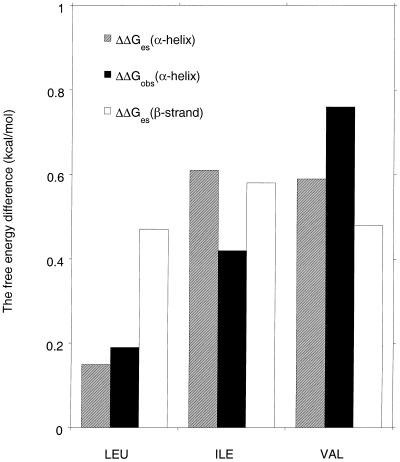
Fig. 4 gives difference ESF values for peptides in the β-strand conformation, in which a single Leu, Ile, Val, or Gly residue is substituted for Ala. Because the ESF values of interior peptide groups do not vary with peptide length in β-strand peptides, a five-residue peptide is chosen as the host, and the substitution is made at the central residue. The largest change in ESF occurs at the site of substitution, but significant changes also may be seen at the two flanking Ala residues and the difference values are summed over all five residues. Val, Ile, and Leu all show substantial, nearly equal, changes in ESF for the β-strand peptides. [Gly shows a change of opposite sign, corresponding to increased solvation; data not shown.] The changes in ESF values are correlated with decreased accessibility of water to the peptide backbone, caused by the amino acid side chain. For the Val, Ile, and Leu mutant peptides, these changes in ESF resemble the changes in peptide NH exchange rate observed by Bai et al. (37) in blocked dipeptides, which have been attributed to steric blocking by the side chain of the access of water to the peptide group.
Figure 4.
Comparison between the measured helix propensity difference (6) (filled bars) and the calculated ESF difference for a mutant peptide versus an alanine host in either the β-strand conformation (empty bars) or the helical conformation (hatched bars). The units are kcal/mol. The comparison is made for Leu, Ile, and Val. The β-strand peptides contain five alanine residues plus N-terminal acetyl and C-terminal N-methylamide blocking groups, and only the central Ala residue is substituted by another amino acid. The ESF values of the five central residues of the mutant β-strand peptide are subtracted from those of the Ala peptide and then summed. The peptide sequence used for calculating ESF values in the helical conformation corresponds to the peptide sequence used for measuring helix propensities (6), which is Ac-K(A)4X(A)4KGY-NH2, where X is either A, L, I, V, or G. In calculating ESF differences between helical peptides, the values for the five central peptide groups (groups 4–8) in the mutant peptide are subtracted from the corresponding values for the Ala peptide and then summed. The side-chain torsion angles are: Leu, χ1–60, χ2 180°; Ile, χ1–60, χ2 180°; Val, χ1 180°; the backbone angles are given in the legends to Figs. 1 and 2.
The mutant peptides in helical conformation have the same sequences as 12-residue peptides studied experimentally (6), and the calculated differences in ESF may be compared directly with the measured differences in helix propensity. The experiments, which were made as a function of temperature, gave results showing that the rank order of helix propensities tends to change with temperature: in some conditions, Val actually has a higher helix propensity than Ala (6). Consequently an enthalpic factor (inferred to be the interaction of water with the peptide group) was predicted to be an important determinant of helix propensities. The calculated ESF values for the helical peptides (Fig. 4) are similar to the calculated β-strand results for Ile and Val, but not for Leu. The experimental changes in helix propensity also are shown in Fig. 4. [Glycine is excluded because of a large contribution from backbone conformational entropy to its helix propensity.]
The observed helix propensity differences are comparable in size to the calculated ESF differences both in the helical and β-strand conformations. The helix propensity differences are known to be correlated with the loss in side-chain entropy on helix formation (found, for example, by Monte Carlo simulation for a flexible helix, ref. 38). The free energy changes caused by the side-chain entropy loss correspond to about 1/3 of the helix-propensity differences between alanine and other nonpolar amino acids, as measured in alanine-based peptides (39). The free energy changes arising from calculated ESF differences in the helical conformation are larger than the changes arising from side-chain entropy loss, but the ESF differences in the random coil conformation must be subtracted (13) to compare calculated ESF differences with observed helix propensity differences. The main conclusion from the calculated ESF differences shown here for mutant peptides is that they are comparable in size, before the random coil term is taken into account, to the observed helix propensity differences. Note that no adjustable parameters are used in calculating the ESF differences besides the peptide geometry and use of the parse parameter set.
Even larger ESF differences in the random coil conformation than shown here for the helix have been found for randomly generated tripeptides (13) in which both the sequences and torsion angles are selected randomly, using a model similar to the all model of Smith et al. (34) (and also using a different set of partial charges than the parse set used here). These calculations have been repeated for blocked tripeptides (AXA) and pentapeptides (AAXAA) where X = A, L, I, V, or G (F.A., unpublished work), by using the coil library of torsion angles (34) and the parse parameter set. The results are similar to the ones given in ref. 13, as regards both the large ESF differences between mutant and all-Ala peptides in the random coil conformation and the decrease in ESF values with increasing peptide length. Srinivasan and Rose (40) have argued recently that the random coil is not random but tends to have segments containing either the α or β conformation.
The calculations given here for mutant peptides suggest an answer to a long-standing puzzle: why are the helix propensity differences measured in alanine-based peptides approximately twice as large as those measured in the natural sequence peptide from RNase T1 or in the RNase T1 protein (41)? The difference between the ESF value of alanine (−7.94 kcal/mol), at an interior peptide group in a β-strand peptide, and valine is approximately twice as large in an all-valine eight-residue peptide (whose ESF is −7.42 kcal/mol, data not shown) as in a single-site mutant, whose ESF is −7.73 kcal/mol. The reason is that neighboring valine residues contribute to the desolvation in an all-valine peptide, whereas neighboring residues have little effect on a valine residue in an alanine-based peptide. Likewise, in a peptide helix the side chains of neighboring residues larger than Ala are likely to contribute to the desolvation of the peptide group at the site of a substitution (6): thus, the amount of desolvation produced by a particular substitution such as Ala→Val will be larger in an alanine-based peptide than in a natural sequence peptide because the peptide group at the site of substitution in the natural sequence peptide already has reduced solvation before the substitution is made.
Concluding Comment
A basic feature of these ESF calculations is that no adjustable parameters are used besides peptide geometry and the parse parameter set; the results are not scaled but instead are compared directly with experiment. The model compounds used to fix the parameters in the parse set do not include peptides, and some changes in the parameters are likely to be made when experimental results for peptides become available. Nevertheless, the main conclusions from this study probably will remain. The conclusions are: (i) solvation of the peptide group in the helix is large enough to be important, both in the enthalpy of helix formation and the free energy of protein folding, (ii) amides are poor models for the solvation of the peptide group in the β-strand conformation, and (iii) side-chain interference with solvation of the peptide group is a major factor affecting helix propensities.
Acknowledgments
We thank Charles Brooks III, Bill DeGrado, David Eisenberg, Bertrand Garcia-Moreno, Pehr Harbury, Barry Honig, Nick Pace, Carol Rohl, George Rose, John Schellman, and Stephen White for discussion and Janet Thornton for use of naccess, and we especially thank B. K. Lee for his suggestions concerning preparation of this paper. This work was supported by the Ministry of Science and Technology of Slovenia.
Abbreviation
- ESF
electrostatic solvation free energy
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200343197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200343197
References
- 1.Baker E N, Hubbard R E. Prog Biophys Mol Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Naim A. J Phys Chem. 1991;95:1437–1444. [Google Scholar]
- 3.Yang A S, Sharp K A, Honig B. J Mol Biol. 1992;227:889–900. doi: 10.1016/0022-2836(92)90229-d. [DOI] [PubMed] [Google Scholar]
- 4.Honig B, Yang A S. Adv Protein Chem. 1995;46:27–58. doi: 10.1016/s0065-3233(08)60331-9. [DOI] [PubMed] [Google Scholar]
- 5.Avbelj F, Moult J. Biochemistry. 1995;34:755–764. doi: 10.1021/bi00003a008. [DOI] [PubMed] [Google Scholar]
- 6.Luo P, Baldwin R L. Proc Natl Acad Sci USA. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholtz J M, Marqusee S, Baldwin R L, York E J, Stewart J M, Santoro M, Bolen D W. Proc Natl Acad Sci USA. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholtz J M, Qian H, York E J, Stewart J M, Baldwin R L. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 9.Sicheri F, Yang D S C. Nature (London) 1995;375:427–431. doi: 10.1038/375427a0. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell J B O, Price S L. J Comp Chem. 1990;11:1217–1233. [Google Scholar]
- 11.Sitkoff D, Sharp K A, Honig B. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 12.Florian J, Warshel A. J Phys Chem B. 1997;101:5583–5595. [Google Scholar]
- 13.Avbelj F. J Mol Biol. 2000;300:1335–1359. doi: 10.1006/jmbi.2000.3901. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Naim A. J Phys Chem. 1978;82:792–803. [Google Scholar]
- 15.Lee B K. Biopolymers. 1991;31:993–1008. doi: 10.1002/bip.360310809. [DOI] [PubMed] [Google Scholar]
- 16.Lee B K. Methods Enzymol. 1995;259:555–557. doi: 10.1016/0076-6879(95)59061-7. [DOI] [PubMed] [Google Scholar]
- 17.Wolfenden R. Biochemistry. 1978;17:201–204. doi: 10.1021/bi00594a030. [DOI] [PubMed] [Google Scholar]
- 18.Wolfenden R, Andersson L, Cullis P M, Southgate C C. Biochemistry. 1981;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 19.Della Gatta G D, Barone G, Elia V. J Solution Chem. 1986;15:157–167. [Google Scholar]
- 20.Tannor D J, Marten B, Murphy R, Friesner R A, Sitkoff D, Nicholls A, Ringnalda M, Goddard W A, Honig B. J Am Chem Soc. 1994;116:11875–11882. [Google Scholar]
- 21.Makhatadze G I, Privalov P L. J Mol Biol. 1993;232:639–659. doi: 10.1006/jmbi.1993.1416. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell J B O, Price S L. Chem Phys Lett. 1991;180:517–523. [Google Scholar]
- 23.Dunitz J D. Science. 1994;264:670. doi: 10.1126/science.264.5159.670. [DOI] [PubMed] [Google Scholar]
- 24.Brant D A, Flory P J. J Am Chem Soc. 1965;87:663–664. [Google Scholar]
- 25.Avbelj F, Moult J. Proteins Struct Funct Genet. 1995;23:129–141. doi: 10.1002/prot.340230203. [DOI] [PubMed] [Google Scholar]
- 26.Avbelj F, Fele L. J Mol Biol. 1998;279:665–684. doi: 10.1006/jmbi.1998.1792. [DOI] [PubMed] [Google Scholar]
- 27.Price S L, Faerman C H, Murray C W. J Comp Chem. 1991;12:1187–1191. [Google Scholar]
- 28.Yang A-S, Honig B. J Mol Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- 29.Sneddon S F, Tobias D J, Brooks C L., III J Mol Biol. 1989;209:817–820. doi: 10.1016/0022-2836(89)90609-8. [DOI] [PubMed] [Google Scholar]
- 30.Soman K V, Karimi A, Case D A. Biopolymers. 1991;31:1351–1361. doi: 10.1002/bip.360311202. [DOI] [PubMed] [Google Scholar]
- 31.Brüschweiler R, Morikis D, Wright P E. J Biomol NMR. 1995;5:353–356. doi: 10.1007/BF00182277. [DOI] [PubMed] [Google Scholar]
- 32.Head-Gordon T, Head-Gordon M, Frisch M J, Brooks C L, III, Pople J A. J Am Chem Soc. 1991;113:5989–5997. [Google Scholar]
- 33.Beachy M D, Chasman D, Murphy R B, Halgren T A, Friesner R A. J Am Chem Soc. 1997;119:5908–5920. [Google Scholar]
- 34.Smith L J, Bolin K A, Schwalbe H, MacArthur M W, Thornton J M, Dobson C M. J Mol Biol. 1996;255:494–506. doi: 10.1006/jmbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- 35.Brooks C L, III, Case D A. Chem Rev. 1993;93:2487–2502. [Google Scholar]
- 36.Tobias D J, Brooks C L., III Biochemistry. 1991;30:6059–6070. doi: 10.1021/bi00238a033. [DOI] [PubMed] [Google Scholar]
- 37.Bai Y, Milne J S, Mayne L, Englander S W. Proteins Struct Funct Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creamer T P, Rose G D. Proteins Struct Funct Genet. 1994;19:85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 39.Rohl C A, Chakrabartty A, Baldwin R L. Protein Sci. 1996;5:2623–2637. doi: 10.1002/pro.5560051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan R, Rose G D. Proc Natl Acad Sci USA. 1999;96:14258–14263. doi: 10.1073/pnas.96.25.14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers Y K, Pace C N, Scholtz J M. Proc Natl Acad Sci USA. 1997;94:2833–2837. doi: 10.1073/pnas.94.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cabani S, Gianni P, Mollica V, Lepori L. J Solution Chem. 1981;10:563–595. [Google Scholar]
- 43.Ben-Naim A, Marcus Y. J Chem Phys. 1984;81:2016–2027. [Google Scholar]
- 44.Chothia C. J Mol Biol. 1976;55:379–400. [Google Scholar]
- 45.Manas, E.S., Getahun, Z., Wright, W. W., DeGrado, W. F. & Vanderkooi, J. M. (2000) J. Am. Chem. Soc., in press.