Abstract
The technology, experimental approaches, and bioinformatics that support proteomic research are evolving rapidly. The application of these new capabilities to the study of neurodegenerative diseases is providing insight into the biochemical pathogenesis of neurodegeneration as well as fueling major efforts in biomarker discovery. Here, we review the fundamentals of commonly used proteomic approaches and the outcomes of these investigations with autopsy and cerebrospinal fluid samples from patients with neurodegenerative diseases.
Keywords: Alzheimer disease, Cerebrospinal fluid, Mass spectrometry, Neurodegeneration, Parkinson disease, Proteomics
INTRODUCTION
Proteomic technologies provide powerful means to systematically profile the peptide or protein constituents of complex mixtures and, in some instances, to use these data to impute protein identity, amount, or both. Although complementary to genomics, there are important differences between gene expression profiling and protein profiling. Perhaps most important is that it is now possible to interrogate essentially the entire transcriptome, athough there is no comparably comprehensive approach to the proteins encoded therein. This may be viewed by some as sufficient justification to concentrate on the transcriptome; however, the quantitative concordance between message and corresponding protein is surprisingly poor (1–3). Moreover, the complex and rich range of chemical modifications that occur to proteins after translation are largely transparent to genomics but are coming into progressively greater focus in proteomics. Another important consideration in the context of this review is the relative stability of protein versus messenger RNA in samples of brain obtained from autopsy.
Human neurodegenerative diseases range from rare to common illnesses. Indeed, some such as Alzheimer disease (AD) and Parkinson disease (PD) pose serious public health challenges that will increase in the coming decades. Many discoveries have been made in the genetic causes and risk factors for several neurodegenerative diseases (4); however, there are relatively few proteomic studies of neurodegenerative diseases to date. The major foci of these investigations have been analysis of protein from autopsy samples in studies of pathogenesis and from cerebrospinal fluid (CSF) in the pursuit of biomarkers. It is important to be aware of some limitations of these initial proteomic studies of human neurodegenerative diseases. Many are limited to comparisons between 1 diseased group and controls and therefore cannot address the important issue of disease specificity. This limitation has been approached by some more recent investigations reviewed later. Another limitation has been a focus on changes observed during clinically diagnosed stages of disease, whereas some recent studies have begun to explore alterations that accompany preclinical disease stages.
FUNDAMENTALS OF PROTEOMICS
Shared Features of Different Proteomic Techniques
Although it is difficult to summarize such a dynamic and rapidly changing scientific area, the various approaches to protein profiling share common features. These include sample preparation, protein or peptide separation, mass spectrometry (MS), and bioinformatic data processing, as well as subsequent confirmation and validation. For the sake of clarity, we will use validation to mean replication of findings by alternative method(s) in the same samples used in profiling and confirmation to mean replication of findings in independent samples.
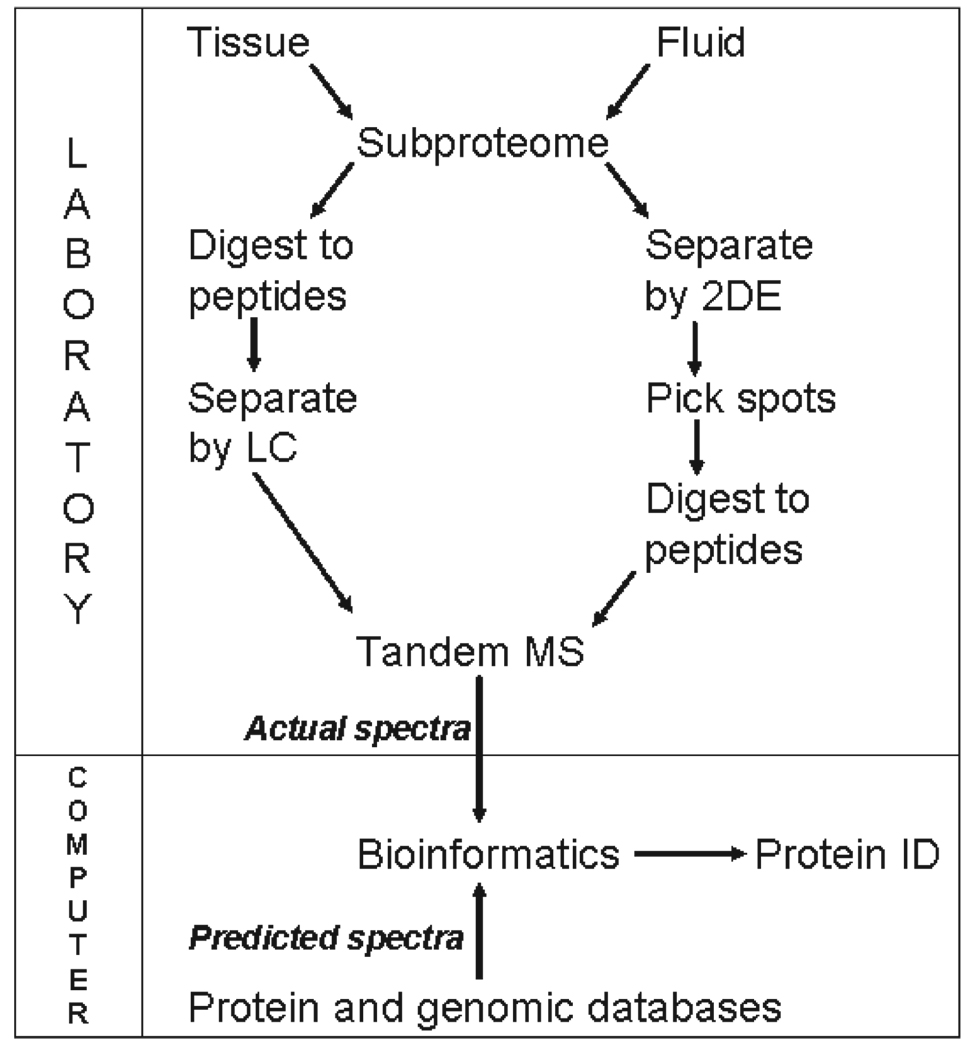
Sample preparation is a major source of variation in the outcomes of proteomic experiments. Again, proteomics differs from genomics in that it is not yet possible to analyze virtually all proteins in a cell, tissue, or body fluid comprehensively, especially in samples with wide ranges of protein concentration. Consequently, the first step in all proteomic experiments is preparation of a subproteome, or a subset of proteins, that is defined by the physical or chemical means used to isolate it from all proteins in tissue or body fluid. Examples to follow have generated subproteomes using gradient detergent extractions to enrich for hydrophobic or membranebound proteins, affinity purification, immunoprecipitation, and laser capture microdissection (Fig. 1). Typical subproteomes are still complex protein mixtures that require further separation. Classically, proteins are separated by 2-dimensional gel electrophoresis (2-DE), and spots detected on the gel by a variety of means are excised and digested to peptides, typically with trypsin. Alternatively, the protein mixture in the subproteome is first digested to peptides, and these are then separated by single or multidimensional liquid chromatography (LC).
FIGURE 1.
Diagram of the steps in typical proteomic experiments that use liquid chromatography (LC) or 2-dimensional gel electrophoresis (2-DE) followed by tandem mass spectrometry (MS-MS). Protein identification (ID) is imputed by bioinformatic tools that compare tandem MS data with protein or genomic databases.
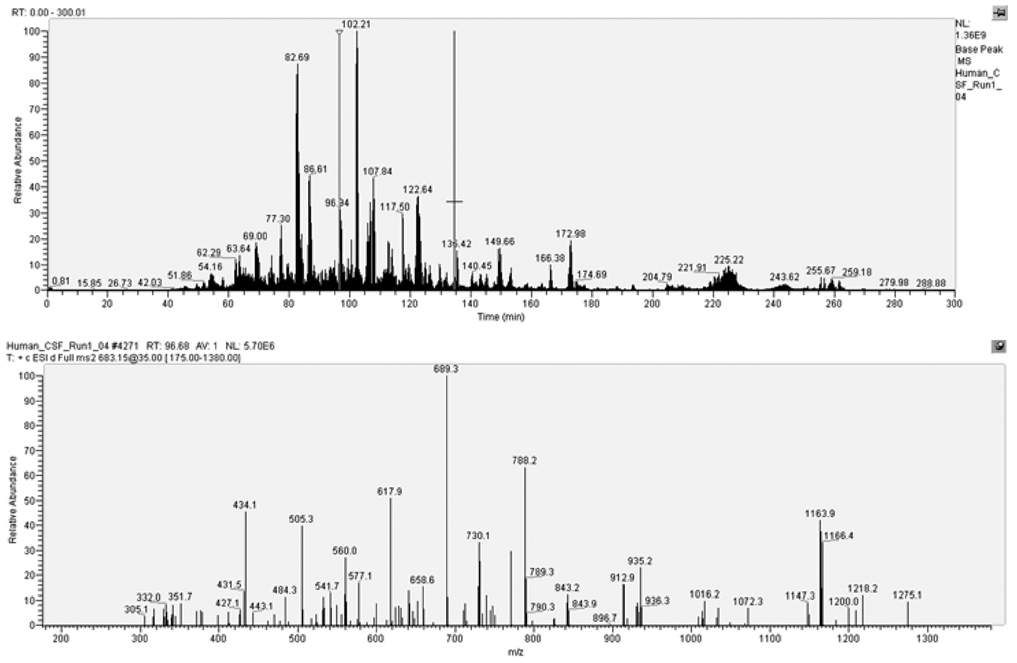
Mass spectrometry is an elegant technique for detecting the mass of charged species. There are many excellent reviews devoted to MS. Here we summarize the basics. The first step in MS is to ionize and vaporize the analytes (i.e. proteins or their [typically] tryptic peptide fragments) without significant fragmentation. This can be achieved by a variety of methods: 2 commonly used approaches are matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI). Matrix-assisted laser desorption ionization mixes the analyte with 1 of several different molecules that facilitate ionization and vaporization. Generating peptide ions from ESI is complex and is thought to involve a process called Coulombic fission. The peptide or protein ions are then introduced into the mass analyzer. Again, there are different types including ion trap, triple quadrupole, time-of flight (TOF), and Fourier transform ion cyclotron that differ in their mechanisms of ion separation, mass accuracy, and resolution. It is critical to appreciate that some, but not all, common proteomic techniques use tandem MS (MS-MS) to analyze the daughter or fragment ions of the parent ions whose mass was determined in first MS dimension. In these collision-induced dissociation tandem MS experiments, the energy applied greatly favors fragmentation of the peptide (amide) bond over amino acid side chains in the parent ion, thereby generating an ensemble of daughter ions, called b ions and y ions, from which the amino acid sequence can be deduced (Fig. 2). Detectors and multipliers generate the signal that is typically plotted as the total ion chromatogram for the first MS dimension, with the x axis being time and the y axis being relative abundance, and mass-to-change ratio (m/z) chromatogram for the daughter ions.
FIGURE 2.
Typical collision-induced dissociation tandem mass spectrometry data that show total ion chromatogram (time vs relative abundance) in the upper panel. The selected ion marked by the red line in the total ion chromatogram is subjected to further fragmentation to yield characteristic daughter ions in the lower mass-to-change ratio (m/z) chromatogram.
The next step is synthesis of the wealth of mass spectral data obtained. A major step forward in the evolution of proteomics was the development of bioinformatic tools to interpret the ever increasing rich data sets generated from advancing technology. Here, the pivotal event was the development of search engines, such as SEQUEST and MASCOT, which correlate actual tandem mass spectra with predicted tandem mass spectra based on species-specific amino acid sequences from protein or genomic databases (5, 6). It is important to appreciate that these different programs do not perform in exactly the same way and can produce different peptide matches and levels of confidence. Moreover, the databases currently searched are incomplete and imperfect. It is for this reason that all results require validation.
Specifics of Different Proteomic Techniques
Classically, experiments isolate a subproteome of interest, separate the protein by 2-DE, excise the spot of interest from the stained gel, digest the proteins contained in each spot with trypsin, and then perform tandem MS with MALDI TOF-TOF. This powerful approach is still extensively used, although 2-DE is labor intensive and has a relatively low dynamic range. Some of these issues have been alleviated partially by advances in technology, especially the use of fluorescent dyes. Proteins in a control sample can be labeled with 1 fluorescent dye and proteins in an experimental sample labeled with a different fluorescent dye. The samples are then mixed, separated by 2-DE, and the relative fluorescent intensities are measured within the spot of interest before digestion and tandem MS. In this way, analysis of fluorescent intensity provides relative quantification, and tandem MS provides protein identification for the spot of interest in the gel.
One response to the time and labor devoted to spot picking was to develop multidimensional LC systems for the separation of peptides that are coupled to tandem MS. In a typical experiment, the subproteome is first digested to peptides, and these are separated by LC and then directly ionized by ESI followed by tandem MS. Alternatively, the separated peptides are spotted directly onto a MALDI plate followed by tandem MS. An advantage of these approaches is that they can be much more automated than the 2-DE method; however, this comes with a price. The major one is that without the gel and fluorescent dyes, there are no quantitative data. Indeed, it is critical to appreciate that the size of the resulting peptide ion peaks does not necessarily bear any quantitative resemblance to the amount of protein in the subproteome; the major sources of variance are ionization efficiency and detector sensitivity. Several groups have ingeniously addressed this shortcoming of high-throughput proteomics by adapting a classical MS technique: the stable isotope dilution assay. In general, stable isotope labeling coupled with MS is more quantitatively accurate than are comparisons of spot densities on 2-DE. All of these techniques require the introduction of a stable isotope label, usually deuterium or 13C into samples that are then mixed and analyzed simultaneously by MS. The isotopically labeled peptide serves as a mass signature for sample source and as the foundation for estimation of relative abundance, and in some instances, the absolute concentration of the ionized peptides. There are several approaches to the introduction of isotope labels to proteins or peptides; among these are isotope-coded affinity tags (7) and isobaric tags for relative and absolute quantification (iTRAQ) (8). Isotope-coded affinity tag was one of the earlier approaches and was limited to labeling cysteinyl thiolates with light (protonated) or heavy (deuterated) probes (Fig. 3). Isobaric tags for relative and absolute quantification now permits differential isotopic labeling of ε-amino lysyl groups with up to 8 different probes. A bioinformatics approach similar to that previously described is used for imputation of protein identity from peptide ion data. Specialized computer software is needed to integrate isotopic data from multiple peptides to estimate protein concentrations. Newer methods for quantification without isotope labels are an area of active investigation (9).
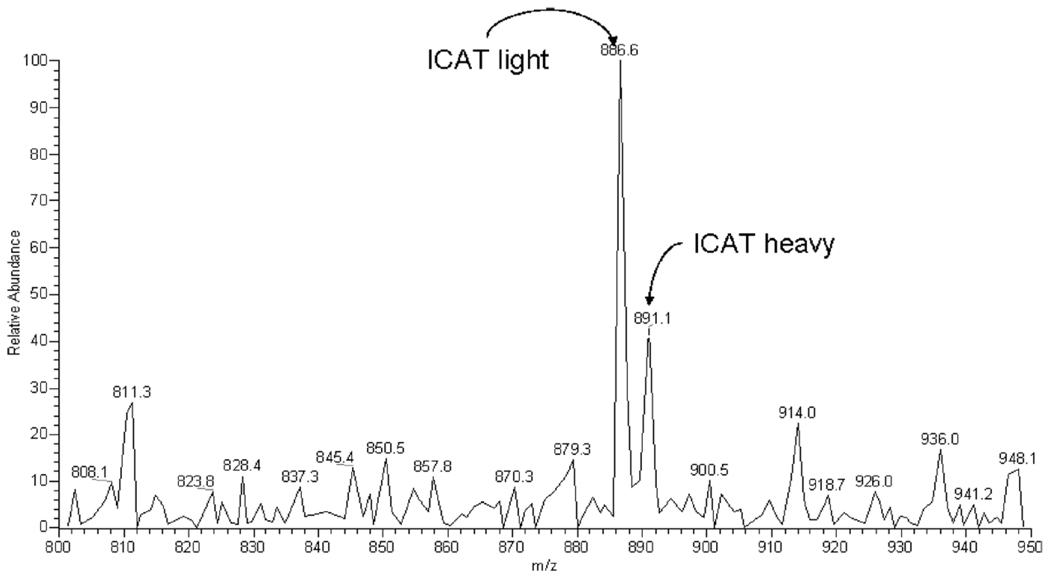
FIGURE 3.
Two subproteomes were generated from human CSF: one was labeled with protonated isotope-coded affinity tags (ICAT) reagent and the other with deuterated ICAT reagent. They were then combined before tandem mass spectrometry. The mass-to-change ratio (m/z) chromatogram shows the relative abundance of ions conjugated to the light (protonated) versus heavy (deuterated) ICAT reagent.
A third approach used in proteomics of neurodegenerative diseases is surface-enhanced laser desorption/ionization (SELDI), also called SELDI-TOF. Surface-enhanced laser desorption/ionization is a variant of MALDI that is commonly used in biomarker discovery. There are few very attractive features of SELDI (10). First, proteins or peptides can be separated or enriched through a variety of plates or chips with immobilized biochemically active materials that preferentially bind proteins based on hydrophobicity, charge interactions, chelation, and other characteristics. In addition, SELDI plates can be activated with immobilized organics that can covalently bind antibodies, other proteins such as receptors, and even nucleic acids. After the subproteome has been incubated with the desired plate, the poorly bound proteins or peptides are washed away, thereby acting like affinity chromatography. The versatility of agents that can be used at this step is a clear strength of SELDI. Another advantage is that SELDI is relatively simple and quick compared with other proteomic techniques. A disadvantage of SELDI as it is commonly used is that only a single MS dimension is used and there is, therefore, no attempt to impute protein identity. Rather, the typical experiment deals with finding those ions that have optimal characteristics and then using other techniques to identify the protein from which the selected ions are derived. Finally, given the nature of the SELDI experiment, it typically is not coupled with stable-isotope dilution techniques; therefore, reproducibility and quantification are problematic (11).
These are not the only approaches to MS. One example, an elegant technique of particular interest to pathologists, is tissue-based MS that can profile proteins in situ from different regions within a tissue specimen (12). The methods previously described, however, are those that have been used to date to investigate human neurodegenerative diseases.
Identifying Protein Modifications
One approach to evaluating protein modifications is in preparation of the subproteome. For example, some investigators have focused on only those spots from 2-DE that have reactivity for protein carbonyls (vide infra). Alternatively, an antibody that recognizes only a certain posttranslational modification may be adhered to a SELDI plate. Another approach has been to analyze tandem MS data for apparent mass shifts not predicted by the peptide sequence. There are several computer programs that perform such searches. Two contrasting approaches are SEQUEST and MASCOT versus P-MOD and InsPecT. The SEQUEST and MASCOT search for specified mass shifts on specified amino acids, for example +16 on M for methionine sulfoxide, across all peptide ions detected. In contrast, P-MOD and InsPectT search the same spectral data in a different manner (13, 14). Here, one provides the programs with the amino acid sequence of a protein of interest, and they list all unexpected masses and a calculated level of confidence.
PROTEOMIC STUDIES OF NEURODEGENERATIVE DISEASES FROM AUTOPSY BRAIN
In this section, we review proteomic studies of autopsy samples from patients with neurodegenerative diseases. Although there are several excellent proteomic investigations of corresponding animal models, we limited our comments to investigations of human tissue and grouped them by the method of subproteome preparation.
Homogenates of Human Brain Regions
Four studies have used 2-DE followed by tandem MS to investigate detergent-extractable proteins in brain regions of patients with AD, and in 1 case, patients with Down syndrome. One study identified 37 proteins with altered levels in AD (15); 11 of these had increased levels in AD and were identified by investigators by tandem MS. No validation was performed. Using a similar approach but not the same extraction protocol, another study identified 15 proteins, the levels of which differed in different AD brain regions (16). Again, no validation was performed. Agreement between these 2 similar studies is low; only increased levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and synaptotagmin I in AD were confirmed. A third similar study identified significantly decreased levels of mitochondrial complex III core protein 1 (17). This finding was not validated. A fourth study of frontal cortex focused on 9 antioxidant proteins and identified significantly increased peroxiredoxin 2 from AD patients compared with controls; parallel studies showed that peroxiredoxin 3 levels were decreased in frontal cortex from Down syndrome and Pick disease but not AD patients (18).
Laser Capture Microdissection of Hallmark Structures
There have been 3 reports of proteomic characterization of hallmark structures of neurodegenerative diseases (i.e. amyloid plaques and neurofibrillary tangles [NFTs] from AD and Lewy bodies [LB] from patients with dementia with LBs [DLB]) that were enriched by laser capture microdissection from human brain samples. Amyloid plaques were localized by thioflavin-S staining, microdissected, and then extracted with detergent; NFTs and LBs were identified by immunohistochemistry (tau-2 for NFTs, α-synuclein [SNCA] for LBs) and solubilized with formic acid; all were then separated with LC followed by MS-MS. A total of 488 proteins were identified from laser capture microdissection of amyloid plaques; several of these were validated by immunohistochemistry. Of these, 26 proteins were enriched in captured amyloid plaques compared with areas without amyloid. Of particular interest was the identification of dynein heavy chain in both human postmortem amyloid plaques as well as in plaques obtained from APPswe/PS1ΔE9 transgenic mice (19).
Seventy-two proteins were identified in NFTs obtained by laser capture microdissection by 2 or more unique peptides, of which 63 had no previously known association with NFTs (20). One of these proteins, GAPDH, localized to NFTs by immunohistochemistry, was validated in the detergent-insoluble fraction from hippocampus of patients with AD and was confirmed by coimmunoprecipitation with PHF-tau. Coincident with this publication was the observation by others that polymorphisms of GAPD may be genetic risk factors for late-onset AD (21). This finding has been corroborated by others, although the overall effect may be small (22). Others have subsequently shown that GAPDH localizes to LBs (23), can be oxidized and aggregated by exposure to Aβ peptides (24), and has increased expression in the hippocampus of AD patients compared with controls (25).
Traditional biochemical or histochemical methods previously identified approximately 70 molecular components of LBs in both the brainstem and cerebral cortex (26); SNCA has been identified as one of the major constituents. A recent study used laser capture microdissection of SNCA-immunoreactive cerebral cortical LBs from patients with DLB coupled with LC/ESI/MS/MS. This platform identified 156 protein with 2 or more unique peptides; of these, 17 had previously been associated with cerebral cortical or brainstem LBs (27). One particular protein, heat shock cognate 71 was previously associated only with brainstem LBs.
Insoluble Proteins in Cerebral Cortex
Accumulation of insoluble proteins is a defining characteristic of several neurodegenerative diseases, including AD and PD (28, 29). Indeed, it was the identification of detergent-insoluble protein such as Aβ and paired helical filament tau that ushered in the modern era of neurodegenerative disease research (30, 31). Conversion to insoluble forms disrupts protein function, can lead to further cellular damage, and may precede the formation of the hallmark inclusions (32). To gain insight into disease pathogenesis, we have focused modern proteomic techniques on this class of pathological protein (33). We used LC-MS-MS and identified in the detergent-insoluble fraction of late-onset AD temporal cortex samples 125 proteins by 2 or more unique peptides that included several proteins critical to Aβ production, components of synaptic scaffolding, and products of genes linked to an increased risk of late-onset AD. Each of 15 candidates were validated by Western blot including GAPDH (33), and the levels of several are altered in hippocampi of AD patients compared with controls (25). Aβ, tau, and 7 of 8 other newly identified detergent-insoluble proteins were confirmed to be increased in temporal cortex by Western blot, not only from patients with late-onset AD but from patients with AD associated with mutations in PSEN1 and PSEN2. All of these except tau were elevated in individuals with mild cognitive impairment, whereas none except Aβ were elevated in aged APPswe mice. These results extend the amyloid hypothesis for AD to include widespread protein insolubility that is not exclusive to Aβ, even early in the course of AD before the onset of dementia.
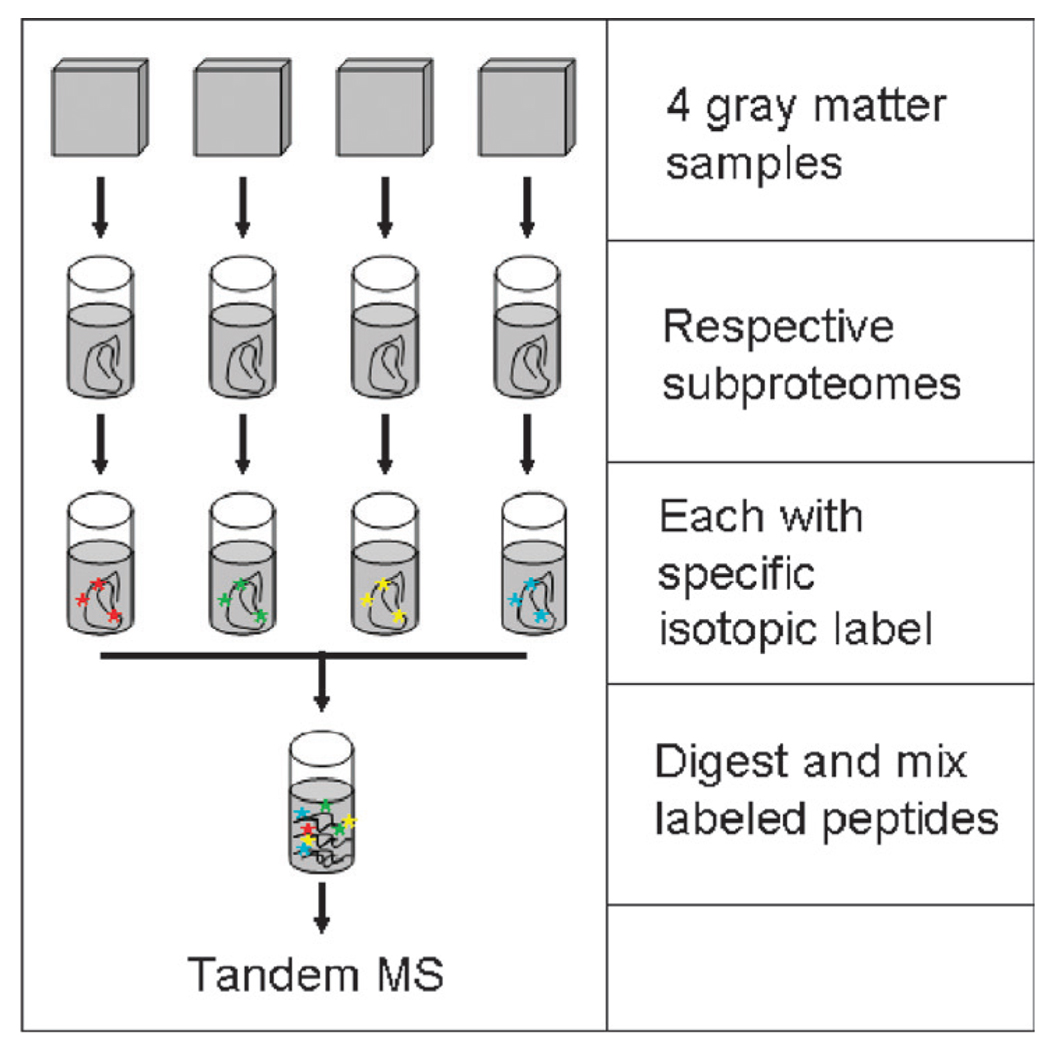
We have used iTRAQ labeling combined with MALDI-TOF-TOF to analyze the detergent-insoluble proteome in frontal cortex from 4 groups of individuals: patients with late-onset AD and their controls from mainland United States and Chamorros from Guam who had Parkinson-dementia complex or their controls (Fig. 4). In addition to the expected increase in abnormal frontal cortical Aβ peptides, tau, ubiquitin, and apolipoprotein E (apoE) in AD, and tau in Parkinson-dementia complex, we identified SNCA as a major detergent-insoluble protein in Parkinson-dementia complex but not AD despite a lack of LBs in corresponding tissue samples (34). The proteomic findings were confirmed by ELISA in frontal and temporal cortices in which SNCA levels in Parkinson-dementia complex patient samples were quantitatively similar to patients with DLB and echo earlier findings of abnormal SNCA immunohistochemical findings in some patients with Parkinson-dementia complex (35–38).
FIGURE 4.
Experimental approach for isobaric tags for relative and absolute quantification labeling of 4 samples. MS, mass spectrometry.
Modified Proteins
Substantial antibody-based evidence supports accumulation of proteins that are pathologically modified by oxidative or nitrative reactions in several neurodegenerative diseases, including AD. One limitation of these earlier studies has been the difficulties in identification of the modified proteins. Proteomic techniques provide approaches to investigate this facet of AD pathogenesis.
Several elegant studies have combined immunochemical detection of protein carbonyls (a form of protein oxidation) in 2-DE followed by tandem MS for protein identification from soluble extracts of AD cerebrum. Proteins pathologically oxidized in AD include glutamine synthase, creatine kinase BB, UCHL1, α-enolase, and DRP-2/CRMP2 (39, 40). At least 3 of these oxidized proteins validated earlier results, including those on UCHL1-validated earlier results (41).
A similar approach coupled immunologic detection of nitrotyrosine residues with tandem MS to identify α-enolase, triosephosphate isomerase, and neuropolypeptide h3 as targets for this type of pathological modification in AD cerebral cortex (40, 42). Moreover, the authors extended their investigations to autopsy samples from individuals who died with amnestic mild cognitive impairment (MCI), a clinically defined condition that largely represents prodromal AD. They showed that modified proteins in amnestic MCI partially overlap with those identified in patients with dementia from AD (43, 44).
One group used affinity-purification with the MC1 monoclonal antibody to isolate a soluble fraction of PHF-tau from AD brain (45). In addition to identifying a large number of phosphorylated sites, they found that soluble PHF-tau is ubiquitin conjugated within its microtubule-binding domain at residues Lys-254, Lys-311, and Lys-353. Polyubiquitin conjugates formed mostly at Lys-48 of ubiquitin, suggesting a failure of the ubiquitin-proteasome system.
Another study investigated methionine oxidation to its sulfoxide in neuron-specific β-III tubulin and Aβ peptides (46). We used P-MOD to identify and map posttranslational modifications using tandem MS data from detergent-insoluble protein fractions from AD temporal cortex. Our results confirmed the direct observations of others by identifying methionine 35 sulfoxides in Aβ peptides and numerous sites of tau phosphorylation. P-MOD mapped several abundant methionine sulfoxides to neuron-enriched β-III tubulin but not α-III tubulin, its heterodimeric partner. These findings point to oxidative modification of β-III tubulin as a potential contributor to the neuronal cytoskeletal disruption in AD.
Lewy Body Disorders
Lewy body disorders are a recent consensus classification of diseases that share the histopathologic hallmark of LB formation (47). This includes several neurodegenerative diseases, the most common of which are PD and DLB. We previously described the proteomic analysis of laser capture microdissected LBs from cerebral cortex of patients with DLB. Basso et al (48) examined protein levels in the substantia nigra pars compacta from 4 patients who died of PD and 4 controls using 2-DE followed by MALDI-TOF-TOF. They identified 44 proteins in their subproteome extracted with urea and detergent. Nine of these showed abundant changes between the 2 groups; several were mitochondrial and reduced oxygen species-scavenging proteins. A recent study compared 3 subcellular fractions of frontal cortex from 4 groups: control individuals and patients with PD who had brainstem, limbic, or frontal cortex LB formation (49). Each sample was labeled with specific iTRAQ reagent before analysis by MALDI TOF/TOF. A total of 1,864 nonredundant proteins were identified; 199 of these displayed significant changes in relative abundance between PD groups and control. One particular mitochondrial protein, mortalin, was further validated by Western blot and found to be decreased in all PD groups as well as in a cellular model of PD (50). Mortalin seems to have several activities, including molecular chaperoning, mitochondrial import, and energy generation, and response to oxidative stress (51).
Other Neurodegenerative Diseases
Although there are several excellent studies of murine and cellular models of amyotrophic lateral sclerosis (ALS), Huntington disease, and prion diseases, we are unaware of any that has examined human brain or spinal cord from patients with these diseases.
PROTEOMIC STUDIES OF HUMAN CSF
Cerebrospinal fluid is widely considered an imperfect but relatively accessible portal into the neurochemical changes that accompany neurological diseases (52–57). Over the past few years, several groups have used proteomics to characterize the human CSF proteome and to profile CSF biomarkers related to a few major neurodegenerative diseases, such as AD, PD, and ALS (58–67). Together, these studies have generated lists of protein candidates associated with each disease that are altered in relative abundance within CSF. Major discrepancies have also been noted among different groups of investigators that are likely related to several factors especially preparation of the subproteome.
CSF Sample Collection and Storage
Human CSF derives from choroid plexus and the extracellular fluid of the brain and spinal cord (68); the volume of approximately 140 to 150 mL in an adult is renewed every 6 to 8 hours (69, 70). It has a rostrocaudal protein concentration gradient that is about 2.5-fold (71). Moreover, CSF production rate varies throughout the day and is altered by aging and some medications (72, 73). Thus, CSF provides a temporally limited window into the neurochemical activity of the CNS that is confounded by regionally varying mixture with a transudate of plasma, circadian variation, changes with age, and alteration by medications. Great care must obviously be taken, therefore, to match fractions of CSF obtained from individuals for study.
In addition to physiological variables, contamination with blood during lumbar puncture is another critical factor. The CSF protein content is approximately 1/200th that of blood, yet the protein profile overlaps extensively with blood (74–76). As a result, even minute contamination with blood can have large effects on the concentrations of CSF proteins. Several methods are available to test for trace contamination of blood in CSF samples, such as red blood cell density and measuring concentrations of exclusive blood proteins in CSF; one such protein is apoB (77). Because it is extremely difficult to obtain CSF samples without some blood contamination, we consider CSF with more than 10 red blood cells per microliter or an apoB serum: CSF concentration ratio less than 6,000 unacceptably contaminated by blood (58, 61, 77). Using relative apoB concentration is particularly helpful with samples that have been previously sedimented.
Although a seemingly trivial and straightforward procedure, CSF sample storage can significantly influence sample integrity. Protein degradation can occur, particularly when samples are kept at −20°C or have undergone multiple freeze-thaw cycles; this may be especially problematic for cystatin C (52, 78). Another unsettled issue related to CSF storage is the addition of protease inhibitors (79, 80). Currently, no recommendation has been made on this issue by the Human Proteome Organization.
Characterization of CSF Proteome
Several laboratories are using tandem MS to define the human CSF proteome. Early experiments used CSF and 2-DE and identified a few hundred proteins (81–84). The number of identified CSF proteins has increased exponentially since the application of LC-based proteomics (58, 61–63, 85–87). Indeed, with improvements in chromatography and MS, the human CSF proteome has been expanded to more than 2,000 proteins (88).
CSF Proteins in Neurodegenerative Diseases
Characterization of the human CSF proteome is 1 step toward identifying protein profiles that can aide in diagnosis and monitoring progression of neurodegenerative diseases. Several groups are pursuing this goal using a variety of platforms.
Surface-Enhanced Laser Desorption/Ionization
Several SELDI-based studies have focused on AD. Carrette and colleagues (52) have used SELDI to compare AD patients with controls and have identified several unique ion peaks that can differentiate disease from controls groups. Several of these putative AD biomarkers have been subsequently purified and identified as cystatin C, β2-microglobulin isoforms, to name a few. Notably, however, cystatin C is not only prone to freeze/thawing artifacts (78), but also seems to be nonspecific to AD as it also is altered in patients with Creutzfeldt-Jakob disease (89), multiple sclerosis (90), and chronic pain (91). One study used SELDI to investigate CSF from patients with AD or frontotemporal dementia versus controls; some peaks were discriminating, but no proteins were identified (92). Another study used SELDI analysis of CSF from 113 patients with MCI who were followed for up to 6 years and 28 controls who were followed for 3 years after lumbar puncture (93). These investigators discovered a panel of 17 candidate biomarkers that could distinguish between patients with stable MCI and patients with MCI who progressed to AD; however, the identity of most of these proteins is not yet known.
Two studies have reported SELDI-based identification of CSF biomarker candidates for ALS. One study identified 30 ion peaks with statistically significant differences between controls and patients with ALS. Transthyretin and cystatin C were identified and validated to be decreased in ALS CSF, whereas the carboxy-terminal fragment of the neuroendocrine protein 7B2 was increased in CSF from ALS patients (94). In another study, 3 candidates were identified by SELDI as being significantly lower in CSF from patients with ALS (n = 36) compared with controls (n = 21). These findings were validated in a separate set of samples, and 2 candidates were identified as cystatin C and a peptic fragment of neurosecretory protien VGF (66).
We are aware of 1 study that used SELDI to investigate CSF from patients with frontotemporal dementia (n = 16) versus controls (n = 12). Ten ion peaks with good signal-to-noise ratio and resolution were significantly differentially expressed in frontotemporal dementia; 5 were increased and 5 were decreased. Five of the ion peaks were purified and identified by tandem MS as a fragment of neurosecretory protien VGF, transthyretin (TTR), conjugated TTR, cystatin C, and a fragment of chromogranin B (95). Another group also has noted the potential for the relative concentrations of conjugated TTR in CSF to discriminate between AD and controls (96).
Two-Dimensional Gel Electrophoresis Tandem MS
Five major studies have used 2-DE coupled with tandem MS in the search for CSF biomarkers for AD. Castano et al (97) examined pooled CSF samples obtained from neuropathologically confirmed AD and nondemented control subjects (n = 43 for both groups) and identified 5 differentially expressed proteins in AD versus controls. Davidsson et al (59) identified 6 proteins and their isoforms that were significantly altered in CSF of AD (n = 15) patients versus controls (n = 12). Hu et al (65) discovered 11 proteins or isomers that are altered significantly in AD patients (n = 2) versus controls (n = 4). Finehout et al (64) reported a panel of 23 spots (21 that have been identified) that could be used to differentiate AD (n = 34) from non-AD (n = 9 normal controls and n = 24 other diseases) with high sensitivity and specificity. Puchades et al (60) compared samples from AD patients and normal control subjects (n = 7 for both groups) revealing 9 proteins to be significantly altered in AD. Only 10 proteins were identified in 2 or more of these studies. These include albumin precursor, apoA1, apoE, α-1β gly-coprotein, α-1 antitrypsin (or precursor), β-2 microglobulin, complement component 3, prostaglandin D synthase, retinal binding protein, and TTR.
Liquid Chromatography-Tandem MS
A third approach is multidimensional LC followed by tandem MS. One group used this technology coupled with isotope-coded affinity tags labeling to investigate specific changes in the proteome associated with aging (61) or AD (62) and identified approximately 400 CSF proteins, many of which had altered relative abundance in AD. The same groups subsequently used iTRAQ labeling to compare the CSF proteome of patients with AD, PD, DLB, and age-matched controls. More than 1,500 CSF proteins were identified; of those, 136, 72, and 101 proteins seemed to be uniquely associated with AD, PD, and DLB, respectively (58). Although these studies validated several of their findings, only a very small fraction of candidate biomarkers has been confirmed across platforms, underscoring the need for testing the use of proteomics-discovered candidates by alternative means in large-scale clinical studies.
Multianalyte Profile
We extended our iTRAQ proteomics discoveries by developing a multianalyte profile (MAP) for the 8 best-performing biomarker candidates and then conducted an independent clinical study using CSF from 95 control subjects, 48 patients with probable AD, and 40 patients with probable PD. Our 8-member MAP agreed with expert diagnosis for 95% of controls, 95% of probable PD, and 75% of probable AD. This MAP (in decreasing order of contribution to classification) consisted of tau, brain-derived neurotrophic factor, interleukin 8, Aβ42, β2-microglobulin, vitamin D-binding protein, apoAII, and apoE. This first large-scale clinical application of a proteomic-discovered MAP suggests a panel of 8 CSF proteins that are highly effective at identifying PD but only modestly effective at identifying AD (98). The MAPs for other candidate CSF biomarkers are pending. Finally, MAPs are not limited to antibody-based platforms. For example, MS-based MAPs for biomarker confirmation or validation are under active investigation (99). This new platform stands out for its precision without a requirement for antibodies.
CONCLUSIONS
The technology, experimental approaches, and bioinformatics that support proteomic research continue to undergo rapid evolution. The application of these new capabilities to the study of neurodegenerative diseases is providing insights into the biochemical pathogenesis of neurodegeneration as well as fueling major efforts in the discovery of biomarkers. Experience gained so far combined with expected advances in the near future will lead to greater throughput, accuracy, and reproducibility. Recent studies previously cited and additional future studies will focus on the critically important issues of disease specificity and changes in the proteome that occur during latent and prodromal stages of disease. Ultimately, the convergence of validated and confirmed data from multiple laboratories will establish richly informative consensus subproteomes for human brain regions and CSF in health and disease.
Acknowledgments
This work was supported by Grants AG05136 and AG029808 from the National Institutes of Health, by the Michael J. Fox Foundation for Parkinson’s Research, by the C.-M. Shaw endowment, and by the Nancy and Buster Alvord Endowment.
REFERENCES
- 1.Ideker T, Thorsson V, Ranish JA, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MD, Yu LR, Conrads TP, et al. Proteome analysis of DNA damage-induced neuronal death using high throughput mass spectrometry. J Biol Chem. 2004;279:26685–26697. doi: 10.1074/jbc.M401274200. [DOI] [PubMed] [Google Scholar]
- 3.Griffin TJ, Gygi SP, Ideker T, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Tsuang DW, Bird TD. Genetics of dementia. Med Clin North Am. 2002;86:591–614. doi: 10.1016/s0025-7125(02)00003-2. [DOI] [PubMed] [Google Scholar]
- 5.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 6.Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Haqqani AS, Kelly JF, Stanimirovic DB. Quantitative protein profiling by mass spectrometry using label-free proteomics. In: Starkey M, Elaswarapu R, editors. Genomics Protocols. vol. 439. Totowa, NJ: Humana Press; 2008. pp. 241–256. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra M, Vonk RJ, Jansen RC. SELDI-TOF mass spectra: A view on sources of variation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:12–23. doi: 10.1016/j.jchromb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Albrethsen J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007;53:852–858. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
- 12.Cornett DS, Reyzer ML, Chaurand P, et al. MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 13.Tanner S, Shu H, Frank A, et al. InsPecT: Identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 14.Hansen BT, Davey SW, Ham AJ, et al. P-Mod: An algorithm and software to map modifications to peptide sequences using tandem MS data. J Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 15.Schonberger SJ, Edgar PF, Kydd R, et al. Proteomic analysis of the brain in Alzheimer’s disease: Molecular phenotype of a complex disease process. Proteomics. 2001;1:1519–1528. doi: 10.1002/1615-9861(200111)1:12<1519::aid-prot1519>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Shiozaki A, Tsuji T, Kohno R, et al. Proteome analysis of brain proteins in Alzheimer’s disease: Subproteomics following sequentially extracted protein preparation. J Alzheimers Dis. 2004;6:257–268. doi: 10.3233/jad-2004-6306. [DOI] [PubMed] [Google Scholar]
- 17.Kim SH, Vlkolinsky R, Cairns N, et al. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer’s disease and Down syndrome. Cell Mol Life Sci. 2000;57:1810–1816. doi: 10.1007/PL00000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krapfenbauer K, Engidawork E, Cairns N, et al. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 19.Liao L, Cheng D, Wang J, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279:37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Woltjer RL, Cimino PJ, et al. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Nowotny P, Holmans P, et al. Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101:15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin PI, Martin ER, Bronson PG, et al. Exploring the association of glyceraldehyde-3-phosphate dehydrogenase gene and Alzheimer disease. Neurology. 2006;67:64–68. doi: 10.1212/01.wnl.0000223438.90113.4e. [DOI] [PubMed] [Google Scholar]
- 23.Olah J, Tokesi N, Vincze O, et al. Interaction of TPPP/p25 protein with glyceraldehyde-3-phosphate dehydrogenase and their co-localization in Lewy bodies. FEBS Lett. 2006;580:5807–5814. doi: 10.1016/j.febslet.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 24.Cumming RC, Schubert D. Amyloid-beta induces disulfide bonding and aggregation of GAPDH in Alzheimer’s disease. FASEB J. 2005;19:2060–2062. doi: 10.1096/fj.05-4195fje. [DOI] [PubMed] [Google Scholar]
- 25.Sultana R, Boyd-Kimball D, Cai J, et al. Proteomics analysis of the Alzheimer’s disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- 26.Wakabayashi K, Tanji K, Mori F, et al. The Lewy body in Parkinson’s disease: Molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology. 2007;27:494–506. doi: 10.1111/j.1440-1789.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- 27.Leverenz JB, Umar I, Wang Q, et al. Proteomic identification of novel proteins in cortical Lewy bodies. Brain Pathol (Zurich, Switzerland) 2007;17:139–145. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba M, Nakajo S, Tu PH, et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 29.Trojanowski JQ, Lee VM. Brain degeneration linked to “fatal attractions” of proteins in Alzheimer’s disease and related disorders. J Alzheimers Dis. 2001;3:117–119. doi: 10.3233/jad-2001-3116. [DOI] [PubMed] [Google Scholar]
- 30.Glenner GG, Wong CW. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophy Res Comm. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 31.Lee VM, Balin BJ, Otvos L, Jr, et al. A68: A major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 32.Skovronsky DM, Lee VM, Trojanowski JQ. Neurodegenerative diseases: New concepts of pathogenesis and their therapeutic implications. Ann Rev Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 33.Woltjer RL, Cimino PJ, Boutte AM, et al. Proteomic determination of widespread detergent-insolubility including Aβ but not tau early in the pathogenesis of Alzheimer’s disease. FASEB J. 2005;19:1923–1925. doi: 10.1096/fj.05-4263fje. [DOI] [PubMed] [Google Scholar]
- 34.Yang W, Woltjer RL, Sokal I, et al. Quantitative proteomics identifies surfactant-resistant alpha-synuclein in cerebral cortex of Parkinsonism-dementia complex of Guam but not Alzheimer’s disease or progressive supranuclear palsy. Am J Pathol. 2007;171:993–1002. doi: 10.2353/ajpath.2007.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winton MJ, Joyce S, Zhukareva V, et al. Characterization of tau pathologies in gray and white matter of Guam parkinsonism-dementia complex. Acta Neuropathol (Berl) 2006;111:401–412. doi: 10.1007/s00401-006-0053-0. [DOI] [PubMed] [Google Scholar]
- 36.Sebeo J, Hof PR, Perl DP. Occurrence of alpha-synuclein pathology in the cerebellum of Guamanian patients with parkinsonism-dementia complex. Acta Neuropathol. 2004;107:497–503. doi: 10.1007/s00401-004-0840-4. [DOI] [PubMed] [Google Scholar]
- 37.Forman MS, Schmidt ML, Kasturi S, et al. Tau and alpha-synuclein pathology in amygdala of Parkinsonism-dementia complex patients of Guam. Am J Pathol. 2002;160:1725–1731. doi: 10.1016/s0002-9440(10)61119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamazaki M, Arai Y, Baba M, et al. Alpha-synuclein inclusions in amygdala in the brains of patients with the parkinsonism-dementia complex of Guam. J Neuropathol Exp Neurol. 2000;59:585–591. doi: 10.1093/jnen/59.7.585. [DOI] [PubMed] [Google Scholar]
- 39.Castegna A, Aksenov M, Aksenova M, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 40.Castegna A, Aksenov M, Thongboonkerd V, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II: Dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 41.Choi J, Levey AI, Weintraub ST, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 42.Butterfield DA, Sultana R. Identification of 3-Nitrotyrosine-modified brain proteins by redox proteomics. Methods Enzymol. 2008;440:295–308. doi: 10.1016/S0076-6879(07)00819-1. [DOI] [PubMed] [Google Scholar]
- 43.Sultana R, Reed T, Perluigi M, et al. Proteomic identification of nitrated brain proteins in amnestic mild cognitive impairment: A regional study. J Cell Mol Med. 2007;11:839–851. doi: 10.1111/j.1582-4934.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butterfield DA, Reed TT, Perluigi M, et al. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: Implications for the role of nitration in the progression of Alzheimer’s disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cripps D, Thomas SN, Jeng Y, et al. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is poly-ubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 46.Boutte AM, Woltjer RL, Zimmerman LJ, et al. Selectively increased oxidative modifications mapped to detergent-insoluble forms of Aβ and beta-III tubulin in Alzheimer’s disease. FASEB J. 2006;20:1473–1483. doi: 10.1096/fj.06-5920com. [DOI] [PubMed] [Google Scholar]
- 47.Lippa CF, Duda JE, Grossman M, et al. DLB and PDD boundary issues: Diagnosis, treatment, molecular pathology, and biomarkers. Neurology. 2007;68:812–819. doi: 10.1212/01.wnl.0000256715.13907.d3. [DOI] [PubMed] [Google Scholar]
- 48.Basso M, Giraudo S, Corpillo D, et al. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 49.Shi M, Jin J, Wang Y, et al. Mortalin: A protein associated with progression of Parkinson disease? J Neuropathol Exp Neurol. 2008;67:117–124. doi: 10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- 50.Jin J, Hulette C, Wang Y, et al. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: Relevance to Parkinson disease. Mol Cell Proteomics. 2006;5:1193–1204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Kaul SC, Deocaris CC, Wadhwa R. Three faces of mortalin: A housekeeper, guardian and killer. Exp Gerontol. 2007;42:263–274. doi: 10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 52.Carrette O, Demalte I, Scherl A, et al. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer’s disease. Proteomics. 2003;3:1486–1494. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- 53.Blennow K. CSF biomarkers for Alzheimer’s disease: Use in early diagnosis and evaluation of drug treatment. Expert Rev Mol Diagn. 2005;5:661–672. doi: 10.1586/14737159.5.5.661. [DOI] [PubMed] [Google Scholar]
- 54.Davidsson P, Sjogren M. The use of proteomics in biomarker discovery in neurodegenerative diseases. Dis Markers. 2005;21:81–92. doi: 10.1155/2005/848676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Ascenzo M, Relkin NR, Lee KH. Alzheimer’s disease cerebrospinal fluid biomarker discovery: A proteomics approach. Curr Opin Mol Ther. 2005;7:557–564. [PubMed] [Google Scholar]
- 56.Galasko D. Biomarkers for Alzheimer’s disease—clinical needs and application. J Alzheimers Dis. 2005;8:339–346. doi: 10.3233/jad-2005-8403. [DOI] [PubMed] [Google Scholar]
- 57.Olsson A, Vanderstichele H, Andreasen N, et al. Simultaneous measurement of {beta}-amyloid(1–42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin Chem. 2005;51:336–345. doi: 10.1373/clinchem.2004.039347. [DOI] [PubMed] [Google Scholar]
- 58.Abdi F, Quinn JF, Jankovic J, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 59.Davidsson P, Westman-Brinkmalm A, Nilsson CL, et al. Proteome analysis of cerebrospinal fluid proteins in Alzheimer patients. Neuroreport. 2002;13:611–615. doi: 10.1097/00001756-200204160-00015. [DOI] [PubMed] [Google Scholar]
- 60.Puchades M, Hansson SF, Nilsson CL, et al. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer’s disease. Brain Res Mol Brain Res. 2003;118:140–146. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Goodlett DR, Peskind ER, et al. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol Aging. 2005;26:207–227. doi: 10.1016/j.neurobiolaging.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Goodlett DR, Quinn JF, et al. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7:125–133. doi: 10.3233/jad-2005-7205. [DOI] [PubMed] [Google Scholar]
- 63.Pan S, Wang Y, Quinn JF, et al. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 64.Finehout EJ, Franck Z, Choe LH, et al. Cerebrospinal fluid proteomic biomarkers for Alzheimer’s disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 65.Hu Y, Malone JP, Fagan AM, et al. Comparative proteomic analysis of intra-and interindividual variation in human cerebrospinal fluid. Mol Cell Proteomics. 2005;4:2000–2009. doi: 10.1074/mcp.M500207-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Pasinetti GM, Ungar LH, Lange DJ, et al. Identification of potential CSF biomarkers in ALS. Neurology. 2006;66:1218–1222. doi: 10.1212/01.wnl.0000203129.82104.07. [DOI] [PubMed] [Google Scholar]
- 67.Bowser R, Cudkowicz M, Kaddurah-Daouk R. Biomarkers for amyotrophic lateral sclerosis. Expert Rev Mol Diagn. 2006;6:387–398. doi: 10.1586/14737159.6.3.387. [DOI] [PubMed] [Google Scholar]
- 68.McComb JG. Recent research into the nature of cerebrospinal fluid formation and absorption. J Neurosurg. 1983;59:369–383. doi: 10.3171/jns.1983.59.3.0369. [DOI] [PubMed] [Google Scholar]
- 69.Han CY, Backous DD. Basic principles of cerebrospinal fluid metabolism and intracranial pressure homeostasis. Otolaryngol Clin North Am. 2005;38:569–576. doi: 10.1016/j.otc.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 70.Bergsneider M. Evolving concepts of cerebrospinal fluid physiology. Neurosurg Clin N Am. 2001;12:631–638. vii. [PubMed] [Google Scholar]
- 71.Huhmer AF, Biringer RG, Amato H, et al. Protein analysis in human cerebrospinal fluid: Physiological aspects, current progress and future challenges. Dis Markers. 2006;22:3–26. doi: 10.1155/2006/158797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nilsson C, Stahlberg F, Thomsen C, et al. Circadian variation in human cerebrospinal fluid production mesured by magnetic resonance imaging. Am J Physiol. 1992;262:R20–R24. doi: 10.1152/ajpregu.1992.262.1.R20. [DOI] [PubMed] [Google Scholar]
- 73.Preston JE. Ageing choroid plexus-cerebrospinal fluid system. Microsc Res Tech. 2001;52:31–37. doi: 10.1002/1097-0029(20010101)52:1<31::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 74.Blennow K, Fredman P, Wallin A, et al. Protein analysis in cerebrospinal fluid. II. Reference values derived from healthy individuals 18–88 years of age. Eur Neurol. 1993;33:129–133. doi: 10.1159/000116919. [DOI] [PubMed] [Google Scholar]
- 75.Blennow K, Fredman P, Wallin A, et al. Protein analyses in cerebrospinal fluid. I. Influence of concentration gradients for proteins on cerebrospinal fluid/serum albumin ratio. Eur Neurol. 1993;33:126–128. doi: 10.1159/000116918. [DOI] [PubMed] [Google Scholar]
- 76.Blennow K, Fredman P, Wallin A, et al. Protein analysis in cerebrospinal fluid. III. Relation to blood-cerebrospinal fluid barrier function for formulas for quantitative determination of intrathecal IgG production. Eur Neurol. 1993;33:134–142. doi: 10.1159/000116920. [DOI] [PubMed] [Google Scholar]
- 77.Osman I, Gaillard O, Meillet D, et al. A sensitive time-resolved immunofluorometric assay for the measurement of apolipoprotein B in cerebrospinal fluid. Application to multiple sclerosis and other neurological diseases. Eur J Clin Chem Clin Biochem. 1995;33:53–58. doi: 10.1515/cclm.1995.33.1.53. [DOI] [PubMed] [Google Scholar]
- 78.Carrette O, Burkhard PR, Hughes S, et al. Truncated cystatin C in cerebrospinal fluid: Technical [corrected] artefact or biological process? Proteomics. 2005;5:3060–3065. doi: 10.1002/pmic.200402039. [DOI] [PubMed] [Google Scholar]
- 79.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hardt M, Thomas LR, Dixon SE, et al. Toward defining the human parotid gland salivary proteome and peptidome: Identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry. 2005;44:2885–2899. doi: 10.1021/bi048176r. [DOI] [PubMed] [Google Scholar]
- 81.Davidsson P, Nilsson CL. Peptide mapping of proteins in cerebrospinal fluid utilizing a rapid preparative two-dimensional electrophoretic procedure and matrix-assisted laser desorption/ionization mass spec-trometry. Biochim Biophys Acta. 1999;1473:391–399. doi: 10.1016/s0304-4165(99)00197-x. [DOI] [PubMed] [Google Scholar]
- 82.Raymackers J, Daniels A, De Brabandere V, et al. Identification of two-dimensionally separated human cerebrospinal fluid proteins by N-terminal sequencing, matrix-assisted laser desorption/ionization–mass spectrometry, nanoliquid chromatography-electrospray ionization-time of flight-mass spectrometry, and tandem mass spectrometry. Electrophoresis. 2000;21:2266–2283. doi: 10.1002/1522-2683(20000601)21:11<2266::AID-ELPS2266>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 83.Sickmann A, Dormeyer W, Wortelkamp S, et al. Towards a high resolution separation of human cerebrospinal fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:167–196. doi: 10.1016/s1570-0232(01)00626-2. [DOI] [PubMed] [Google Scholar]
- 84.Finehout EJ, Franck Z, Lee KH. Towards two-dimensional electrophoresis mapping of the cerebrospinal fluid proteome from a single individual. Electrophoresis. 2004;25:2564–2575. doi: 10.1002/elps.200406012. [DOI] [PubMed] [Google Scholar]
- 85.Maccarrone G, Milfay D, Birg I, et al. Mining the human cerebrospinal fluid proteome by immunodepletion and shotgun mass spectrometry. Electrophoresis. 2004;25:2402–2412. doi: 10.1002/elps.200305909. [DOI] [PubMed] [Google Scholar]
- 86.Ramstrom M, Ivonin I, Johansson A, et al. Cerebrospinal fluid protein patterns in neurodegenerative disease revealed by liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:4010–4018. doi: 10.1002/pmic.200400871. [DOI] [PubMed] [Google Scholar]
- 87.Wenner BR, Lovell MA, Lynn BC. Proteomic analysis of human ventricular cerebrospinal fluid from neurologically normal, elderly subjects using two-dimensional LC-MS/MS. J Proteome Res. 2004;3:97–103. doi: 10.1021/pr034070r. [DOI] [PubMed] [Google Scholar]
- 88.Xu J, Chen J, Peskind E, et al. Characterization of proteome of human cerebrospinal fluid. Int Rev Neurobiol. 2006;73:29–98. doi: 10.1016/S0074-7742(06)73002-1. [DOI] [PubMed] [Google Scholar]
- 89.Sanchez JC, Guillaume E, Lescuyer P, et al. Cystatin C as a potential cerebrospinal fluid marker for the diagnosis of Creutzfeldt-Jakob disease. Proteomics. 2004;4:2229–2233. doi: 10.1002/pmic.200300799. [DOI] [PubMed] [Google Scholar]
- 90.Irani DN, Anderson C, Gundry R, et al. Cleavage of cystatin C in the cerebrospinal fluid of patients with multiple sclerosis. Ann Neurol. 2006;59:237–247. doi: 10.1002/ana.20786. [DOI] [PubMed] [Google Scholar]
- 91.Mannes AJ, Martin BM, Yang HY, et al. Cystatin C as a cerebrospinal fluid biomarker for pain in humans. Pain. 2003;102:251–256. doi: 10.1016/S0304-3959(02)00403-7. [DOI] [PubMed] [Google Scholar]
- 92.Simonsen AH, McGuire J, Podust VN, et al. A novel panel of cerebrospinal fluid biomarkers for the differential diagnosis of Alzheimer’s disease versus normal aging and frontotemporal dementia. Dement Geriatr Cogn Disord. 2007;24:434–440. doi: 10.1159/000110576. [DOI] [PubMed] [Google Scholar]
- 93.Simonsen AH, McGuire J, Hansson O, et al. Novel panel of cerebrospinal fluid biomarkers for the prediction of progression to Alzheimer dementia in patients with mild cognitive impairment. Arch Neurol. 2007;64:366–370. doi: 10.1001/archneur.64.3.366. [DOI] [PubMed] [Google Scholar]
- 94.Ranganathan S, Williams E, Ganchev P, et al. Proteomic profiling of cerebrospinal fluid identifies biomarkers for amyotrophic lateral sclerosis. J Neurochem. 2005;95:1461–1471. doi: 10.1111/j.1471-4159.2005.03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruetschi U, Zetterberg H, Podust VN, et al. Identification of CSF biomarkers for frontotemporal dementia using SELDI-TOF. Experimental Neurology. 2005;196:273–281. doi: 10.1016/j.expneurol.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 96.Biroccio A, Del Boccio P, Panella M, et al. Differential post-translational modifications of transthyretin in Alzheimer’s disease: A study of the cerebral spinal fluid. Proteomics. 2006;6:2305–2313. doi: 10.1002/pmic.200500285. [DOI] [PubMed] [Google Scholar]
- 97.Castano EM, Roher AE, Esh CL, et al. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer’s disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 98.Zhang J, Sokal I, Peskind ER, et al. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129:526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan S, Rush J, Peskind ER, et al. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7:720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]