Abstract
PI3-kinase and PTEN are major positive and negative regulators, respectively, of the PI3-kinase pathway, which regulates growth, survival, and proliferation. These key signaling components are two of the most frequently mutated proteins in human cancers, resulting in unregulated activation of PI3K signaling and providing irrefutable genetic evidence of the central role of this pathway in tumorigenesis. PTEN regulates PI3K signaling by dephosphorylating the lipid signaling intermediate PIP3, but PTEN may have additional phosphatase-independent activities, as well as other functions in the nucleus. In this review, we highlight current work showing cancer-relevant complexities in the regulation of PTEN and PI3K activity, potential novel functions for PTEN, and feedback regulation within the pathway. The significance and complexity of PI3K signaling make it an important but challenging therapeutic target for cancer.
Keywords: PIK3CA, tumor suppressor, phosphatase, mTOR, mutations
INTRODUCTION
The phosphatidylinositol 3–kinase (PI3K) pathway is evolutionarily conserved from yeast to mammals. In higher eukaryotes the PI3K pathway regulates diverse cellular processes, including metabolism, survival, proliferation, apoptosis, growth, and cell migration; it also participates in specialized context-dependent functions (1).
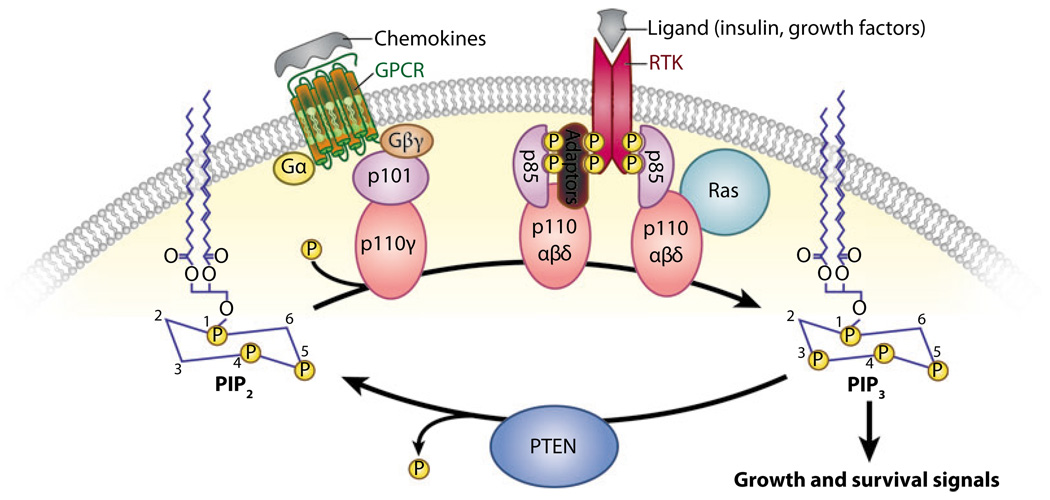
PI3Ks are a family of intracellular lipid kinases that phosphorylate the 3′-hydroxyl group of phosphatidylinositols and phosphoinositides. PI3Ks are classified into three groups (classes I, II, and III) based on structure and substrate specificity (1). Class I PI3Ks primarily phosphorylate phosphatidylinositol-4,5-bisphosphate (PIP2) to generate the lipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) (Figure 1). Members of this group are further subdivided according to the signaling receptors that activate them. Class IA PI3Ks are activated by growth factor receptor tyrosine kinases (RTKs), whereas class IB PI3Ks are activated by G protein–coupled receptors (GPCRs) (1). Members of class IA are heterodimers of a regulatory subunit (with three isoforms: p85α, p85β, and p55γ) and a p110 catalytic subunit (also with three isoforms: α, β, and δ). Class IB members consist of a p101-regulatory subunit (of which two potential homologous subunits have been reported: p84 and p87PIKAP) and a p110γ catalytic subunit. In mammals, class IA PI3Ks transduce signals from insulin and growth factors to regulate proliferation, survival, growth, and glucose homeostasis (1).
Figure 1.
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is an antagonist of class I phosphatidylinositol 3—kinase (PI3K) signaling. In response to extracellular stimuli (e.g., insulin, growth factors, chemokines), the catalytic subunit of PI3K (p110) is recruited to receptor tyrosine kinases (RTKs) or G protein—coupled receptors (GPCRs) at the membrane through its regulatory subunit (p85 or p101), where it phosphorylates phosphatidylinositol-4,5 bisphosphate (PIP2) to generate phosphatidylinositol-3,4,5 trisphosphate (PIP3). PTEN is a lipid phosphatase that antagonizes the action of PI3K by dephosphorylating PIP3 at position D3 to generate PIP2.
Class II and class III PI3Ks use phosphatidylinositol (PI) as a substrate to generate PI-3-P. Class II PI3Ks bind clathrin in coated pits, suggesting a function in membrane trafficking and receptor internalization (2). There is one class III PI3K in mammals, VPS34, which represents the only mammalian PI3K that is also conserved in yeast. VPS34 acts as a sensor of the available amino acids and signals to mammalian target of rapamycin (mTOR) to regulate cell growth and autophagy in response to low nutrient pools (3, 4).
Although signaling through all classes of PI3K is connected to key growth-regulatory processes, thus far a central role in cancer has been demonstrated selectively for class IA PI3Ks. Class IA PI3Ks transduce signals downstream of oncogenic RTKs, and PIK3CA, encoding the class IA PI3K catalytic subunit p110α, is the only PI3K gene identified with common mutations in human cancer. Therefore, we focus our discussion on the cancer relevance of signaling through class IA PI3Ks.
SIGNALING THROUGH THE PI3-KINASE PATHWAY
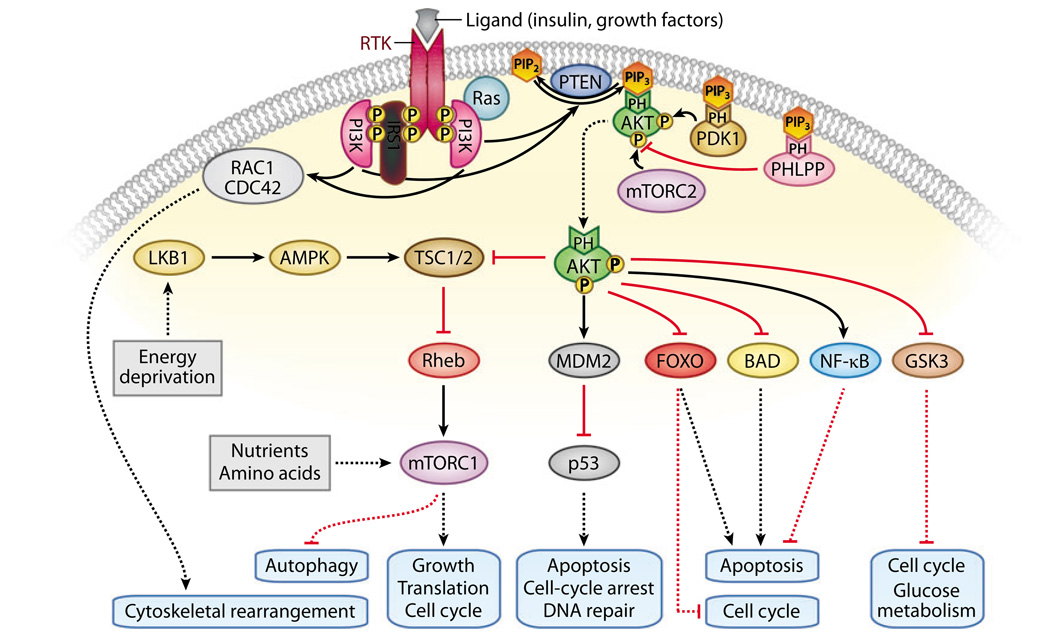
A wealth of knowledge regarding PI3K regulation has come from genetic studies in Caenorhabditis elegans and Drosophila melanogaster, both of which have a single class IA PI3K controlling growth and metabolism downstream of a unique insulin-like receptor (1). Although this information has illuminated our understanding of PI3K signaling, the pathway is more complex in mammals due to the evolution of multiple family members for some effectors in the pathway and the diversity of growth factors that transmit their signals through PI3K in context-dependent fashions. In response to extracellular cues, the catalytic subunits of class I PI3Ks are recruited to activated membrane receptors through their regulatory subunits. Activated PI3Ks catalyze the formation of PIP3 from PIP2, and the lipid phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome 10) directly opposes the activity of PI3Ks by dephosphorylating PIP3 into PIP2 (Figure 1), thus acting as the central negative regulator of PI3K. PIP3 transduces activating signals by binding to the pleckstrin homology (PH) domains of proteins, thereby recruiting them to the membrane (1). The serine-threonine kinase AKT (also known as PKB), is a centrally important downstream effector of PIP3 (Figure 2). AKT is recruited to the membrane via PIP3 binding ofitsPH domain and is fully activated following phosphorylation either by PDK1 (3-phosphoinositidedependent kinase) at threonine 308 (5) and at serine 473 by the rapamycin-insensitivemTOR complex (mTORC2) (6) or potentially by other kinases in some contexts (7).
Figure 2.
The phosphatidylinositol 3—kinase (PI3K) signaling pathway. Activated receptor tyrosine kinases (RTKs) recruit and activate PI3K, leading to increased phosphatidylinositol-3,4,5-trisphosphate (PIP3) levels. PIP3 recruits many proteins to the membrane by binding to their pleckstrin homology (PH) domains, including the serine/threonine kinases AKT, 3-phosphoinositide-dependent kinase (PDK1), and the phosphatase PH domain and leucine rich repeat protein phosphatase (PHLPP). Membrane-bound AKT is rendered fully active through its phosphorylation by PDK1 and the rapamycin-insensitive mammalian target of rapamycin (mTOR) complex (mTORC2), and it is inactivated when dephosphorylated by PHLPP. Activated AKT may phosphorylate a range of substrates, thereby activating or inhibiting these targets and resulting in cellular growth, survival, and proliferation through various mechanisms. PI3K can also regulate downstream targets, such as RAC1/CDC42, in an AKT-independent manner. Activation of the rapamycin-sensitive mTOR complex (mTORC1) is inhibited by the tuberous sclerosis complex (TSC1 and TSC2) 1 and 2, which can be regulated by AKT as well as through PI3K-independent signaling from LKB1 and AMP-activated protein kinase (AMPK). Abbreviations: GSK, glycogen synthase kinase; NF-κB, nuclear factor—κ B; PIP2, phosphatidylinositol-4,5 bisphosphate; RAC1, Ras-related C3 botulinum toxin substrate 1.
FOXO and TSC2 (tuberous sclerosis complex 2) are two key substrates of AKT that are conserved in insulin signaling in invertebrates. AKT-mediated phosphorylation of the transcription factor FOXO can increase proliferation and survival by causing FOXO to be retained in the cytoplasm, preventing it from activating transcription of cell-cycle-regulatory genes such as p27Kip1 and proapoptotic genes such as FasL and Bim (1). TSC1 (hamartin) and TSC2 (tuberin) form a complex that inhibits activity of the small G protein Rheb. AKT-mediated phosphorylation of TSC2 relieves its inhibition of Rheb activity, leading to activation of the rapamycin-sensitivemTOR complex mTORC1. The TSC complex is also activated under nutrient-/energy-poor conditions by the action of the serine/threonine ki-nase LKB1/STK11 (serine/threonine protein kinase 11) and AMPK (AMP-activated protein kinase), leading to the attenuation ofmTORC1 signaling. mTORC1 activity promotes growth through upregulation of protein synthesis, at least in part through modulation of two important components of the protein synthesis machinery, 4E-BP1 (eukaryotic translation initiation factor 4E–binding protein 1) and p70S6 kinase (8).
PTEN, TSC1, TSC2, and LKB1 are all tumor-suppressor genes that negatively regulate mTORC1 activity, and their inherited mutation results in distinct familial syndromes with some shared clinical features including cancer predisposition and multiple hamartomas (9, 10). AKT can also phosphorylate a wide array of additional substrates that also influence growth, proliferation, and survival (11). AKTmediated phosphorylation inhibits the activities of some proteins such as the proapoptotic protein BAD and glycogen synthase kinase 3 (GSK3), which modulates glucose metabolism as well as cell-cycle-regulatory proteins (12). For other substrates such as MDM2, which promotes degradation of the tumor-suppressor p53, or the transcription factor nuclear factor–kappa B (NF-κB), AKT-mediated phosphorylation enhances activity (13). AKT can regulate multiple targets that promote aerobic glycolysis, a metabolic feature of many cancer cells (14). The factors that determine which collection of AKT substrates is targeted in response to different PI3K-activating signals remain somewhat unclear, although they may be determined in part by (a) the selective expression of specific substrates in particular cell types and (b) cross-talk with other signaling pathways. In addition to the host of downstream targets of PI3K signaling that drive cancer-relevant processes such as cell-cycle progression and survival, PIP3 also influences cell motility by signaling through Rac and Cdc42 (15) (Figure 2), a connection that may be relevant to tumor cell invasion as well as normal developmental roles for PI3K signaling.
LINKING THE PI3-KINASE PATHWAY TO CANCER
A clear link between the PI3K pathway and cancer was established in the 1980s, when the pathway’s lipid kinase activity was associated with two viral oncoproteins, the src protein of Rous sarcoma virus and the middle-T protein of polyoma virus (16, 17). Binding of SH2-containing p85 to phosphotyrosines on the viral oncoproteins recruits the p110 catalytic subunit into these oncogenic cellular complexes, leading to activation of the pathway (18). Similarly, PI3K is activated by RTKs, including several oncogenic growth factor receptors such as epidermal growth factor receptor, platelet-derived growth factor receptor, and mesenchymalepithelial transition factor, thereby illustrating the participation of this pathway in transducing cancer-relevant cues (19–21). Definitive evidence for the oncogenicity of PI3K was provided by the isolation of a constitutively active p110α isoform from the genome of the oncogenic avian retrovirus ASV16 (22). Most importantly, somatic mutations in human cancer specifically target p110α and PTEN at very high frequency, resulting in increased activity of the PI3K signaling pathway and providing clear genetic evidence for a central role for PI3K in human cancer (Table 1).
Table 1. Incidence of PTEN and PIK3CA mutations in human cancers.
| Primary tumor tissue | Percentage of tumors with mutation/number of samples | |||
|---|---|---|---|---|
| PIK3CA | PTEN | |||
| % | # | % | # | |
| Prostate | 29 | 7 | 14 | 371 |
| Breast | 27 | 987 | 6 | 561 |
| Endometrium | 23 | 199 | 38 | 1467 |
| Colon | 15 | 1128 | 9 | 344 |
| Urinary tract | 17 | 162 | 9 | 142 |
| Upper aerodigestive tract | 10 | 229 | 4 | 529 |
| Ovary | 8 | 670 | 8 | 574 |
| Stomach | 8 | 362 | 5 | 446 |
| Liver | 7 | 253 | 5 | 354 |
| Esophagus | 7 | 124 | 1 | 94 |
| Pancreas | 6 | 66 | 1 | 67 |
| Central nervous system | 5 | 808 | 20 | 2758 |
| Hematopoietic and lymphoid tissue | 4 | 510 | 6 | 866 |
| Lung | 3 | 537 | 8 | 548 |
| Skin | 3 | 149 | 17 | 555 |
| Thyroid | 2 | 186 | 5 | 591 |
Values taken from http://www.sanger.ac.uk/genetics/CGP/cosmic.
Activating Mutations in the p110α Catalytic Subunit of PI3-Kinase
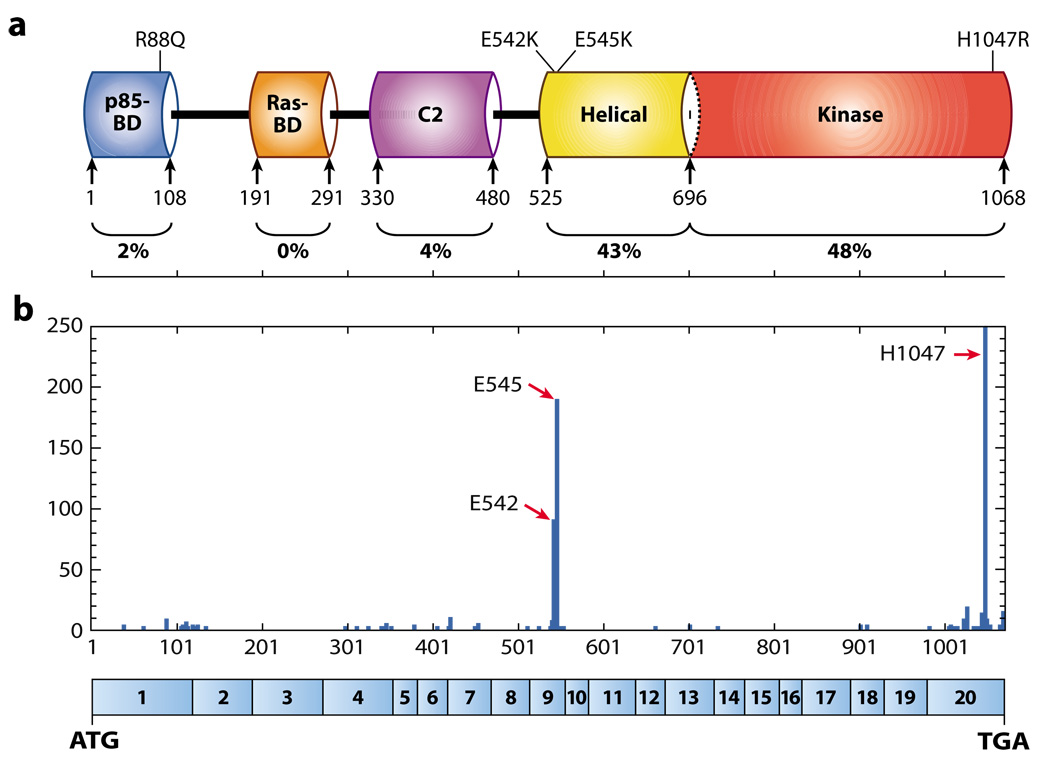
Among the four different isoforms of the p110 catalytic subunit of PI3K, PIK3CA—the gene encoding the p110α catalytic subunit—is the only gene frequently mutated in human cancer (23). PIK3CA is frequently amplified in head and neck, cervical, gastric, and lung cancers (1). To determine the potential involvement of point mutations in activation of PI3K pathway members, Samuels and coworkers (23) evaluated the sequences encoding the kinase domains of eight PI3K genes and eight PI3K-like genes from a large collection of colorectal carcinomas and identified frequent mutations in PIK3CA. They then evaluated the entire coding sequence of the gene and found three hot spot mutations at the highest frequency. Subsequently, somatic point mutations in PIK3CA have been found in a significant fraction of commonly occurring human tumors (Table 1). The highest incidence of PIK3CA mutations was seen in prostate, breast, endometrium, and colon cancers, which are common in the population. However, PIK3CA mutations were also found in a significant fraction of other tumor types (Table 1). With few exceptions, most of those mutations are missense substitutions [see the COSMIC (Catalogue of Somatic Mutations in Cancer) database: http://www. sanger.ac.uk/genetics/CGP/cosmic]. Strikingly, around 80% of PIK3CA mutations are one of the three hot spot mutations identified in the original study: E542K and E545K in the helical domain and H1047R in the kinase domain (24) (Figure 3). These mutations show increased PI3K activity in vitro (23, 25, 26), lead to growth factor–independent activation of AKT (25, 27), and induce transformation of fibroblasts and mammary epithelial cells (25, 26, 28). The identification of hot spot regions for mutation may lead to an underestimate of the total frequency of PIK3CA mutations, as some sequencing studies focus only on the hot spot regions, not the full open reading frame, and therefore may fail to detect the full spectrum of mutations in this gene.
Figure 3.
p110α protein structure and mutation distribution. (a) p110α is characterized by five functional domains: p85-regulatory subunit—binding domain (p85-BD), Ras-binding domain (Ras-BD), C2 domain, helical domain, and kinase catalytic domain. The percentage of mutations identified in each domain is shown at bottom. (b) Distribution of cancer-specific mutations in PIK3CA and their relative frequency of occurrence in the functional domains. The three hot spots for mutations (E542, E545, H1047) are depicted. Values are taken from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://www.sanger.ac.uk/genetics/CGP/cosmic) and include single substitutions and complex mutations. Amino acid numbers are listed along the x axis, with the corresponding PIK3CA exon structure encoding p110α shown in blue boxes below. The numbers of mutations are listed along the y axis. Abbreviations: ATG, start codon; TGA, stop codon.
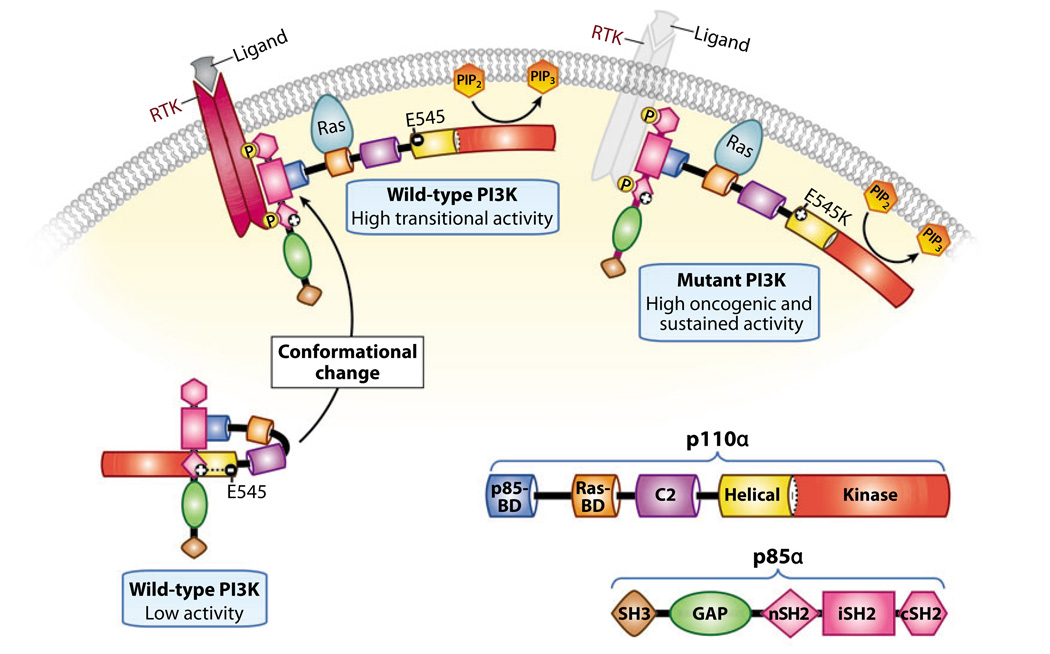
Structural studies of p110α predicted five main domains: an N-terminal adaptor-binding domain that binds to the p85-regulatory subunit, a Ras-binding domain, a C2 domain, a helical domain, and a C-terminal kinase domain (24, 29) (Figure 3a). Two elegant structural studies provided insights into how certain p110α mutations promote cancer cell growth and survival through activation of the PI3K pathway (Figure 4). First, Miled et al. (30) solved the crystal structure of the adaptorbinding domain and the helical domain of p110α in a complex with the p85α inter-Src homology 2 (iSH2) and N-terminal inter-Src homology 2 (nSH2) domains. They studied the E545K helical-domain oncogenic mutant and found that this mutation disrupts an inhibitory charge-charge interaction with the p85 nSH2 domain. Second, Huang et al. (31) reported the crystal structure of a complex between the full-length p110α protein and a polypeptide containing the p110α binding domains of p85α. The structure identified a contact between the C2 domain of p110α and the iSH2 domain of p85, suggesting a role for iSH2 in lipid binding. It also shed light on the molecular mechanisms by which PIK3CA oncogenic mutations affect PI3K activity (Figure 4). These findings have revealed new mechanisms for activating PI3K and have provided the basis for the design of therapeutic agents that specifically target the p110α mutated enzyme and spare the normal one, thus preventing potential harmful side effects.
Figure 4.
Mechanistic model of enhanced activation induced by the E545K mutation in p110α. Based on the crystal structure of p110α, the following model has been proposed. The wild-type catalytic p110α subunit is stabilized by binding to the p85α-regulatory subunit through its p85 binding domain (BD). In addition, its activity is maintained at a low state through interaction of its negatively charged helical domain with the positively charged N-terminal inter-Src homology 2 (nSH2) domain of p85α. Receptor tyrosine kinase (RTK) activation and its subsequent phosphorylation recruits the p85α/p110α complex to the membrane, causing conformational changes that relieve the charge-inhibitory interactions of p85α on p110α and lead to high transitional p110α activity. In the presence of the E545K mutation, the negative charge is converted to a positive charge, resulting in the abrogation of the p85α/p110α charge-inhibitory interactions, sustained p110α activity at the membrane, and ultimately delayed dissociation of the p85α subunit from the phospho-RTK. Abbreviations: cSH2, C-terminal inter-Src homology 2; iSH2, inter-Src homology 2; PI3K, phosphatidylinositol 3—kinase; PIP2, phosphatidylinositol-4,5-bisphosphate; PIP3, phosphatidylinositol-3,4,5-trisphosphate. Adapted from Lee et al. (31a).
A novel in vivo model to dissect protein function provided strong evidence for the significance of the Ras-binding domain in p110α. A mouse knockin model introduced a mutation into the endogenous Pik3ca locus, which encodes an enzymatically active p110α that cannot interact with Ras. This model demonstrated the requirement of the Ras/p110α interaction for both developmental and malignant growth factor signaling in Ras-induced lung and skin cancer (32).
Infrequent Mutations in AKT
AKT represents a key node in the PI3K pathway and is located at a crucial crossroads of various upstream signaling events. For example, the PIK3CA hot spot mutations usually induce constitutive growth factor–independent phosphorylation of AKT (24). Similarly, lack of PTEN leads to elevated levels of phosphorylated AKT (1, 33). In mammals, there are three AKT isoforms encoded by highly related genes. Infrequent AKT1 gene amplification has been reported in various human cancers, including a single gastric carcinoma, glioblastomas, and gliosarcomas (34–36). AKT2 amplification was identified in head and neck squamous cell carcinoma (30%) and in pancreatic (20%), ovarian (12%), and breast (3%) cancers (37–40). Carpten et al. (41) identified a transforming E17K PH-domain mutation in AKT1 in breast (8%), colorectal (6%), and ovarian (2%) cancers. This E17K-AKT1 mutant exhibits transforming activity in vitro and in vivo owing to its constitutive PIP3-independent recruitment to the membrane. However, identification of frequent missense mutations causing increased activity of any of the three AKT isoforms has been elusive.
Inactivating Mutations of PTEN in Sporadic Tumors
The most extensive evidence for the involvement of the PI3K pathway in human cancer stems from studies of the PTEN tumorsuppressor gene. In the 1990s, two groups independently searching for tumor suppressors on chromosome 10q23, which is frequently deleted in advanced cancers, identified a new phosphatase termed PTEN, also known as MMAC (mutated in multiple advanced cancers) (42–44). As an antagonist of PI3K signaling, impaired PTEN function leads to PIP3 accumulation in cells and to unrestrained activation of its downstream signals (1, 33, 45). PTEN loss of function occurs in a wide spectrum of human cancers through mutations, deletions, transcriptional silencing, or protein instability at a frequency that can rival p53 alterations in particular settings (46). A lack of redundancy of PTEN might explain the high frequency of mutations. Despite its potential serine, threonine, and tyrosine phosphatase activity, the lipid phosphatase function of PTEN has been shown to be the major driving force in tumor suppression (1, 33). A cancer-derived mutation, G129E, which abrogates the lipid phosphatase activity of PTEN but spares its protein phosphatase activity, results in inactivation ofPTEN tumor-suppressor functions in vitro (47).
PTEN somatic mutations occur in a large percentage of human cancers, with the highest numbers found in endometrium, central nervous system, skin, and prostate cancers (Table 1). Mechanisms of inactivation ofPTEN are well highlighted in sporadic cancers. In the central nervous system, loss of 10q, including PTEN, is found in 70% of glioblastomas and marks the transition to the most aggressive grade of astrocytic tumors. Somatic mutation in the second allele of PTEN, which results in biallelic inactivation, occurs in 25% to 40% of glioblastomas (33, 48). In endometrial cancer, PTEN mutations are detected in complex atypical hyperplasia (premalignant lesions in the progression of endometrial neoplasia), suggesting an initiation role in endometrial carcinomas (49). Cowden syndrome (CS) patients and mice harboring germline mutations in PTEN are predisposed to endometrial carcinomas but not to glioblastomas, consistent with PTEN’s functions in initiation and progression, respectively, in these two tumor types (9, 50).
There is clear genetic evidence that mutations in multiple components of the PI3K path-way are not necessarily redundant. Although activating mutations in p110α and loss of PTEN function both enhance PI3K signaling, these mutations do not serve equivalent functions. In endometrial cancer, mutations in PTEN and PIK3CA both occur frequently and often concomitantly within the same tumor, indicating a potential additive or synergistic effect (51). Intriguingly, the two mutations confer distinct selective advantages because PTEN mutations are found at similar frequencies in hyperplastic precursor lesions and carcinomas, whereas PIK3CA mutations selectively occur in carcinomas, showing a dramatic association between PIK3CA mutation and tumor invasion (52). Thus, the coexistence of PTEN and PIK3CA mutations in endometrial carcinoma indicates that the PI3K pathway can be activated by alterations in multiple genes; it also suggests that those two genes may have additional nonoverlapping consequences during endometrial tumorigenesis (52).
Inherited Mutation in PTEN Supports a Two-Hit Model of Inactivation
Analysis of PTEN mutations shows that it fulfills all criteria of the two-hit hypothesis of tumor suppression: Inherited mutation of PTEN causes cancer predisposition, and both alleles are inactivated (two hits) in sporadic tumors and in tumors from patients with germline mutations (53). Inherited mutations in PTEN are associated with a wide and diverse clinical spectrum of disorders, collectively known as PTEN hamartoma tumor syndromes, that are characterized by the development of benign tumors (hamartomas) (9, 54). These syndromes are CS, Bannayan–Riley–Ruvalcaba syndrome, Proteus syndrome, and Proteus-like syndrome. These disorders share phenotypic similarities but are clinically distinct, representing a highly variable spectrum of abnormalities. There is no correlation between specific PTEN mutations and particular phenotypes, and identical mutations have been observed in association with the clinical presentation of different syndromes (54). PTEN germline mutations are found in 80% of CS cases (54). CS is an autosomal dominant disorder characterized by multiple hamartomas and a risk of breast, thyroid, and endometrial carcinomas. Renal cell carcinoma, certain brain tumors, and melanoma may occur at increased frequency in CS, although this association is less clear (9).
The Role of PTEN Haploinsufficiency
Loss of heterozygosity for PTEN occurs at a much higher frequency than biallelic inactivation in sporadic tumors. It is possible that PTEN inactivation may be underestimated by missing homozygous deletions, promoter mutations, or methylation and silencing of the PTEN promoter (9, 55, 56). These mechanisms have all been demonstrated to inactivate PTEN, but have been studied much less extensively than point mutations. However, it remains unclear whether haploinsufficiency of PTEN provides a selective growth advantage in tumors lacking a second hit in the remaining PTEN allele, or whether an alternative tumor-suppressor gene on chromosome 10q may be the target of allelic deletions. Clearly, the substantial incidence of biallelic PTEN inactivation has yielded genetic evidence that it provides a strong selective advantage; therefore, haploinsufficiency for PTEN might contribute to tumorigenesis only in certain cellular contexts, where loss of the second allele is not required for a selective advantage, or it may even create a selective disadvantage. Evidence for a role of Pten haploinsufficiency was demonstrated in a mouse model of prostate cancer in which the dosage of Pten was inversely related to the severity of the tumor phenotype (57). In what contexts does PTEN haploinsufficiency contribute to tumorigenesis? The different growth factor signal inputs to particular cell types may create specific settings wherein subtypes of cells are more sensitive to smaller changes in PI3K signaling. Alternatively, given that other cancer-associated mutations also serve to activate PI3K signaling, it is possible that PTEN haploinsufficiency may provide a selective growth advantage specifically when combined with another mutation impacting the pathway.
REGULATION OF PTEN-MEDIATED TUMOR SUPPRESSION
PTEN can be regulated through multiple mechanisms, including posttranslational modifications, subcellular localization, and binding partners—all of which potentially impact PTEN levels and/or function.
PTEN-Regulatory Domains
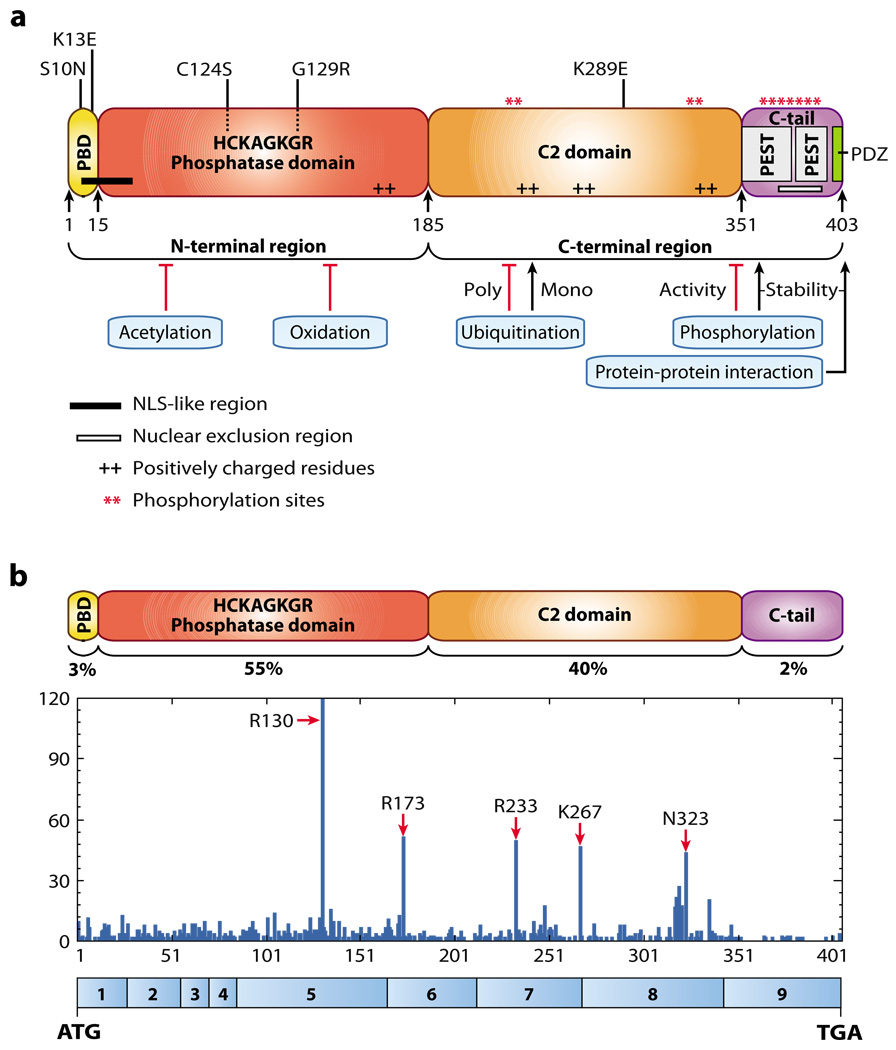
The crystal structure of PTEN revealed two important globular domains, the phosphatase domain and the C2 domain (58). Human PTEN encompasses 403 amino acids and is characterized by five functional domains (Figure 5a): a short N-terminal PIP2-binding domain, a phosphatase domain, a C2 domain, and a C-terminal tail containing PEST (proline, glutamic acid, serine, threonine) sequences and a postsynaptic density protein–Drosophila disc large tumor suppressor–zonula occludens 1 protein (PDZ)–interaction motif. The diversity of tumor-associated mutations occurring in all domains of PTEN strongly suggests that these different domains are physiologically relevant to PTEN-related tumorigenesis. It has been suggested that cytosolic PTEN is found in an inactive conformation with the PIP2-binding domain masking its catalytic site, whereas binding to PIP2 at the plasma membrane would result in an open active conformation of the PTEN protein (59). Tumor-derived mutations identified in this motif—such as K13E, found in glioblastoma and endometrial carcinoma, and S10N, found in lymphoma (60, 61)—argue in favor of the importance of this motif.
Figure 5.
PTEN (phosphatase and tensin homolog deleted on chromosome 10) protein structure and mutation distribution. (a) PTEN is composed of 403 amino acids and is characterized by five functional domains: a phosphatidylinositol-4,5-bisphosphate (PIP2)-binding domain (PBD), a phosphatase domain highlighted by cancer-specific mutations in its catalytic core, a C2 domain with a putative ubiquitination residue associated with cancer, two PEST (proline, glutamic acid, serine, threonine) domains for degradation, and a PDZinteraction motif for protein-protein interactions. Also depicted are an N-terminal nuclear localization signal-like region, a C-terminal nuclear exclusion signal, and several charged residues and phosphorylation sites important for subcellular localization and stability. Several mechanisms regulating the activity of PTEN are shown. (b) Distribution of cancer-specific mutations found in PTEN and their percentage of occurrence in the functional domains. Values are taken from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://www.sanger.ac.uk/genetics/CGP/cosmic") and include single substitutions and complex mutations. Amino acid numbers are listed along the x axis with the corresponding PTEN exon structure shown in blue boxes below, and the numbers of mutations are listed along the y axis. Abbreviations: ATG, start codon; NLS, nuclear localization signal; PDZ, postsynaptic density protein—Drosophila disc large tumor suppressor—zonula occludens 1 protein; TGA, stop codon.
The majority of the missense mutations found in human tumors and CS occur in the phosphatase domain (54) (Figure 5b) and affect the catalytic activity of PTEN, thus underscoring the importance of its phosphatase activity in tumor suppression. The C2 domain harbors basic residues essential for binding to anionic lipids in the plasma membrane, as demonstrated in vitro (58). Altering these residues in vitro diminishes the interaction of PTEN with lipids and ultimately its growth-suppressing activity (62).
In addition to missense mutations, a number of nonsense and frameshift mutations yield truncated PTEN proteins lacking the C-terminal tail and the PDZ-interaction motif (see http://www.sanger.ac.uk/genetics/CGP/cosmic). These mutants retain an intact phosphatase domain and most of the C2 domain, suggesting that the truncated forms of PTEN would be biochemically active. However, these mutants fail to suppress the PI3K pathway when expressed in cells (62–65). Numerous studies have shown that the C-terminal tail and the PDZ-interaction motif are important for PTEN stability and for recruitment to the membrane, respectively (59). The Cterminal tail of PTEN contains multiple sites for phosphorylation, which may regulate its stability, activity, and recruitment to the membrane (59). A specific mutation in the phosphatase domain (C124S) has also been shown to increase PTEN recruitment to the membrane, indicating that both the catalytic domain and the C-terminal tail can influence PTEN localization (59, 66).
The C-terminal tail of PTEN also binds to a number of proteins that may be linked to tumorigenesis (67–70). Binding of the oncogene MSP58 to the PTEN C-terminal domain results in the suppression of MSP58mediated transformation that does not require PTEN catalytic activity (71). Protein interacting to the C-terminus-1 (PICT-1) interacts with PTEN and promotes C-terminaltail phosphorylation, resulting in stabilization of PTEN (69). PICT-1 maps to chromosome 19q13.3, a region frequently deleted in glioma and neuroblastoma (72–75). In neuroblastoma, reduced PICT-1 expression is associated with downregulation of PTEN protein but not transcript, consistent with a role in PTEN stability (68). In addition, PTEN can be recruited to the membrane through binding of its PDZ-interaction motif with PDZ-containing membrane-anchored proteins (76). Therefore, PTEN mutants disrupting the C-terminal tail or the PDZ-interaction motif may show altered stability or may fail to be properly recruited to the membrane or to specific protein complexes. Additional studies are needed to determine the relevance of these specific interactions to PTEN function in tumor suppression as well as in normal physiological settings.
Posttranslational Regulation
PTEN is modified, and likely regulated, by multiple posttranslational mechanisms including phosphorylation, acetylation, oxidation, and ubiquitination (Figure 5a). See Tamguney & Stokoe (55) for a detailed review of these mechanisms.
Phosphorylation of the C-terminal tail stabilizes the PTEN protein but reduces its activity toward PIP3 by repelling it from the membrane and by inhibiting interactions with PDZ domain–containing proteins (Figure 6). In contrast, dephosphorylation of the C-terminal tail causes increased activity accompanied by rapid degradation, thus keeping PTEN activity under tight control. Multiple kinases including CK2, GSK3β, PICT-1, and Rock are capable of phosphorylating the PTEN–C-terminal tail (55).
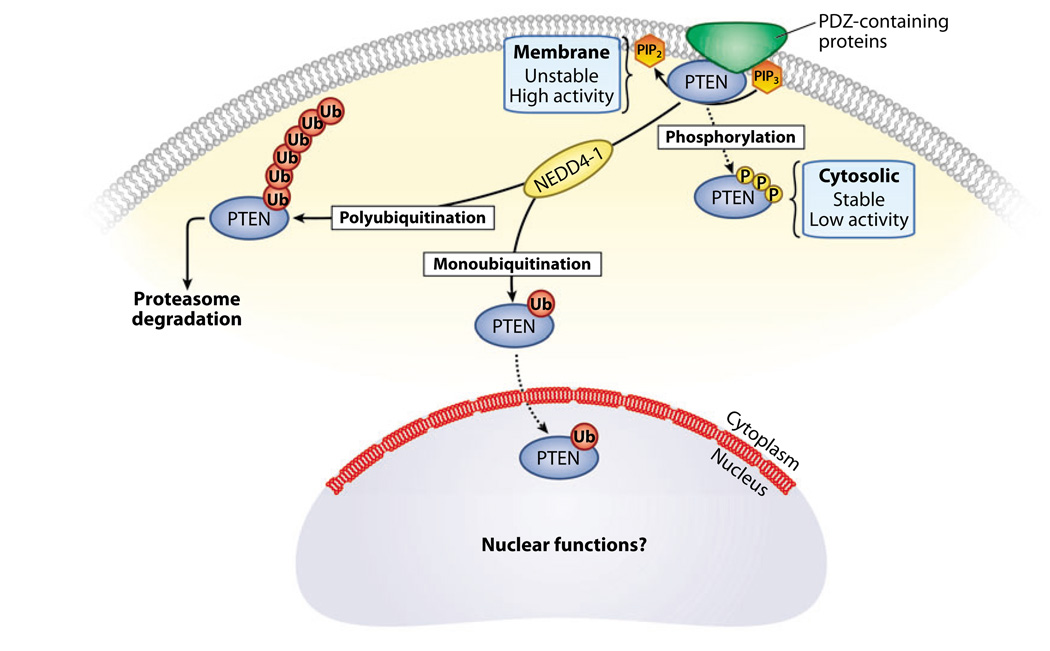
Figure 6.
Regulation of PTEN (phosphatase and tensin homolog deleted on chromosome 10) protein stability and localization. PTEN can be regulated by several mechanisms, including phosphorylation and ubiquitination, that govern its stability and activity through its subcellular localization. When phosphorylated at the C terminus, PTEN adopts a closed conformation that increases its stability but decreases its activity. Conversely, dephosphorylation of PTEN leads to its recruitment to the membrane, where it may interact with PDZ (postsynaptic density protein—Drosophila disc large tumor suppressor—zonula occludens 1 protein) domain-containing proteins through its PDZ interaction motif. Membrane-associated PTEN is more active but less stable. In addition, PTEN turnover can be regulated by NEDD4–1-mediated ubiquitination. Polyubiquitinated PTEN remains in the cytoplasm and is targeted for degradation by the proteasome, whereas monoubiquitination of PTEN appears to regulate import of PTEN into the nucleus, where its function remains unclear. Abbreviations: PIP2, phosphatidylinositol-4,5-bisphosphate; PIP3, phosphatidylinositol-3,4,5 trisphosphate.
The histone acetyltransferase p300/CBPassociated factor interacts with and acetylates PTEN on lysines 125/128, thus negatively regulating PTEN activity by interfering with its catalytic specificity toward PIP3 (77). Reactive oxygen species oxidize residue C124 in the catalytic domain of PTEN, resulting in its inactivation through an intramolecular disulfide bond with C71 (78).
PTEN stability can be regulated further by ubiquitination (67) (Figure 6). PTEN contains two PEST domains associated with ubiquitindependent degradation. Wang et al. (67) identified NEDD4–1 as an E3 ligase that ubiquitinates PTEN at several lysine residues including K289, a residue also targeted by mutation in a CS patient.
Thus, a multitude of posttranslational modifications can modulate PTEN activity. However, in general PTEN is a relatively stable protein, and the physiological stimuli that induce its turnover, phosphorylation, or dephosphorylation are unknown. Therefore, the functional relevance of these different regulatory mechanisms in tumorigenesis remains to be thoroughly elucidated in more physiological settings.
Subcellular Localization
PTEN is well characterized as a cytosolic protein that is recruited to the membrane, where it acts to dephosphorylate PIP3. PTEN may be directed to the membrane via interactions with many membrane-anchored proteins, including MAGI, PAR-3, MAST, SAST, NHERF, and NEP; through its PDZ-interaction motif; and through PIP2 binding (Figure 6) (55, 76). PTEN-membrane recruitment may be very transient, as several milliseconds is sufficient to dephosphorylate PIP3 and antagonize PI3K signaling (59).
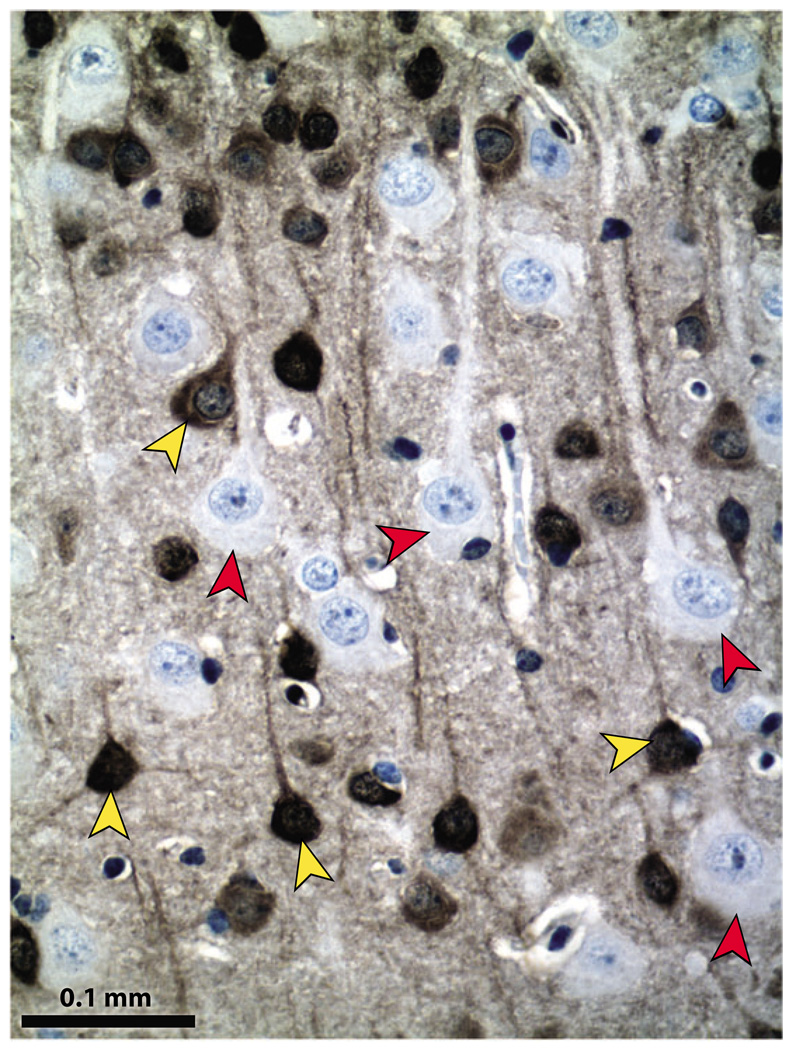
The subcellular localization of PTEN was initially reported to be cytosolic based on the use of green fluorescent protein–fusion proteins. However, differing results among multiple reports show PTEN localization distributed between nuclear and cytoplasmic compartments or exclusively in one of the two compartments. This variation in localization may be due to different cell types or contexts, mutations in PTEN that influence localization and technical issues including use of epitope tags, overexpressed proteins, or differences in antibodies to endogenous protein. There are a number of clear examples in which PTEN can be readily visualized in both compartments. For example, in mature neurons in vivo, PTEN is present in the nucleus, soma, and neuronal projections (Figure 7).
Figure 7.
Pten (phosphatase and tensin homolog deleted on chromosome 10) protein localizes to multiple subcellular compartments. Pten conditional knockout brains contain Pten-positive and Pten-negative neurons, providing internal controls for immunohistochemical detection of Pten. In mature neurons of the cerebral cortex, Pten is present in the nucleus, the soma, and the neuronal projections (yellow arrowheads). Pten-deficient neurons show global hypertrophy, and the absence of Pten is seen clearly in all cell compartments (red arrowheads). Immunohistochemical detection of Pten was performed with anti-Pten antibody from cell signaling (#9559), peroxidase-labeled secondary antibody, diaminobenzidine (DAB) substrate, and hematoxylin counterstain.
The regulation and significance of PTEN subcellular localization is an area of active investigation. In some tumors, subcellular localization of the PTEN protein seems to mediate its activity; exclusion of PTEN from the nucleus may correlate with the degree of neoplastic transformation (79, 80), suggesting an important nuclear function for PTEN in tumor suppression. PTEN possesses two putative nuclear localization signals (NLS) that are required for major vault protein–mediated nuclear import (81). The C2 domain of PTEN is required for this interaction (82). In addition, Gil et al. (83) revealed the presence of multiple nuclear exclusion motifs and different NLS sequences that control PTEN localization in a RAN-dependent manner.
However, Liu et al. (84) showed that PTEN can enter the nucleus by simple diffusion. Trotman et al. (85) demonstrated that monoubiquitination regulates PTEN nuclear import and consequently its tumor-suppressing function (Figure 6). These authors (85) showed that PTEN harboring a germline K289E mutation in intestinal polyps of a CS patient was excluded from the nucleus, although it retained its catalytic activity. This suggests that PTENK289E represents a loss-of-function mutant that is defective in nucleocytoplasmic shuttling and monoubiquitination by NEDD4–1 (85). A positive correlation between nuclear PTEN localization and low tumor grade in colon cancer samples suggests that nuclear exclusion of PTENK289E might compromise PTEN tumorsuppressor function in human cancer. In this model, the regulation of PTEN ubiquitination could strongly influence PTEN function, with monoubiquitination maintaining nuclear tumor-suppressor activity and polyubiquitination reducing PTEN levels and compromising tumor-suppressor activity. Therefore, it will be of particular interest to determine how the proportion of PTEN monoubiquitination versus polyubiquitination is regulated.
The precise role of PTEN in the nucleus and its contribution to tumor suppression is an emerging area of study. Nuclear PTEN may regulate PI3K signaling in the nucleus as well as at the cytoplasmic membrane, as underscored by the presence of key components of the PI3K pathway in both the cytoplasm and nucleus, including PIP2, PIP3, PI3Ks, PDK1, AKT, and PTEN (86) (Figure 6). However, in vitro studies have also suggested potential roles for nuclear PTEN in chromosomal stability, DNA-damage responses, and cell-cycle regulation (87). The use of knockin mouse models to dissect the function of physiologically expressed, selectively mutated proteins that maintain Pten phosphatase activity but disrupt Pten localization should provide powerful insights into the roles of nuclear Pten in different tissues and in normal development and tumorigenesis.
ADDITIONAL FUNCTIONS OF PTEN
Genomic Integrity
Increased genomic instability provides a powerful mechanism to increase mutation frequencies and to accelerate the accumulation of multiple mutations required for cancer initiation and progression. Puc et al. (88) showed that PTEN loss leads to extensive double strand breaks through increased AKT-mediated cytoplasmic sequestration of the kinase CHK1, resulting in altered G2/S arrest in response to DNA damage. The authors analyzed human breast carcinoma samples and observed decreased nuclear CHK1 in tumor cells lacking functional PTEN (88). A surprising study suggested two distinct nuclear functions through which PTEN loss may promote genomic instability (89). Thus, PTEN involvement in the DNA-damage response has been suggested for both cytoplasmic and nuclear PTEN. Further studies are needed to evaluate the mechanisms connecting PTEN to genomic integrity.
Cell Migration
PI3K signaling plays important roles in migration in both development and cancer. In mammalian cells, the PI3K pathway regulates membrane ruffling, cell motility, and cell spreading by mediating key downstream effectors such as Rho, Rac1, and cdc42 (Figure 2) (13, 90). PIK3CA mutations induce increased cell migration and invasion ability in tumor cells (27), and overexpression of PTEN can inhibitglioma cell migration, although surprisingly this activity is independent of PTEN phosphatase activity, raising the possibility of PI3K pathway–independent effects of PTEN on migration (91, 92). Paradoxically, Pten deletion in the developing nervous system shows the failure of neurons to complete migration (93–95); thus, there are examples of Pten loss of function or gain of function inhibiting migration in different settings. Therefore, it is clear that the PI3K pathway can impact cell motility and invasion, which may contribute to invasive properties of tumor cells.
Stem Cell Self-Renewal
Stem cell self-renewal pathways are altered in cancer, and therapeutic targeting of cancer stem cells has begun to draw much attention. Two groups demonstrated a key role of Pten in the self-renewing activity of normal and leukemic hematopoietic stem cells (HSCs) (96, 97). Loss of Pten results in the generation of leukemic cells, accompanied by depletion of normal HSCs. The authors of these studies sugested that Pten normally maintains HSCs in a quiescent state, whereas in a Pten-deficient environment HSCs are driven through the cell cycle, leading to depletion of their reserves (96, 97). Inhibition of PI3K signaling by rapamycin blocks the generation of leukemia-initiating cells and restores the normal self-renewal capacity of HSCs (96, 97). In contrast, Pten loss in the nervous system increases the pool of selfrenewing neural stem cells, in addition to promoting their deregulated proliferation (95, 98).
Cellular Senescence and Interactions with p53
Analysis of a mouse model of prostate cancer showed senescent cells in tumors induced by Pten deficiency and an associated induction of p53 (99). Combined deletion of Pten and p53 in prostate is synergistic, causing much more aggressive tumors that arose with decreased latency and no evidence of senescence. This suggests that loss of p53 abrogates senescence induced by PTEN deficiency and that in some situations, complete loss of PTEN may create a selective disadvantage by inducing senescence. This may explain in part why approximately 70% of primary human prostate tumors fail to inactivate both copies of PTEN (99).
PTEN and p53 regulation may interact at multiple levels. Several studies have demonstrated that PTEN can activate p53 through direct and indirect protein-protein interactions, including phosphatase-dependent and -independent mechanisms (100–102). In addition, PTEN loss may decrease p53 levels via AKT-mediated phosphorylation of MDM2, which increases MDM2 nuclear translocation and subsequent ubiquitination and degradation of p53. Finally, PTEN may also influence MDM2 promoter activity (103, 104). These studies suggest that loss of PTEN function is associated with decreased p53 function, therefore mutations in the two tumor suppressors are predicted to be somewhat redundant. However, other groups have demonstrated the opposite, showing that PTEN loss serves to activate p53 (99, 105). A simple model to explain the functional connections between PTEN and p53 does not emerge from this collection of studies, suggesting that the relationship between these two tumor suppressors, which are of central importance in human cancer, may be heavily influenced by cell-type and tumor-type context.
MOUSE MODELS TO EVALUATE PTEN FUNCTION IN VIVO
Consistent with its central role in the regulation of a growth-regulatory pathway that is critical for normal development and is important in cancer, homozygous deletion of Pten in the mouse germline causes embryonic lethality, and heterozygous deletion causes tumor predisposition (106–108). A large number of Ptenconditional knockout mice have been generated to evaluate Pten function in a physiological setting in different cell and tissue types. These models have provided clear evidence for important roles for Pten in tumor suppression, as well as regulation of cell size, proliferation, cell migration during development, and specialized functions in specific tissues such as the immune and nervous systems (33, 50). The generation of knockin mice to introduce specific mutations that selectively ablate the various modes of regulation and alternative functions for PTEN (outlined above) will provide critical insights into which regulatory mechanisms show the greatest physiological relevance to PTEN function in normal development and tumorigenesis.
THERAPEUTIC TARGETING OF THE PI3-KINASE PATHWAY
Cancer-specific human mutations, functional connections to oncogenic signaling, and robust animal models provide compelling evidence that perturbation of PI3K signaling through multiple mechanisms contributes substantially to human cancer. The development and testing of selective inhibitors of PI3K signaling for cancer therapeutics are exciting and intense areas of investigation (109). Although the pathway presents an obvious target for therapeutic intervention, complexities in signaling regulation pose substantial challenges in therapeutic design.
Feedback Regulation of PI3-Kinase Signaling Through mTOR
A number of basic research studies and clinical trials have investigated the potential of the selective mTOR inhibitor rapamycin (or its analogs) to block the downstream consequences of aberrant PI3K signaling. Although rapamycin selectively blocks the activity of the mTORC1 complex downstream of AKT and TSC, rapamycin treatment may have one of several effects on the upstream regulation of AKT (Figure 8). In some situations, such as short-term treatment of cultured cells and longterm in vivo administration at minimal doses required to suppress mTOR activity in brain, rapamycin and its analogs show selective inhibition of the downstream effectors of mTOR without affecting activation of AKT upstream (110, 111). However, longer-term administration may lead to the sequestration of newly synthesized mTOR before it has entered the rapamycin-resistant complex mTORC2, which is largely responsible for phosphorylation of Ser473 of AKT (111). Thus, rapamycin treatment could inhibit activity of both mTOR and AKT, suggesting that it may be an effective therapeutic blockade of PI3K signaling. However, there is a third possible outcome: Feedback inhibition of PI3K signaling may be relieved by rapamycin treatment, presumably by inhibiting the downstream activation of S6K, which can serve as a negative regulator of PI3K signaling through phosphorylation and inhibition of insulin receptor substrate 1 (IRS1) (112). In this situation, inhibition of mTOR relieves a feedback inhibition of PI3K and results in enhanced PI3K signaling to AKT. As discussed above, AKT has a host of substrates independent of mTOR signaling that may strongly contribute to oncogenesis. Therefore, enhanced AKT activation, even in the contextof repressed mTOR activity, could effectively worsen the situation. Indeed this feedback mechanism has been documented in clinical trials of cancer patients treated with mTOR inhibitors, as well as in cancer cell lines (113). How each tumor responds to mTOR inhibition remains unclear. Their responses may be related to the other mutations contained in the tumor, as there is abundant cross-talk between PI3K signaling and other signaling pathways, including p53, RAS/mitogen-activated protein kinase, tumor growth factor beta, NFκB, WNT, and others (109).
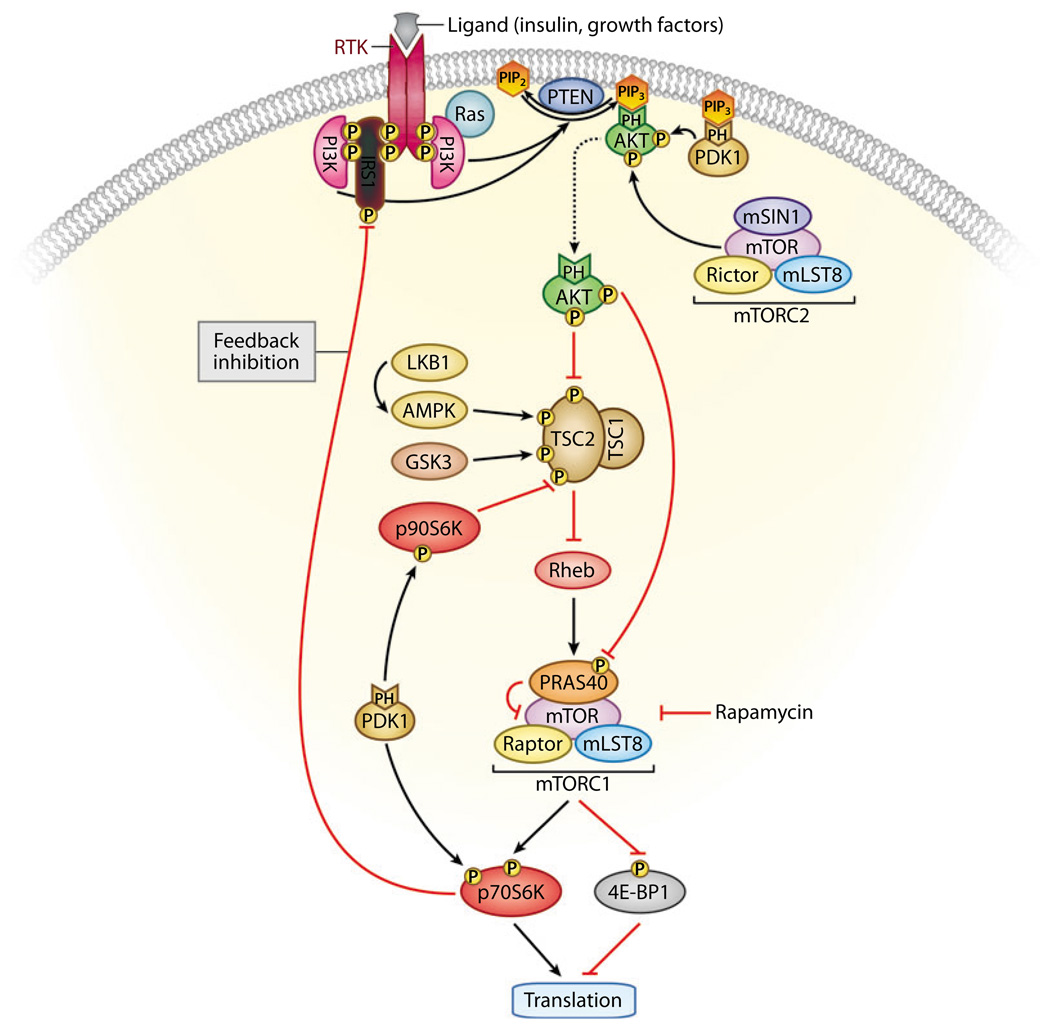
Figure 8.
Feedback inhibition of the phosphatidylinositol 3—kinase (PI3K) pathway. Activated AKT regulates cellular growth through mammalian target of rapamycin (mTOR), a key player in protein synthesis and translation. mTOR forms part of two distinct complexes known as mTORC1 (which contains mTOR, Raptor, mLST8, and PRAS40) and mTORC2 (which contains mTOR, Rictor, mLST8, and mSIN1). mTORC1 is sensitive to rapamycin and controls protein synthesis and translation, at least in part, through p70S6K and eukaryotic translation initiation factor 4E—binding protein 1 (4E-BP1). AKT phosphorylates and inhibits tuberous sclerosis complex 2 (TSC2), resulting in increased mTORC1 activity. AKT also phosphorylates PRAS40, thus relieving the PRAS40 inhibitory effect on mTOR and the mTORC1 complex. mTORC2 and 3-phosphoinositide-dependent kinase (PDK1) phosphorylate AKT on Ser473 and Thr308, respectively, rendering it fully active. mTORC1-activated p70S6K can phosphorylate insulin receptor substrate 1 (IRS1), resulting in inhibition of PI3K activity. In addition, PDK1 phosphorylates and activates p70S6K and p90S6K. The latter has been shown to inhibit TSC2 activity through direct phosphorylation. Conversely, LKB1-activated AMP-activated protein kinase (AMPK) and glycogen synthase kinase 3 (GSK3) activate the TSC1/TSC2 complex through direct phosphorylation of TSC2. Thus, signals through PI3K as well as through LKB1 and AMPK converge on mTORC1. Inhibition of mTORC1 can lead to increased insulin receptor—mediated signaling, and inhibition of PDK1 may lead to activation of mTORC1 and may, paradoxically, promote tumor growth.
Genetic models also demonstrate the significance of feedback regulation in the PI3K pathway. Tsc2+/− mice develop benign hemangiomas with sporadic loss of the wild-type Tsc2 allele and inactive Akt owing to a highly active mTOR/p70S6K pathway, which serves to negatively regulate PI3K through feedback inhibition. However, Pten+/−;Tsc2+/− mice develop aggressive hemangiomas because Pten deletion overrides the negative feedback loop and activates Akt (114, 115). The feedback inhibition of PI3K signaling limits oncogenesis triggered by PI3K-independent defects in Tsc-mediated activation of mTor, such as loss-of-function mutations in TSC1, TSC2, or LKB1. In contrast, inactivation of PTEN effectively deregulates both the upstream and downstream branches of the PI3K pathway. This difference in feedback regulation provides a logical explanation for why PTEN is the only one of these familial tumor-suppressor genes to be frequently targeted in sporadic cancers, even though all of these tumor suppressors converge on mTOR signaling downstream (Figure 8). This example also serves to highlight the need for biomarkers that will help identify which effectors are responsible for the primary defect, which will aid us in predicting how feedback may complicate tumor response.
Other PI3-Kinase Pathway Inhibitors
To address therapeutic failures caused by feedback response to selective inhibitors, one promising alternative is to target the PI3K pathway at multiple key nodes, such as p110α and mTOR. Knight & Shokat (116) have developed multiple inhibitors of PI3K, including a dual inhibitor of p110α and mTOR termed PI-103. PI-103 effectively blocks proliferation of glioma and other tumor cells in vitro as well as in xenografts in vivo (117, 118). Compounds to selectively inhibit other main effectors within the PI3K pathway, including PDK1 and AKT, are currently under development (109). It is likely that additional complexities of PI3K-pathway regulation will be revealed with extensive testing of each inhibitor. Further, tissue- and tumor-type specificity will also be a likely issue for consideration. An animal model of mammary tumorigenesis has shown unexpected diversity in the activity of different AKT family members. Ablation of Akt1 inhibits two animal models of mammary tumorigenesis, whereas ablation of Akt2 accelerates tumorigenesis (119). In this situation, it is difficult to predict whether a pan-AKT inhibitor would inhibit or accelerate tumor growth. Detailed studies to understand the tumor-type specificity of each PI3K effector will ultimately be required to provide the greatest insights into how to tailor therapeutic blockades for the greatest likelihood of success.
CONCLUSIONS
The explosion of research on PI3K signaling in normal development, metabolism, and cancer has revealed a network of signaling effectors with intricate regulation of individual signaling components as well as additional complexity due to the existence of multiple family members with context-specific functions. The current challenge in the field is to place these findings within a physiological framework to understand how and when different modes of regulation are engaged and how they are relevant to cancer. In-depth analysis of mutations in human tumors, elegant in vivo mouse models, and functional assays using biochemical, cellular, and whole-animal approaches are required to bring us to the next level of understanding.
SUMMARY POINTS
Deregulation of the PI3K pathway through selective mutation of PIK3CA and/or PTEN occurs in a large proportion of human cancers.
PTEN is frequently inactivated by biallelic mutation, but it may also contribute to cancer through haploinsufficiency.
In addition to its clear role as a cytoplasmic PIP3 phosphatase, PTEN may have phosphatase-independent functions and additional functions in the nucleus.
Regulation of the PI3K pathway is complicated due to feedback mechanisms as well as cross-talk with other critical pathways important in human cancer.
The significance of the PI3K pathway in cancer makes it an important therapeutic target.
FUTURE ISSUES
Many regulatory mechanisms to modulate PTEN activity have been identified with exciting in vitro studies. These findings must be extended to in vivo model systems to understand how and when different modes of regulation are engaged in a physiological context and how they are relevant to cancer.
The development of multiple inhibitors for different components of the PI3K pathway may allow for tailored therapy and will provide powerful molecular tools to dissect the regulation of this centrally important signaling pathway.
A more detailed understanding of how and when feedack regulation occurs will be required for successful therapeutic targeting of the PI3K pathway.
ACKNOWLEDGEMENTS
We apologize to authors whose work or relevant references could not be cited due to page limitations. We thank members of the Baker laboratory for helpful comments and discussions, and we thank Dr. Melissa Fraser of the Baker lab for the Pten IHC shown in Figure 7. N.C. is a recipient of a postdoctoral fellowship from the Canadian Institutes of Health Research. S.J.B. is supported by the American Lebanese and Syrian Associated Charities and grants from the National Institutes of Health.
Glossary
- PI3K
phosphatidylinositol 3–kinase
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
- RTK
receptor tyrosine kinase
- mTOR
mammalian target of rapamycin
- TSC
tuberous sclerosis complex
- Tumor-suppressor gene
gene targeted by loss-of-function mutations in cancer; mutations in these genes are usually recessive
- Hamartoma
benign malformation containing an abnormal proportion or mixture of cells normally found in the tissue from which it grows
- Somatic mutation
mutation acquired in somatic cells; not found in the germline
- Knockin
technical approach in mouse genetic engineering in which the endogenous wild-type gene is replaced with a mutated version, allowing physiological regulation of a mutated gene in vivo
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3–kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 2.Gaidarov I, Smith ME, Domin J, Keen JH. The class II phosphoinositide 3–kinase C2α is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell. 2001;7:443–449. doi: 10.1016/s1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 3.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurmser AE, Emr SD. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 2002;158:761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 6. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148.First report showing phosphorylation of Akt at serine 473 by the rictor–mTOR complex.
- 7.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 8.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Zbuk KM, Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 10.Schreibman IR, Baker M, Amos C, McGarrity TJ. The hamartomatous polyposis syndromes: a clinical and molecular review. Am. J. Gastroenterol. 2005;100:476–490. doi: 10.1111/j.1572-0241.2005.40237.x. [DOI] [PubMed] [Google Scholar]
- 11.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen P, Frame S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 13.Vivanco I, Sawyers CL. The phosphatidylinositol 3–kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 14.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 16.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 17.Sugimoto Y, Whitman M, Cantley LC, Erikson RL. Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc. Natl. Acad. Sci. USA. 1984;81:2117–2121. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otsu M, Hiles I, Gout I, Fry MJ, Ruiz-Larrea F, et al. Characterization of two 85 kD proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 19.Escobedo JA, Navankasattusas S, Kavanaugh WM, Milfay D, Fried VA, Williams LT. cDNA cloning of a novel 85 kD protein that has SH2 domains and regulates binding of PI3-kinase to the PDGFβreceptor. Cell. 1991;65:75–82. doi: 10.1016/0092-8674(91)90409-r. [DOI] [PubMed] [Google Scholar]
- 20.Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J. Interaction of phosphatidylinositol 3–kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol. Cell Biol. 1992;12:981–990. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klippel A, Escobedo JA, Fantl WJ, Williams LT. The C-terminal SH2 domain of p85 accounts for the high affinity and specificity of the binding of phosphatidylinositol 3–kinase to phosphorylated platelet-derived growth factor β receptor. Mol. Cell Biol. 1992;12:1451–1459. doi: 10.1128/mcb.12.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 23. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502.Demonstrated high frequency of cancer-specific PIK3CA mutations in humans.
- 24.Vogt PK, Kang S, Elsliger MA, Gymnopoulos M. Cancer-specific mutations in phosphatidylinositol 3–kinase. Trends Biochem. Sci. 2007;32:342–349. doi: 10.1016/j.tibs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3–kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 27.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110a and p110(3 phosphatidylinositol 3–kinases in human mammary epithelial cells. Proc. Natl. Acad. Sci. USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3–kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 30.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3–kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 31.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, et al. The structure of a human p110a/p85a complex elucidates the effects of oncogenic PI3Ka mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 31a.Lee JY, Engelman JA, Cantley LC. PI3K charges ahead. Science. 2007;317:206–207. doi: 10.1126/science.1146073. [DOI] [PubMed] [Google Scholar]
- 32.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, et al. Binding of ras to phosphoinositide 3–kinase p110α is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 33.Chow LM, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Staal SP. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc. Natl. Acad. Sci. USA. 1987;84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knobbe CB, Reifenberger G. Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3′ -kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol. 2003;13:507–518. doi: 10.1111/j.1750-3639.2003.tb00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Actor B, Cobbers JM, Buschges R, Wolter M, Knobbe CB, et al. Comprehensive analysis of genomic alterations in gliosarcoma and its two tissue components. Genes Chromosom. Cancer. 2002;34:416–427. doi: 10.1002/gcc.10087. [DOI] [PubMed] [Google Scholar]
- 37.Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int. J. Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama K, Nakayama N, Kurman RJ, Cope L, Pohl G, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. CancerBiol. Ther. 2006;5:779–785. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 39.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 41.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 42. Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356.Seminal discovery of PTEN as the tumor-suppressor gene on 10q23, which is frequently mutated in advanced human cancers.
- 43. Li J, Yen C, Liaw D, Podsypanina K, Bose S, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943.Seminal discovery of PTEN as the tumor-suppressor gene on 10q23, which is frequently mutated in advanced human cancers.
- 44.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor β. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 45. Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375.Showed that PTEN is a lipid phosphatase that regulates PIP3levels.
- 46.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 47.Myers MP, Pass I, Batty IH, Van Der Kaay J, Stolarov JP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 49.Di Cristofano A, Ellenson LH. Endometrial carcinoma. Annu. Rev. Pathol. 2007;2:57–85. doi: 10.1146/annurev.pathol.2.010506.091905. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki A, Nakano T, Mak TW, Sasaki T. Portrait of PTEN: messages from mutant mice. Cancer Sci. 2008;99:209–213. doi: 10.1111/j.1349-7006.2007.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–10673. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- 52. Hayes MP, Wang H, Espinal-Witter R, Douglas W, Solomon GJ, et al. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin. Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375.Demonstrated that the occurrence of PTEN and PIK3CA mutations is associated with different stages of tumor progression in endometrial cancer.
- 53. Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64.First demonstration of germline mutations of PTEN associated with cancer predisposition.
- 54.Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 55.Tamguney T, Stokoe D. New insights into PTEN. J. Cell Sci. 2007;120:4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- 56.Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059.Showed that PTEN dosage is related to tumor severity in a mouse model of prostate cancer, providing experimental evidence of haploinsufficiency for PTEN.
- 58.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 59.Vazquez F, Devreotes P. Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction. Cell Cycle. 2006;5:1523–1527. doi: 10.4161/cc.5.14.3005. [DOI] [PubMed] [Google Scholar]
- 60.Minaguchi T, Yoshikawa H, Oda K, Ishino T, Yasugi T, et al. PTEN mutation located only outside exons 5, 6, and 7 is an independent predictor of favorable survival in endometrial carcinomas. Clin. Cancer Res. 2001;7:2636–2642. [PubMed] [Google Scholar]
- 61.Gronbaek K, Zeuthen J, Guldberg P, Ralfkiaer E, Hou-Jensen K. Alterations of the MMAC1/PTEN gene in lymphoid malignancies. Blood. 1998;91:4388–4390. [PubMed] [Google Scholar]
- 62.Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, Hanafusa H. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000;60:7033–7038. [PubMed] [Google Scholar]
- 63.Tolkacheva T, Chan AM. Inhibition of H-Ras transformation by the PTEN/MMAC1/TEP1 tumor suppressor gene. Oncogene. 2000;19:680–689. doi: 10.1038/sj.onc.1203331. [DOI] [PubMed] [Google Scholar]
- 64.Koul D, Jasser SA, Lu Y, Davies MA, Shen R, et al. Motif analysis of the tumor suppressor gene MMAC/PTEN identifies tyrosines critical for tumor suppression and lipid phosphatase activity. Oncogene. 2002;21:2357–2364. doi: 10.1038/sj.onc.1205296. [DOI] [PubMed] [Google Scholar]
- 65.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl. Acad. Sci. USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das S, Dixon JE, Cho W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, et al. NEDD4–1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okahara F, Itoh K, Nakagawara A, Murakami M, Kanaho Y, Maehama T. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol. Biol. Cell. 2006;17:4888–4895. doi: 10.1091/mbc.E06-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okahara F, Ikawa H, Kanaho Y, Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 70.Kim RH, Peters M, Jang Y, Shi W, Pintilie M, et al. DJ-1,a novel regulator ofthe tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 71.Okumura K, Zhao M, Depinho RA, Furnari FB, Cavenee WK. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc. Natl. Acad. Sci. USA. 2005;102:2703–2706. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith JS, Tachibana I, Pohl U, Lee HK, Thanarajasingam U, et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics. 2000;64:44–50. doi: 10.1006/geno.1999.6101. [DOI] [PubMed] [Google Scholar]
- 73.Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 74.Mora J, Cheung NK, Chen L, Qin J, Gerald W. Loss of heterozygosity at 19q13.3 is associated with locally aggressive neuroblastoma. Clin. Cancer Res. 2001;7:1358–1361. [PubMed] [Google Scholar]
- 75.Hartmann C, Johnk L, Kitange G, Wu Y, Ashworth LK, et al. Transcript map of the 3.7-Mb D19S112-D19S246 candidate tumor suppressor region on the long arm of chromosome 19. Cancer Res. 2002;62:4100–4108. [PubMed] [Google Scholar]
- 76.Bonifant CL, Kim JS, Waldman T, et al. NHERFs,NEP,MAGUKs,andmore:interactionsthatregulate PTEN. J. Cell Biochem. 2007;102:878–885. doi: 10.1002/jcb.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB. PCAF modulates PTEN activity. J. Biol. Chem. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 78.Seo JH, Ahn Y, Lee SR, Yeol Yeo C, ChungHur K. The major target of the endogenously generated reactive oxygen species in response to insulin stimulation is phosphatase and tensin homolog and not phosphoinositide—3 kinase (PI—3 kinase) in the PI—3 kinase/Akt pathway. Mol. Biol Cell. 2005;16:348–357. doi: 10.1091/mbc.E04-05-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomaki P, et al. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002;161:439–447. doi: 10.1016/S0002-9440(10)64200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, et al. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 2000;157:1097–1103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung JH, Ginn-Pease ME, Eng C. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res. 2005;65:4108–4116. doi: 10.1158/0008-5472.CAN-05-0124. [DOI] [PubMed] [Google Scholar]
- 82.Yu Z, Fotouhi-Ardakani N, Wu L, Maoui M, Wang S, et al. PTEN associates with the vault particles in HeLa cells. J. Biol. Chem. 2002;277:40247–40252. doi: 10.1074/jbc.M207608200. [DOI] [PubMed] [Google Scholar]
- 83.Gil A, Andres-Pons A, Fernandez E, Valiente M, Torres J, et al. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell. 2006;17:4002–4013. doi: 10.1091/mbc.E06-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu F, Wagner S, Campbell RB, Nickerson JA, Schiffer CA, Ross AH. PTEN enters the nucleus by diffusion. J. Cell Biochem. 2005;96:221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- 85.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 87.Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. J. Cell Sci. 2008;121:249–253. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- 88.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, et al. Lack of PTEN equesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 89.Shen WH, Balajee AS, Wang J, Wu H, Eng C, et al. Essential role for nuclear PTENinmaintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 90.Liliental J, Moon SY, Lesche R, Mamillapalli R, Li D, et al. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 91.Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 92.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 93.Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, et al. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat. Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 94.Marino S, Krimpenfort P, Leung C, Van Der Korput HA, Trapman J, et al. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- 95.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 96. Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747.Demonstration of a potential role of PTEN in long-term hematopoietic stem cell maintenance and in leukemia stem cells.
- 97. Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703.Demonstration of a potential role of PTEN in long-term hematopoietic stem cell maintenance and in leukemia stem cells.
- 98.Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, et al. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc. Natl. Acad. Sci. USA. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou M, Gu L, Findley HW, Jiang R, Woods WG. PTEN reverses MDM2-mediated chemotherapy resistance by interacting with p53 in acute lymphoblastic leukemia cells. Cancer Res. 2003;63:6357–6362. [PubMed] [Google Scholar]
- 101.Li AG, Piluso LG, Cai X, Wei G, Sellers WR, Liu X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell. 2006;23:575–587. doi: 10.1016/j.molcel.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 102.Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and-independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- 103.Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem. Sci. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- 104.Chang CJ, Freeman DJ, Wu H. PTEN regulates Mdm2 expression through the P1 promoter. J. Biol. Chem. 2004;279:29841–29848. doi: 10.1074/jbc.M401488200. [DOI] [PubMed] [Google Scholar]
- 105.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol. Cell Biol. 2007;27:662–677. doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 1998;19:348–355. doi: 10.1038/1235.Generation of mouse Pten knockout models validating the tumor-suppression function in vivo and unraveling a crucial role in development.
- 107. Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563.Generation of mouse Pten knockout models validating the tumor-suppression function in vivo and unraveling a crucial role in development.
- 108. Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5.Generation of mouse Pten knockout models validating the tumor-suppression function in vivo and unraveling a crucial role in development.
- 109.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 110.Kwon CH, Zhu X, Zhang J, Baker SJ. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:12923–12928. doi: 10.1073/pnas.2132711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 112. Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, et al. Absence of S6K1 protects against age-and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866.Unraveled the presence of a feedback inhibitory loop by S6K1 upon the insulin/IGF pathway.
- 113.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19:1773–1778. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma L, Teruya-Feldstein J, Behrendt N, Chen Z, Noda T, et al. Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression. Genes Dev. 2005;19:1779–1786. doi: 10.1101/gad.1314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Knight ZA, Shokat KM. Chemically targeting the PI3K family. Biochem. Soc. Trans. 2007;35:245–249. doi: 10.1042/BST0350245. [DOI] [PubMed] [Google Scholar]
- 117.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3—kinases. Cancer Res. 2007;67:5840–5850. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 119.Maroulakou IG, Oemler W, Naber SP, Tsichlis PN. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle-T transgenic mice. Cancer Res. 2007;67:167–177. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]