Abstract
Whole genome sequencing of the model organisms has created increased demand for efficient tools to facilitate the genome annotation efforts. Accordingly, we report the further implementations and analyses stemming from our publicly available P{wHy} library for Drosophila melanogaster. A two-step regime—large scale transposon mutagenesis followed by hobo-induced nested deletions—allows mutation saturation and provides significant enhancements to existing genomic coverage. We previously showed that, for a given starting insert, deletion saturation is readily obtained over a 60-kb interval; here, we perform a breakdown analysis of efficiency to identify rate-limiting steps in the process. Transrecombination, the hobo-induced recombination between two P{wHy} half molecules, was shown to further expand the P{wHy} mutational range, pointing to a potent, iterative process of transrecombination–reconstitution–transrecombination for alternating between very large and very fine-grained deletions in a self-contained manner. A number of strains also showed partial or complete repression of P{wHy} markers, depending on chromosome location, whereby asymmetric marker silencing allowed continuous phenotypic detection, indicating that P{wHy}-based saturational mutagenesis should be useful for the study of heterochromatin/positional effects.
AMONG the variety of genetic model organisms, an ongoing concern is the global annotation of the genome in terms of function. In Drosophila melanogaster, transposon-related methods have existed for this purpose from early on (Cooley et al. 1988; Spradling et al. 1995; Hacker et al. 2003). Despite the thousands of insertions that have accumulated among the various collections for Drosophila, appreciable numbers of genomic intervals could benefit from additional transposon mutations. A significant portion of the P insertions also land outside of the gene, resulting in failure to disrupt the phenotype. Since transposon mutagenesis has been among the most heavily used techniques for genetic analysis, this undersaturation can pose challenges to the gene annotations. Over the last several years, the Drosophila field has benefitted from the technical advancements in reverse genetics—including homologous gene targeting and RNAi (Kennerdell and Carthew 1998; Rong and Golic 2000). Homologous gene targeting appears to be most applicable to particular genes rather than genomewide studies, and the Drosophila RNAi libraries are based on the principle of knockdown, not knockout mutation.
Previously this laboratory developed the compound element P{wHy} to aid the study of gene structure and function and to improve the genome coverage by transposon-based saturating mutagenesis; a regional proof of principle was demonstrated in Drosophila, where neighboring P{wHy} elements separated by as much as 60 kb could deletion saturate for the detection of every intervening transcriptional unit (Huet et al. 2002). Other prominent transposon-based tools for systematic deletion have been offered in recent years (Artavanis-Tsakonas 2004; Ryder et al. 2007; Paré et al. 2009). Unlike P{wHy}, however, these systems either (1) depend on having starting transposon pairs (instead of a single insertion) already bounding the locus of interest, which can sometimes hinder detailed resolution, or (2) they do not immediately allow self-contained production of a range of very small to very large deletions across the genome. Significantly, the P{wHy} system is distinguished by a combination of advantages, such as broad distribution, fine granularity (the tendency of local hobo jumping to yield various deletion endpoints), and high frequency (the efficiency at producing many deletions). In addition, accumulated experience with this system suggested the possibility of more advanced P{wHy}-based strategies for achieving very large deletions—without being combined with other libraries—and for operating in environments of strong positional silencing. This has led to our demonstrating augmented utility of P{wHy} in large-scale functional annotation of the Drosophila genome. Here we report these analyses for the extensive, publicly available P{wHy} collection, principally for the second and third chromosomes.
MATERIALS AND METHODS
The P{wHy} collection:
This collection is derived from a hybrid transposon molecule described in Huet et al. (2002), which depicts the primary mechanism. Briefly, P{wHy} is a compound element comprised of P-transposon carrier arms and a central deleter transposon, hobo, which is flanked by white and yellow genes. Flanking deletions are obtained by introducing a source of hobo transposase, followed by recombination between the original and second copy of hobo; the direction of the deletion is indicated by the particular P{wHy} marker lost. The genetic schemes and strains for the basic manipulation of P{wHy} transposition are described in Huet et al. (2002).
The collection currently consists of >800 inserts and was generated by interchromosomal remobilization of an X-associated starting insertion, using the standard P-transposition methods. The resulting P{wHy} insertions in this collection thus reside predominantly on chromosomes II and III, with a more limited number of insertions on chromosome IV. The fly strains containing these autosomal insertions are available through the Drosophila Stock Center at Bloomington, Indiana.
Mapping of P{wHy} insertions:
The P{wHy} insertion mapping has been archived through FlyBase/Bloomington Stock Center (http://flystocks.bio.indiana.edu/Browse/insertions/PwHy-top.htm) (under the collections title “Deletion Generators”) and the Drosophila Gene Disruption Project (Bellen et al. 2004); the present study also reflects substantial mapping and annotation running through 2008 in the sequence database and FlyBase. Transposons were mapped by extraction of Drosophila adult DNA (Huang et al. 2000), followed by short genome walking with iPCR (Ochman et al. 1988; Triglia et al. 1988)—using P-related primers (Bellen et al. 2004)—and with the Universal Fast Walking method (Myrick and Gelbart 2002). Basic Local Alignment Search Tool (BLAST) alignments (Altschul et al. 1990) were performed against the GenBank database and the Berkeley Drosophila Genome Project (BDGP) database (Adams et al. 2000; Celniker et al. 2002). Only insertions whose locations could be rigorously assigned at the molecular level were considered for inclusion in the current P{wHy} collection.
Transrecombination:
y−w−; P{5′wHy}14F06W or 14H10W/SM6a; P{hsH\T-2}/+ males were crossed to y−w−;P{3′wHy}02B10Y/SM6a virgin females and progeny were heatshocked on days 2, 4, and 6 for 30 min at 37°. F1 males were collected and crossed to yw;Gla/SM6a virgin females and progeny were screened for the presence of both the w+ and y+ markers.
Molecular and genetic tests of transrecombinant strains:
PCR tests of the 5′ 14F06 and 14H10 insertion sites and the 3′ 02B10 insertion site were performed as described in Mohr and Gelbart (2002) with Pendout2 and genomic DNA-specific primers. Long-range PCR detection of an intact P{wHy} element was performed with the primers ASR5 and SER5 at 68° and ASR6 and SER6 at 55° with LA-Taq (Takara Mirus Bio): ASR5, ctgccgataggtcagatgtcgtggc; SER5, agcttatggtgaagtgttgccaggc; ASR6, ccctttattaagatttcacacagatcagcc; SER6, ccacatgacgtatgagcaagtggca.
Test for positional silencing of P{wHy} insertions:
An eye mosaicism test was performed to determine if the repression of the white marker in a particular P{wHy} insertion was due to positional silencing, rather than to damage to the transposon itself . Briefly, virgin females of the line w*;+;P{Δ2-3}99B Sb/TM6—which constitutively expresses P transposase (Robertson et al. 1988)—were crossed to males of the candidate P{wHy} strain. The integrity of the transposon at the starting insertion site, and thus positional silencing at that site, would be indicated by the remobilization-dependent appearance of mottled pigmentation (mosaicism) in the eyes of the F1 progeny.
RESULTS
Current status of P{wHy} distribution along the autosomes:
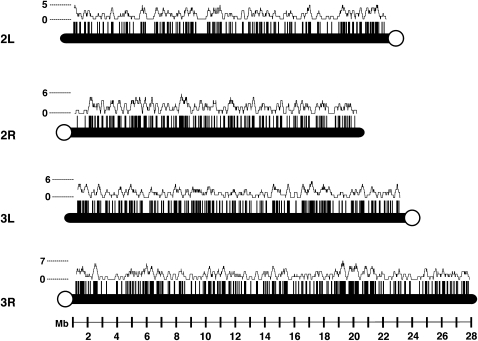
At present, the P{wHy} library is composed of 810 insertions (Figure 1) selected from a larger, starting collection of >2000 lines. The chief criteria for selection were (i) certainty of mapping on the chromosome, (ii) nonredundancy of insertion locus, (iii) even spacing of the inserts throughout the genome, and (iv) enhancement of the overall genome coverage provided by the established mutational collections, based on P{wHy} deletion and transrecombinational properties (see below). The average distance between each pair of inserts is 120 kb.
Figure 1.—
Distribution of the current P{wHy} library on the autosomes. Each vertical tick along a chromosome arm represents an insertion site. The corresponding line graph follows the density of inserts by scanning the genome in 10,000 base increments, using a window of 200,000 bases.
Because the library was generated on the basis of the interchromosomal transposition of a P{wHy} element starting on the first chromosome, the inserts resulting from the expansion reside predominantly on the second and third chromosomes, with only a very few inserts on the fourth chromosome.
The distribution of P{wHy} inserts is as follows: 178 are on the left arm of chromosome II (2L); 194 inserts are on 2R; 212 inserts are on 3L; 251 inserts are on 3R; and just 3 inserts are on the fourth chromosome. At the time of this writing, 810 of these 838 lines have been archived at the Bloomington Stock Center. There are a relative few genome regions of sharply reduced P{wHy} density—i.e., where there is significantly greater than the average separation of 120 kb between inserts. An interval on 2L, at nucleotide coordinates corresponding to cytology 29E–30B10, is the largest region devoid of inserts. However, a query of FlyBase (http:www.flybase.org) reveals that there are at least 100 transposon insertions from the established insertion collections in this interval, including a number of elements from the extensive Rorth collection (Rorth et al. 1998). This observation would immediately indicate that the absence of P{wHy} in this region is simply a statistical fluctuation and not a generalized exclusion of P transposition.
Efficiency of generating P{wHy}-based deletion sets:
We sought to characterize the overall efficiency of P{wHy} in making deletions suitable for genomic annotations. The lines indicated in Table 1 were analyzed statistically for this purpose. The hobo-mediated deletions were first molecularly screened against P{wHy}-confined hobo mobilization by PCR test as earlier described (Huet et al. 2002). New hobo flanking sequence for “genomic events,” having deletion endpoints lying outside of P{wHy}, was successfully determined 67% of the time (Table 2), using iPCR or UFW mapping (see materials and methods); that a higher percentage was not achieved tended to reflect sequencing interference rather than failure to synthesize the mapping amplicon. We also observed that 87.4% of these endpoints were nonredundant. Thus, only a minority of the genomic events were intractable to the mapping techniques employed.
TABLE 1.
P{wHy} lines used for statistical analysis of deletion events
| P{wHy} line | Cytogical location | Accession number | Insertion site position in bp |
|---|---|---|---|
| P{wHy}B401C03 | 34C | AE003640.1 | 167628–35 |
| P{wHy}01D01 | 58D | AE003456.1 | 201422–29 |
| P{wHy}01D09 | 46A | AE003832.1 | 241470–77 |
| P{wHy}01F04 | 52C | AE003808.1 | 196444–51 |
| P{wHy}02H09 | 39C | AE003669.2 | 239782–89 |
TABLE 2.
Molecular mapping of genomic events
| Deletion sets
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 01C03
|
01D01
|
01D09
|
01F04
|
02H09
|
|||||||
| w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | Total (%) | |
| Total genomic events | 16 | 11 | 65 | 215 | 233 | 37 | 18 | 9 | 50 | 10 | 664 |
| Events sequenced | 13 | 7 | 53 | 164 | 153 | ND | 13 | ND | 36 | 8 | 445 (67) |
| Mapping class | |||||||||||
| Candidate deletions | 9 | 4 | 47 | 119 | 127 | ND | 9 | ND | 31 | 4 | 350 (78.9) |
| Elsewhere in genome | 1 | 1 | 2 | 16 | 16 | 2 | 1 | 0 | 39 (8.8) | ||
| Events confined to P{wHy} | 0 | 2 | 4 | 7 | 1 | 0 | 1 | 2 | 17 (3.8) | ||
| Multiple events | 3 | 0 | 0 | 8 | 1 | 2 | 1 | 2 | 17 (3.8) | ||
| Insertion into repeats | 0 | 0 | 0 | 7 | 6 | 0 | 0 | 0 | 13 (2.9) | ||
| Other | 0 | 0 | 0 | 7 | 2 | 0 | 0 | 0 | 9 (2) | ||
Elsewhere in genome, deletion endpoint found in region unrelated to starting site; multiple events, hobo appears in multiple copies; ND, no data. 01D01 w+y− and 01 D09 w−y+ deletion sets have been presented in Huet et al. (2002) and are shown here for comparison.
The resulting deletions were further examined by stepwise analysis. Mobilization events fell into six categories: (1) “candidate deletions,” composed of successfully mapped, and otherwise unproblematic flanking sequence; (2) “elsewhere on the genome,” in which the hobo element lies very far (>800 kb, or on a different chromosome) from the starting insertion site; (3) events confined to P{wHy}; (4) “multiple events,” in which hobo occurs in multiple copies; (5) “insertion into repeats,” for which hobo is found within repetitive elements, so as to confound mapping; and (6) “other,” representing complex rearrangements, such as reversal of hobo orientation.
We found that 72.5% of deletions were <60 kb in size (Table 3), which we previously determined (Huet et al. 2002, which showed experiments sizing the extent of every event in a deletion set, including the less saturated 0.1–0.4 Mb range) to be the range within which deletion saturation can be effectively achieved using P{wHy}. Table 4 presents efficiencies for each stage of the P{wHy} strategy. There are wide differences among the various steps: <1% of 02H09 mobilization crosses yielded w+y− derivatives, whereas 50% of the mapped derivatives generated candidate deletions. We observed that the steps of lesser or greater efficiency were consistently so among different starting lines, indicating that the hobo mobilization and the recovery of genomic events are the two key rate-limiting steps of the P{wHy} process.
TABLE 3.
Nonredundant deletions of <60 kb
| Deletions
|
||
|---|---|---|
| Deletion set | >60 kb | <60 kba (%) |
| 01C03 w−y+ | 0 | 9 (100) |
| 01C03 w+y− | 0 | 4 (100) |
| 01D01 w−y+ | 8 | 27 (77) |
| 01D01 w+y− | 26 | 74 (74) |
| 01D09 w−y+ | 35 | 78 (69) |
| 01F04 w−y+ | 0 | 8 (100) |
| 02H09 w−y+ | 1 | 25 (96) |
| 02H09 w+y− | 2 | 2 (50) |
| Total | 75 | 198 (72.5) |
01D01 w+y− and 01 D09 w−y+ deletion (nonredundant and genetically validated) sets have been presented in Huet et al. (2002) and are shown here for comparison.
Threshold distance for nested deletion saturation.
TABLE 4.
Stepwise efficiency of creating deletions using the P{wHy} mobile element
| Set of deletions
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 01C03
|
01D01
|
01D09
|
01F04
|
02H09
|
|||||||
| Steps of the procedure | w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | w−y+ | w+y− | Total |
| Total crosses | 387 | 387 | 1870 | 1870 | 1539 | 1539 | 1015 | 1015 | 3442 | 3442 | 8253 |
| hobo mobilization | 37 (10) | 19 (5) | 285 (15) | 363 (19) | 404 (26) | 168 (11) | 58 (6) | 22 (2) | 96 (3) | 19 (1) | 1471 |
| Survival and fertile | 34 (92) | 17 (89) | 272 (95) | 351 (97) | 354 (88) | 152 (90) | 40 (69) | 17 (77) | 96 (100) | 19 (100) | 1352 |
| Genomic events | 16 (47) | 11 (65) | 65 (24) | 215 (61) | 233 (66) | 37 (24) | 18 (45) | 9 (53) | 50 (52) | 10 (53) | 664 |
| Mapped derivatives | 13 (81) | 7 (64) | 53 (82) | 164 (76) | 153 (66) | ND | 13 (72) | ND | 34 (68) | 8 (80) | 445 (72) |
| Candidate deletions | 9 (69) | 4 (57) | 47 (89) | 119 (73) | 127 (83) | ND | 9 (69) | ND | 31 (91) | 4 (50) | 350 (78) |
| Nonredundant deletions | 9 (100) | 4 (100) | 35 (74) | 105 (88) | 114 (90) | ND | 8 (89) | ND | 26 (84) | 4 (100) | 305 (87) |
| Validated deletions | ND | 4 (100) | ND | 100 (95) | 113 (99) | ND | nd | ND | ND | ND | 217 (95) |
| Deletions <60 kb | 9 (100) | 4 (100) | 27 (77) | 74 (74) | 78 (69) | ND | 8 (100) | ND | 25 (96) | 2 (50) | 228 |
Total crosses indicates the number of crosses set up for each set. All other entries represent the number of the chromosomes belonging to the indicated step. The w+y− class includes a minority that is phenotypically w−y−, but structurally confirmed as w+y−; deletions are validated by genetic complementation test. The percentage of the deletions relative to the number of deletions of the previous step is in parentheses, when known; ND, no data. Total is the overall number of chromosomes for each step. 01D01 w+y− and 01 D09 w−y+ deletion sets have been presented in Huet et al. (2002) and are shown here for comparison.
Large deletions by transrecombination:
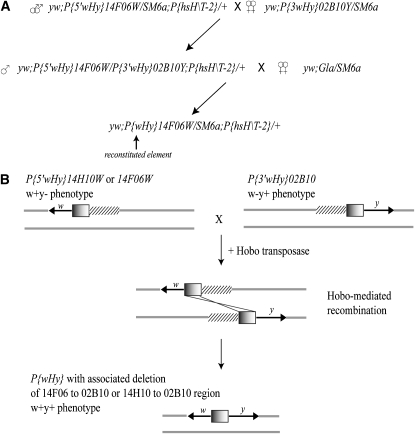
The ability to use P{wHy} to generate deletions of a very large, specific size would contribute to the utility of the P{wHy} tool, since with respect to deletions much larger than 60 kb, the original procedure deviates noticeably from a strictly flat distribution (Huet et al. 2002). We reasoned that hobo-transposase mediated recombination between P{wHy}-derived P{5′wHy} and P{3′wHy} elements could accomplish our goal (Figure 2). The size of the associated deletion is determined by the distance between the starting P{wHy} insertions. P{5′wHy} and P{3′wHy} elements derived from insertions 101 kb and 27 kb apart were generated in a previous study (Mohr and Gelbart 2002). To test if transrecombination results in efficient generation of larger deletions, we placed P{5′wHy}14F06W and P{5′wHy}14H10W chromosomes in trans to P{3′wHy}02B10Y chromosomes in the presence of hobo transposase. Next, we screened for and recovered the w+y+ phenotypic class indicative of successful transrecombination (Table 5).
Figure 2.—
Transrecombination between P{5′wHy} and P{3′wHy} elements. (A) Strategy for generation of transrecombinants with P{5′wHy}14F06W and P{3′wHy}02B10Y elements. (B) Model of transrecombination. Hatched bars indicate the presence of deletions on the P{5′wHy} and P{3′wHy} chromosomes. Note that the 5′ insertion site of the P{5′wHy} element and the 3′ insertion site of the P{3′wHy} element define the extents of the deletion in the resulting transrecombinant and that reconstitution of a functional whole element permits restarting the original P{wHy} deletion scheme anew.
TABLE 5.
Transrecombination between P{5′wHy} and P{3′wHy} elements
| A. Size of starting deletions | |||
|---|---|---|---|
| P{5′wHy} deletions | P{3′wHy} deletions | ||
| P{5′wHy}14F06W-05 | 737 bp | P{3′wHy}02B10Y-10 | 412 bp |
| P{5′wHy}14F06W-10 | 21,850 bp | P{3′wHy}02B10Y-14 | 56,058 bp |
| P{5′wHy}14H10W-10 | 893 bp | ||
| B. Efficient recovery after Hobo transposase-mediated transrecombination | |||
| Starting elements
|
w+y+ events/crosses
|
||
| 14F06 to 02B10 (101 kbp) | |||
| P{5′wHy}14F06W-05, P{3′wHy}02B10Y-10 | 3/18 | ||
| P{5′wHy}14F06W-05, P{3′wHy}02B10Y-14 | 1/15 | ||
| P{5′wHy}14F06W-10, P{3′wHy}02B10Y-10 | 1/24 | ||
| P{5′wHy}14F06W-10, P{3′wHy}02B10Y-14 | 2/13 | ||
| 14H10 to 02B10 (27 kbp) | |||
| P{5′wHy}14H10W-10, P{3′wHy}02B10Y-10 | 6/31 | ||
| P{5′wHy}14H10W-10, P{3′wHy}02B10Y-14 | 3/30 | ||
| Total | 16/131 | ||
Genetic and molecular tests were used to determine if the resulting w+y+ strains had intact P{wHy} elements and associated deletions as expected. PCR analyses confirmed the 5′ and 3′ P{wHy} insertion sites and the presence of an intact P{wHy} element in the transrecombinant strains. Genetic analysis confirmed that the deletions behave as expected in complementation tests with available genetic reagents in the region (Table 6; Mohr and Gelbart 2002). Together these data suggest that transrecombination provides an efficient method for generating deletions with an intact P{wHy} element (Table 6, Figure 2).
TABLE 6.
Transrecombinants have the predicted pattern of complementation
| 101-kbp deletion (14F06 to 02B10) | 27-kbp deletion (14H10 to 02B10) | |||
|---|---|---|---|---|
| Df(2R)14H10W-15 | + | + | + | + |
| Df(2R)02B10Y-11 | − | − | − | − |
| eIF3-S814F06 | − | − | + | + |
| Df(2R)14H10Y-45 | − | − | + | + |
+, complement; −, fail to complement.
An indicator for positional silencing:
Although the white marker in P{wHy} is usually quite easily scored, a number of lines exhibited diminished expression of this gene. A subset of these flies had no detectable eye pigmentation (e.g., line DG37311 on 2L), and were scored y+w−. Such lines comprise ∼10% of the starting collection and include small groups that are locally clustered on the genome. (Fly stocks representing such regions were included in the companion submissions to the Bloomington Stock Center.) A smaller number of lines, ∼5% of the total, were scored y−w+. In our deletion experiments we also detected a small percentage of derivatives that were phenotypically y−w−, but were structurally y−w+ (Table 4). These observations are reminiscent of position effects on transposon markers previously reported by those investigating chromatin and long-range silencing (Balasov 2002; Yan et al. 2002).
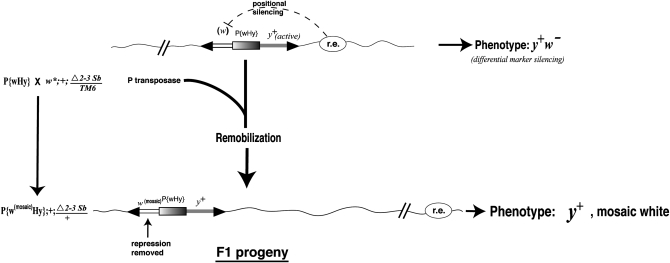
We took advantage of the ability of the P{wHy} to remobilize upon introduction of a stable source of P transposase (Robertson et al. 1988). An eye mosaicism test was performed on a sample of the y+w− flies to rule out the possibility that the P{wHy} element in such lines may have been damaged (e.g., during the expansion for making the P{wHy} library). We chose lines DG37311, DG37606, and DG37710 and reasoned that if the P{wHy} elements were still intact, (i) they should be capable of somatic remobilization and (ii) the expression of the white gene should reappear in some of the progeny, depending on where the remobilized element landed. This would result in a classic mosaicism, a patchy pigmentation based on multiple independent transposition events in a given eye.
We performed the cross diagrammed in Figure 3, in which the flies under examination were mated to the transposase-bearing flies in a w* (nonpigmented) genetic background. We obtained mottled eye color within the F1 progeny, indicating that the P{wHy} element was structurally intact and that the gene silencing observed when the transposon was at its original location in line DG37311, DG37606, or DG37710 was due to positional silencing. It is noteworthy that the initial silencing was differential with respect to each transposon marker, allowing continued phenotypic detection from start to finish. These results suggest that P{wHy} is likely to be useful as a general tool for evaluating positional silencing (see discussion).
Figure 3.—
Crosses for mosaicism test of positional silencing. Appearance of mottled eye color arising from somatic remobilization in F1 progeny. Asymmetric marker silencing allowed phenotypic detection throughout these experiments.
DISCUSSION
We present in this report a large scale P{wHy}-based disruption study using P-remobilized inserts (Huet et al. 2002) that, in turn, have the ability to produce a range of small-to-very-large hobo-induced deletions from any particular starting insertion site. This insertion-then-deletion potential had been further established by Weiler (2007) and Suyama et al. (2009), as well as by Mohr and Gelbart (2002), who achieved the systematic disruption of chromosome region 54D–55B using P{wHy}.
The P{wHy} collection of this study presents a reasonably comprehensive distribution of insertions throughout the genome (Figure 1). Such a distribution reflects, in large part, the ability to select the inserts on the basis of the distance between adjacent P{wHy} elements, and not on the basis of whether the element had inserted strictly within a gene. This flexibility of choice is inherent to the “deleter” component of the P{wHy} transposon. However, for the present form of this system, it is cautioned that P{wHy} inserts should not be immediately expected to produce hobo-mediated nested deletion series across (the relatively few) intervals that are prohibited from disruption because of haploinsufficiency in the region—although in principle, one should be able to generate compensatory regional duplications, in addition to deletions, by the P{wHy} transrecombination scheme, to evade lethality. It should also be possible to establish a collection of P{wHy} inserts on X by standard P remobilization of any of the current second or third chromosome inserts, and then screening for transposition to the first chromosome.
Straightforward quantitative estimates can be applied to the P{wHy} collection. By introducing the hobo transposase (Huet et al. 2002), fine-grained nested deletions over a range of 60 kb in either direction from the original insertion site can be readily obtained. It follows that each starting insertion has a total (fine-grained) “reach” of 120 kb.
We observe that a deletion set can be produced from a single initial insertion, obviating the need for a starting pairwise configuration for any locus, in contrast to the prevalent DrosDel system (Ryder et al. 2007). That is, in terms of completeness of genome coverage DrosDel can ultimately delete the vast majority of genes, whereas P{wHy} offers an appreciably high deletion range per insert, while providing exceptional resolution. Distinctively, when P{wHy} is used for transrecombination (discussed below), a completely functional element is regenerated, and thus even further deletions can be made by the original P{wHy} method (without the need for a third element at the beginning)—altogether bringing substantial capability to this system.
With >800 nonredundant strains in the P{wHy} collection, the entire insertion library has a potential genomic coverage of ∼100,000 kb (100 Mb of “coverage potential”), i.e., the majority (∼60%) of the Drosophila genome. Coverage in the 3-Mb Adh region is a useful illustration, since the available P insertions outside of this study had been estimated to mutate only a minority of the predicted protein coding genes centered about Adh over this range (Ashburner et al. 1999). The current P{wHy} collection reported here provides 21 starting insertions in this region, thus yielding a density sufficient for ∼75% coverage; in particular, we find genome disruption down to within 60 kb, or less, of the Adh gene on chromosome 2L, on the basis of starting insertions and local deletion potential. In practice, the highest resolution for phenotypic annotation with this system is accomplished by probing the region of interest with overlapping, scanning deletions between two neighboring insertions on the order of 60 kb apart (Huet et al. 2002). Applying this criteria would somewhat reduce the effective genomic coverage below the 60% level since, for many of the starting insertions, the nearest P{wHy} neighbor is >60 kb away. Of course, an ideal P{wHy} collection would consist of 3000 inserts evenly spaced throughout the genome, since that would bring any two neighboring inserts uniformly to within ∼60 kb of each other, permitting the complete resolution of all of the transcription units in the cell. However, on the basis of our experience, we estimate that obtaining this number of useful insertions would require a starting collection of >8000 strains, given the likelihood of locational redundancies.
In our gene functional studies, we found that the range of the P{wHy}-associated deletions, and thus the potential genomic coverage, could be augmented by hobo-mediated recombination—or transrecombination—between a pair of widely separated P{wHy} half molecules. The fact that each half molecule from such a pair is deleted for a different marker permitted a screen on the basis of the reconstitution to a doubly marked, complete P{wHy} element. The ability to create more substantial deletions of at least 100 kb in this manner at once suggests an iterative scheme for traversing even larger genomic intervals by sequential deletion–reconstitution–deletion among neighboring P{wHy} elements, giving rise to megabase-sized deletions, similar in length to the classical Drosophila deficiencies. This advantage—coordinated delivery of large-deletion/finely-resolved analyses—would be useful in many areas of the genome with unusual genetic or control element behavior, but where the molecular signatures are not readily predicted.
A subset of the P{wHy} collection was strongly repressed for the phenotype of one of the transposon markers, indicating the possibility of site-dependent interactions of this transposon with the chromosome. The positional silencing of transposon and nontransposon genetic elements has been well documented, particularly with regard to the modulation of white or yellow gene expession as an indicator (Zhang and Spradling 1994; Balasov 2002; Yan et al. 2002). Our observation of genetic derepression in remobilization-induced mosaicism demonstrates that the P{wHy} transposon should be applicable to the molecular dissection of chromosomal repressor elements/silencer domains. Here, P{wHy} benefits particularly from having two visible markers, making possible the detection of both the original insertion and the silencing. Specifically, due to the differential susceptibility to silencing observed between white and yellow (the latter usually less sensitive), we were often able to track P{wHy} from the start, despite complete white marker suppression. We believe that it will be feasible to finely map the location and extent of such repressive domains by scanning the regions of interest with nested hobo-induced deletions, using restored expression of the single remaining P{wHy} marker as a readout for disruption of the repressor (Figure 3). We envision this as a phenotype reversal assay—beginning with (repressed) y+w− flies, and ending with (postdeletion) y−w+ progeny, which could conceivably be employed for the detailed molecular analysis of centric heterochromatic regions, where positional silencing effects are frequently encountered (reviewed in Dillon and Festenstein 2002; Grewal and Moazed 2003). It is probable that increasing attention will be paid to the gene regulation in the heterochromatic regions, since they comprise about a third of the Drosophila genome, and the close study of these segments could yield important clues on such issues as chromosome architecture.
Acknowledgments
We are indebted to Rob Kulathinal, Haiyan Zhang, and FlyBase for database support. We thank Kathleen Matthews, Robert Levis, Joseph Carlson, Roger Hoskins, and the Gene Disruption Project for curation and data submission. We are grateful to Todd Martin and Yevgenya Kraytsberg for technical assistance and to Beverley Matthews for critical reading of the manuscript. We thank Gerald Rubin for encouragement and guidance. This work was funded by grants from the National Institute of General Medical Sciences (GM28669) and the National Human Genome Research Institute (HG000739) (to W.M.G.).
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., 2004. Accessing the Exelixis collection. Nat. Genet. 36 207. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., S. Misra, J. Roote, S. E. Lewis, R. Blazej et al., 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics 153 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov, M. L., 2002. Genetic factors controlling white gene expression of the transposon A(R) 4-24 at a telomere in Drosophila melanogaster. Genome 45 1025–1034. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Luo, Y. He, J. W. Carlson et al., 2004. The BDGP Genome Disruption Project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3:RESEARCH0079.1–0079.14. [DOI] [PMC free article] [PubMed]
- Cooley, L., R. Kelley and A. Spradling, 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239 1121–1128. [DOI] [PubMed] [Google Scholar]
- Dillon, N., and R. Festenstein, 2002. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18 252–258. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and D. Moazed, 2003. Heterochromatin and epigenetic control of gene expression. Science 301 798–802. [DOI] [PubMed] [Google Scholar]
- Hacker, U., S. Nystedt, M. P. Barmchi, C. Horn and E. A. Wimmer, 2003. piggyBac-based insertional mutagenesis in the presence of stably integrated P elements in Drosophila. Proc. Natl. Acad. Sci. USA 100 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. M., E. J. Rehm and G. M. Rubin, 2000. Recovery of DNA sequences flanking P-element insertions: inverse PCR and plasmid rescue, pp. 429–437 in Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Huet, F., J. T. Lu, K. V. Myrick, L. R. Baugh, M. A. Crosby et al., 2002. A deletion-generator compound element allows deletion saturation analysis for genomewide phenotypic annotation. Proc. Natl. Acad. Sci. USA 99 9948–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 1017–1026. [DOI] [PubMed] [Google Scholar]
- Mohr, S. E., and W. M. Gelbart, 2002. Using the P{wHy} hybrid transposable element to disrupt genes in region 54D–55B in Drosophila melanogaster. Genetics 162 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrick, K. V., and W. M. Gelbart, 2002. Universal Fast Walking for direct and versatile determination of flanking sequence. Gene 284 125–131. [DOI] [PubMed] [Google Scholar]
- Ochman, H., A. S. Gerber and D. L. Hartl, 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré, A. C., D. M. Dean and J. Ewer, 2009. Construction and characterization of deletions with defined end points in Drosophila using P-elements in trans. Genetics 181 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics 118 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, G. M. Rubin et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125 1049–1057. [DOI] [PubMed] [Google Scholar]
- Ryder, E., M. Ashburner, R. Bautista-Llacer, J. Drummond, J. Webster et al., 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., D. M. Stern, I. Kiss, J. Roote, T. Laverty et al., 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama, R., A. Jenny, S. Curado, W. Pellis-van Berkel and A. Ephrussi, 2009. The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development 136 95–105. [DOI] [PubMed] [Google Scholar]
- Triglia, T., M. G. Peterson and D. J. Kemp, 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16: 8186. [DOI] [PMC free article] [PubMed]
- Weiler, K. S., 2007. E(var)3–9 of Drosophila melanogaster encodes a zinc finger protein. Genetics 177 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C. M., K. W. Dobie, H. D. Le, A. Y. Konev and G. H. Karpen, 2002. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics 161 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., and A. C. Spradling, 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91 3539–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]