Abstract
Telomeres are essential features of linear genomes that are crucial for chromosome stability. Telomeric DNA is usually replenished by telomerase. Deletion of genes encoding telomerase components leads to telomere attrition with each cycle of DNA replication, eventually causing cell senescence or death. In the Saccharomyces cerevisiae strain W303, telomerase-null populations bypass senescence and, unless EXO1 is also deleted, this survival is RAD52 dependent. Unexpectedly, we found that the S. cerevisiae strain S288C could survive the removal of RAD52 and telomerase at a low frequency without additional gene deletions. These RAD52-independent survivors were propagated stably and exhibited a telomere organization typical of recombination between telomeric DNA tracts, and in diploids behaved as a multigenic trait. The polymerase-δ subunit Pol32 was dispensable for the maintenance of RAD52-independent survivors. The incidence of this rare escape was not affected by deletion of other genes necessary for RAD52-dependent survival, but correlated with initial telomere length. If W303 strains lacking telomerase and RAD52 first underwent telomere elongation, rare colonies could then bypass senescence. We suggest that longer telomeres provide a more proficient substrate for a novel telomere maintenance mechanism that does not rely on telomerase, RAD52, or POL32.
TELOMERES, the ends of eukaryotic chromosomes, are crucial for genome stability and the complete replication of genetic information encoded on linear chromosomes (reviewed in Chakhparonian and Wellinger 2003). The distal portion of Saccharomyces cerevisiae chromosomes is composed of ∼350–500 bp of degenerate repeat sequences [(TG1–3)n] (Walmsley et al. 1984), with a G-rich single-stranded 3′ extension that varies in length during the cell cycle (Wellinger et al. 1993; Larrivee et al. 2004). This organization is essential for the binding of telomere-specific proteins and ensures that the telomere is not repaired as a double-strand break (d'Adda di Fagagna 2008). Subtelomeres contain two distinct types of repeats (Chan and Tye 1983a,b). Tandem arrays of up to four Y′ elements are embedded between telomeric TG1–3 repeat tracts on half to two-thirds of yeast telomeres. The Y′ element contains a unique XhoI recognition site; digestion of genomic DNA with this enzyme yields a characteristic terminal restriction fragment (TRF) of ∼1.2 kbp in wild-type cells. The other subtelomeric feature, the core X element, is found on all yeast chromosomes and is separated from Y′ sequences by telomeric TG1–3 repeats. Together, these regions contribute to the stability, replication, and maintenance of yeast telomeres.
Due to the semiconservative nature of DNA replication, telomeres shorten with each replication cycle (as first predicted by Watson 1972 and Olovnikov 1973). In the absence of telomerase, a reverse transcriptase that replenishes telomeric sequences, telomere shortening can lead to a critically short telomere length after several cell divisions. When a subset of telomeres reaches this so-called “critical” threshold, the cell enters a nonproliferative state termed senescence (Lundblad and Szostak 1989; Lundblad and Blackburn 1993; Lendvay et al. 1996). Deletion of any of the genes encoding telomerase subunits leads to an ever shorter telomere (EST) phenotype and cell death within 60–80 generations (Lendvay et al. 1996). However, a small subset of cells in the arrested population is able to maintain viability by replenishing telomere DNA via a recombination-based mechanism (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Chen et al. 2001). These so-called “survivors” are not always stably propagated and telomeres may continue to shorten over time, with subsequent lengthening when telomeres become very short (Teng et al. 2000).
The generation of survivors almost always depends on RAD52-dependent homologous recombination. The two broad classes of survivors that have been described to date (type I and type II) differ in the sequence amplified at chromosome ends and the proteins required for recombination (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Chen et al. 2001). In type I survivors, telomere maintenance involves amplification of subtelomeric Y′ sequences and acquisition of Y′ sequences on all telomeres (Lundblad and Blackburn 1993; Le et al. 1999), and survival depends on Rad51, Rad52, Rad54, Rad55, and Rad57 (Le et al. 1999; Teng and Zakian 1999). The telomeric DNA exhibits a characteristic XhoI terminal restriction fragment (TRF) distribution, although of a smaller size than in wild-type cells due to shorter TG1–3 telomeric DNA tracts. Chromosomes of type I survivors are longer than in wild-type cells, likely due to the amplification of the Y′ element, and appear heterogeneous when analyzed by pulsed-field gel electrophoresis (Liti and Louis 2003). In type II survivors, telomeres are maintained by TG1–3 amplification (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Teng et al. 2000), which depends on Rad52, the MRX complex (Mre11, Rad50, and Xrs2), Sgs1, and Rad59 (Le et al. 1999; Teng et al. 2000; Chen et al. 2001; Huang et al. 2001; Johnson et al. 2001; Tsukamoto et al. 2001). When digested with XhoI, telomeric DNA exhibits discrete fragments of various sizes due to the amplification and propagation of differing telomere lengths on each chromosome end. Type I and type II telomere maintenance pathways appear to be closely related to the break-induced replication mechanism (BIR) that repairs chromosomal double-strand breaks (DSBs) (reviewed in McEachern and Haber 2006). The replication protein Pol32 is required for recovery of both types of survivors, suggesting that these pathways may depend on recombination-dependent DNA replication (Lydeard et al. 2007). Cells may survive via changes in telomeric DNA structure that permit recombination, rather than the accumulation of extragenic suppressors (reviewed in McEachern and Haber 2006).
The precise frequency of survivor generation has not been accurately determined, although frequencies of one survivor in ≤104 cells/generation have been reported (Lundblad and Blackburn 1993; McEachern and Haber 2006). This low frequency suggests that the ability to survive is not determined by a single genetic locus. For example, Makovets et al. (2008) used a mating-based analysis to demonstrate that a survivor haploid could exert dominance over a senescent haploid. This finding confirms that the acquisition of extragenic suppressors is unnecessary for telomerase-independent survival (Lundblad and Blackburn 1993). Also, Zubko and Lydall showed that the survival of cdc13-1 cells at 36° segregates as a multigenic trait; however, the presence of suppressor mutations has not been ruled out (Zubko and Lydall 2006).
Previously, survivors in S. cerevisiae had not been recovered in cells lacking telomerase and RAD52 unless EXOI or SGS1 was also absent (Maringele and Lydall 2004; Lee et al. 2008). Chromosomes in exo1Δ survivors are linear but have lost telomeric and subtelomeric sequences, resulting in atypical chromosome sizes. These survivors also exhibit large inverted and duplicated repeats (palindromes) at chromosome ends, which probably originate from small inverted repeats (Maringele and Lydall 2004; Lee et al. 2008).
Here, we report that the S. cerevisiae S288C strain can survive, at a low frequency, in the absence of telomerase and RAD52, without any other known genetic alterations. These RAD52-independent survivors could be propagated for several generations and exhibited a type II-like (e.g., telomeric DNA amplification) pattern. Furthermore, the propensity to escape senescence appeared to depend on telomere length. Telomere elongation in telomerase- and RAD52-deficient strains increased the frequency of survival and even permitted survival in a W303 est2Δ rad52Δ strain (which under normal circumstances undergoes rapid senescence in the absence of telomerase and RAD52). RAD52-independent survivors arose in the absence of genes known to affect RAD52-dependent cell survival, and, in diploids, survival behaved as a multigenic trait.
MATERIALS AND METHODS
Yeast strains:
W303 MATa and MATα strains were obtained from M. Tyers (MT234, MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 URA3 GAL+ psi+ ssd1-d2 rad5-535; and MT235, MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 GAL+ psi+ ssd1-d2 rad5-535). S288C MATa and MATα strains were obtained from M. Tyers (BY4741, MATa his3Δ leu2Δ met15Δ ura3Δ; and BY4742, MATα his3Δ leu2Δ lys2Δ ura3Δ).
Yeast manipulations:
Replacement of the Kanr cassette for selection of ORF deletion strains was performed in either haploid or diploid cells, using a PCR-based replacement protocol as described in Brachmann et al. (1998) and Longtine et al. (1998), followed by selection on appropriate medium. Gene disruption was confirmed by PCR, enzymatic digestion of the PCR products, and Southern blot analysis. Genomic DNA of a tlc1Δ strain was a gift from D. Durocher. Transformation of yeast was performed according to the lithium acetate method (Gietz and Woods 2006). For the telomere elongation experiments, W303 or S288C heterozygous diploids (est2Δ/EST2 rad52Δ/RAD52 or tlc1Δ/TLC1 rad52Δ/RAD52) were transformed with the plasmid pVL1107 (Leu+) encoding a Cdc13-Est2 fusion protein [obtained from D. Durocher, originally a gift of V. Lundblad (Evans and Lundblad 1999)]. After dissection and identification, the appropriate haploid was propagated in the absence of leucine for the indicated number of passages. Following telomere elongation, colonies that had lost the plasmid after growth in rich media were identified. Standard genetic procedures were used for sporulation of diploids, microdissection of asci, and identification of haploids (Guthrie and Fink 1991).
Senescence assays on plates were performed according to Maringele and Lydall (2004) and Lebel et al. (2006). Briefly, cells were isolated from freshly dissected tetrads (passage 1) and propagated on YPD plates. After incubation for 4 days at 30°, single colonies were picked from the plate (passage 2) and repropagated on YPD plates to obtain passage 3, etc. This procedure was performed for a total of six serial propagations (the equivalent of 140 generations). Plates of each passage were stored at 4° until all propagations were completed. Growth over six serial propagations (24 days of serial growth for each genotype) was represented by a “summary senescence” plate (as shown in Figures 1, 2, 4, 6, and 7): an isolated colony from each of the individual propagation plates was repropagated onto a sector of a single YPD plate and incubated at 30° for 4 days.
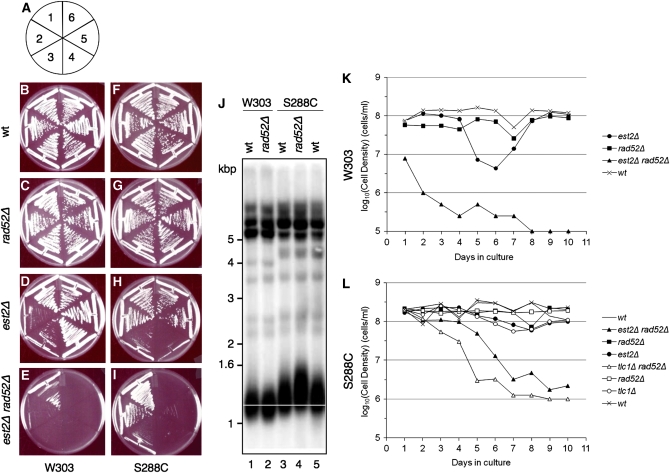
Figure 1.—
The majority of haploid colonies generated from W303 est2∷NAT/EST2 rad52∷URA3/RAD52 and S288C est2∷NAT/EST2 rad52∷URA3/RAD52 strains exhibit an expected senescent phenotype. (A) Schematic of the senescence assays on plates, from passage 1 to 6 (see materials and methods). Senescence phenotypes for the haploid colonies (genotypes indicated at left) resulting from the sporulation of W303 (B–E) and S288C (F–I) diploids are shown. (J) Wild-type S288C telomeres are longer than W303 telomeres. Genomic DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. (K and L) W303 est2Δ rad52Δ (K) and S288C tlc1Δ rad52Δ and est2Δ rad52Δ (L) lose viability in liquid growth assays. Cells were picked from a fresh dissection plate, inoculated into YPD media, and grown to saturation (1–2 × 108 cells/ml). Every 24 hr, cell density was measured using a hemacytometer, and the culture was diluted to 105 cells/ml.
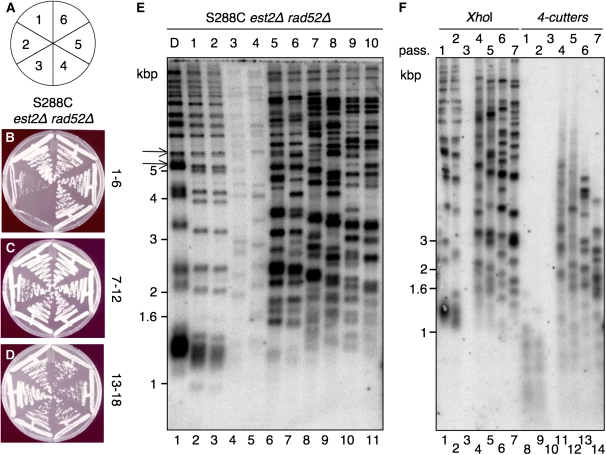
Figure 2.—
Rare S288C est2Δ rad52Δ colonies continue to proliferate and exhibit a terminal restriction fragment (TRF) pattern typical of telomeric tract recombination. (A) Schematic of the senescence assays on plates, from passage 1 to 6 (see details in materials and methods). (B–D) Typical S288C est2Δ rad52Δ survivors (or tlc1Δ rad52Δ; data not shown) can be propagated for several generations. (E) Telomere Southern blot on genomic DNA isolated from single S288C est2∷NAT rad52∷KAN colonies from passages 1–10 (20–200 generations). DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. Lane 1, diploid S288C est2∷NAT/EST2 rad52∷KAN/RAD52 (D); lanes 2–11, haploid S288C est2Δ rad52Δ at increasing passages. Black arrows at left indicate Y′ elements. (F) Telomere Southern blot of S288C est2Δ rad52Δ survivors at increasing passages. Genomic DNA was digested with either XhoI (lanes 1–7) or a mix of AluI, HinfI, HaeIII, and MspI (lanes 8–14), and the membrane was hybridized to a telomeric probe. For each panel, marker sizes are indicated at left in kilobase pairs.
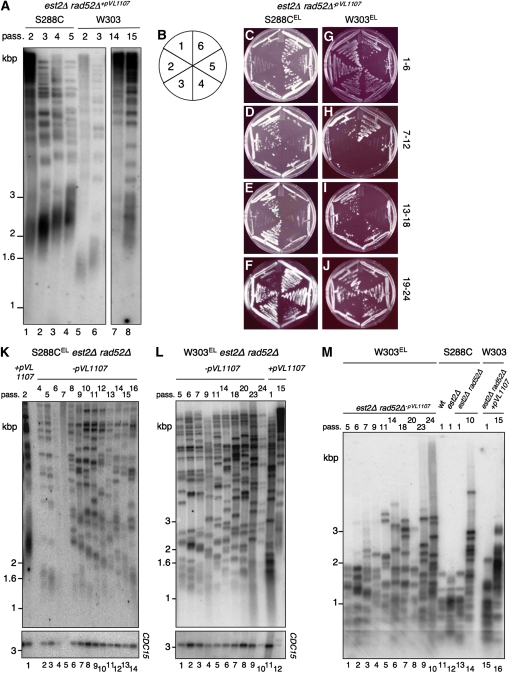
Figure 4.—
Telomere length correlates with the incidence of escape from senescence. (A) Telomere elongation in S288C est2Δ rad52Δ and W303 est2Δ rad52Δ cells containing a plasmid encoding a Cdc13-Est2 fusion protein (pVL1107): telomere Southern blot on genomic DNA from cells containing pVL1107 propagated for several passages (pass.; every 2 days) on SD−LEU plates. Genomic DNA was digested with XhoI. The membrane was hybridized to a telomeric probe. All DNA samples were analyzed on one gel; W303 passages 4–13 were omitted from the image. (B) Schematic summary of the senescence assays on plates (see materials and methods). Summary senescence assays of cells with elongated telomeres (EL) following removal of pVL1107 are shown: S288CEL est2Δ rad52Δ-pVL1107 (C–F) and W303EL est2Δ rad52Δ-pVL1107 (G–J). Telomere Southern blot on genomic DNA isolated from S288CEL est2Δ rad52Δ-pVL1107 (K) or W303EL est2Δ rad52Δ-pVL1107 (L) cells is shown. DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. (K) Lane 1, S288C est2Δ rad52Δ after 2 passages with pVL1107 (passage at which plasmid was removed); lanes 2–14, S288CEL est2Δ rad52Δ at increasing passages after plasmid loss. Bottom panel: blot was stripped and rehybridized to a CDC15 probe as a loading control. Note the underrepresentation of CDC15 in lanes 4 and 5 due to poor growth at these passages. (L) Lanes 11 and 12, W303 est2Δ rad52Δ after 1 and 15 (passage at which plasmid was removed) passages with pVL1107, respectively. Lanes 1–10, W303EL est2Δ rad52Δ at increasing passages after plasmid loss. Bottom panel: blot was stripped and rehybridized to a CDC15 probe as a loading control. The telomeric signal intensity in lanes 10 and 12 should be interpreted in light of the underrepresentation of CDC15. (M) Southern blot of genomic DNA digested with a mixture of the restriction endonucleases (AluI, HinfI, HaeIII, and MspI). The membrane was hybridized to a telomeric probe. Lanes 1–10, W303EL est2Δ rad52Δ at increasing passages after plasmid loss; lanes 11–13, S288C (wt, est2Δ, and est2Δ rad52Δ, each at passage 1); lane 14, S288C est2Δ rad52Δ survivor at passage 10; lanes 15 and 16, W303 est2Δ rad52Δ+pVL1107 at passages 1 and 15 (pVL1107 was removed at passage 15). For each panel, marker sizes are indicated at left in kilobase pairs.
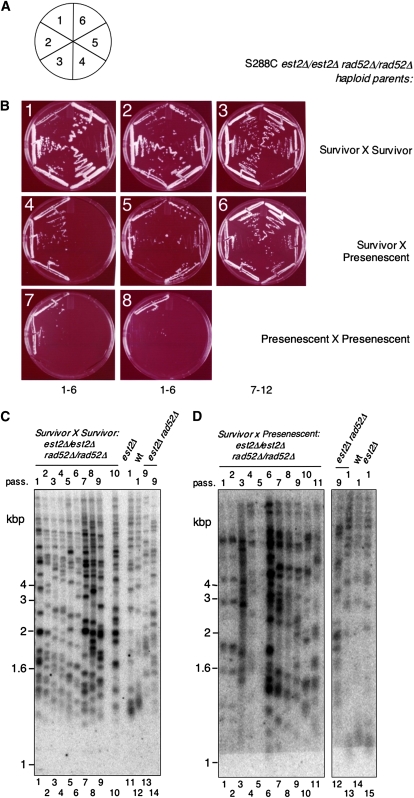
Figure 6.—
RAD52-independent survival in diploids generated from RAD52-independent haploid survivors. Freshly dissected S288C est2Δ rad52Δ haploids (“presenescent”) and S288C est2Δ rad52Δ haploid survivors (“survivor”) were mated to generate est2Δ∷est2Δ rad52Δ∷rad52Δ diploid strains. (A) Schematic of the senescence assays on plates, from passage 1 to 6 (see materials and methods). (B) Summary senescence assays of representative diploids grown on plates for the indicated number of passages. B1 and B2, two representative viable diploids from a survivor × survivor cross (passages 1–6), which all survive later passages (e.g., B2–B3). B4 and B5, representative diploids from a survivor × presenescent cross, some of which do not sustain growth beyond 3 passages (e.g., B4), while others remain viable beyond 12 passages (e.g., B5–B6). B7 and B8, two representative diploids from presenescent × presenescent crosses (no diploids sustain growth; therefore there are no data for passages 7–12). See also Figure S4. (C) Telomere Southern blot of genomic DNA. Two S288C est2Δ rad52Δ haploid survivors at passage 9 (lanes 13 and 14) were mated to produce an est2∷NAT/est2∷NAT rad52∷URA3/rad52∷KAN diploid (lanes 1–10, at indicated passages). Lanes 11 and 12, S288C est2Δ and wild-type haploids. DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. (D) Telomere Southern blot of genomic DNA from a “survivor × presenescent” diploid that escaped senescence. An S288C est2Δ rad52Δ haploid survivor (passage 9; lane 12) was mated to a presenescent est2Δ rad52Δ haploid (passage 1; lane 13) to produce an est2∷NAT/est2∷NAT rad52∷URA3/rad52∷KAN diploid (survivor × presenescent) (lanes 1–11, at indicated passages). Lanes 14 and 15, S288C haploids at passage 1 (wt, est2Δ). DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. All samples were analyzed on one gel; lanes between 11 and 12 were omitted. For each panel, marker sizes are indicated at left in kilobase pairs.
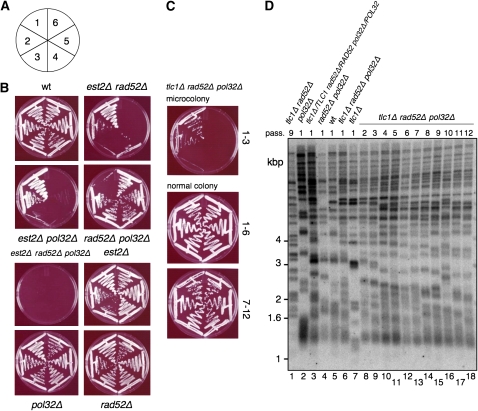
Figure 7.—
Investigation of the potential role of POL32 in RAD52-independent survival. (A) Schematic of the senescence assays on plates, from passages 1 to 6 (see materials and methods). (B) Summary senescence assays of the indicated genotype obtained from sporulation and microdissection of the diploid S288C est2∷NAT/EST2 rad52∷URA3/RAD52 pol32∷KAN/POL32. (C) Summary plates of the senescence of tlc1Δ rad52Δ pol32Δ haploids obtained from the diploid generated by mating an S288C tlc1Δ rad52Δ survivor (at passage 9) to a haploid pol32Δ colony. Microcolonies could not be propagated for >3 passages (e.g., top panel), whereas colonies of regular size could be propagated for at least 12 passages (middle and bottom panels). (D) Telomere Southern blot of DNA of a tlc1Δ rad52Δ pol32Δ viable haploid colony (lanes 6 and 8–18). This haploid was obtained from a diploid (lane 3) generated by mating an S288C tlc1Δ rad52Δ survivor (passage 9; lane 1) to a pol32Δ haploid (lane 2). Lanes 4–7, haploids derived from the heterozygous diploid in lane 3. DNA was digested with XhoI and the membrane was hybridized to a telomeric probe. Marker sizes are indicated at left in kilobase pairs.
To approximate the frequency of escape from senescence, at least 50 colonies were propagated on YPD plates for 4 days at 30°. After 3 passages, the majority of colonies had ceased proliferating and only RAD52-independent survivors were recovered. Surviving colonies were propagated stably for >10 serial passages. To generate type II survivors, freshly dissected est2Δ haploid cells were serially diluted in liquid media to a density of 105 cells/ml every 24 hr for 10 days. RAD52 was subsequently deleted in type II survivor cells by mating with rad52Δ cells, followed by dissection and selection of est2Δ rad52Δ spores. Senescence assays were performed (following 50 individual colonies per experiment) as described above.
Liquid growth assays were performed according to Chen et al. (2001) with modifications. Briefly, at least three colonies per genotype were isolated from freshly dissected tetrads and grown in YPD to saturation (1–2 × 108 cells/ml). Every 24 or 48 hr, cell density was measured using a hemacytometer. The culture was then diluted with fresh liquid YPD to a density of 105 cells/ml. Cells were harvested and genomic DNA was extracted for telomere length analysis.
To mate a RAD52-independent survivor with a freshly dissected haploid (presenescent), cells of opposite mating types were mixed on a YPD plate and incubated for 6–8 hr at 30°. Diploids were selected for the presence of all deletion markers on appropriate media and confirmed as described above.
Telomere Southern blot:
Genomic DNA was isolated, digested with XhoI, resolved through a 0.75% w/v agarose gel in 1× TBE, and transferred onto a nylon membrane. The membrane was hybridized to a radiolabeled yeast telomeric oligo (5′-CACACCCACACCCACACC-3′) to detect terminal restriction fragments (TRFs) (Lebel et al. 2006). As a loading control, a 1.76-kbp fragment of the CDC15 locus was amplified as described in Foster et al. (2006) and Zubko and Lydall (2006), labeled, and used to probe genomic DNA digested with XhoI to reveal a single hybridization fragment at ∼3 kbp.
In-gel hybridization:
The assay was performed as described in Zubko and Lydall (2006) with minor modifications. Genomic DNA was digested with XhoI for 4 hr and then incubated at 65° for 20 min. Radiolabeled and purified oligo (5′-CACACCCACACCCACACC-3′) (100,000 cpm) was added to the DNA and incubated at 37° for 15 min followed by 30 min on ice. Samples were subjected to electrophoresis through 0.75% w/v agarose in 0.5× TBE overnight at 30 V. The gel was dried and exposed to a phosphorimager screen (Molecular Dynamics). To detect double-stranded telomere DNA, the same samples were resolved on an agarose gel and transferred to a membrane under denaturing conditions, as above. Where indicated, genomic DNA was treated with exonuclease I (New England Biolabs, Beverly, MA) according to the manufacturer's instructions and as described previously (Wellinger et al. 1993). Samples were spotted onto nylon membrane using a vacuum apparatus, or digested with XhoI and subjected to electrophoresis as described above, and then probed with strand-specific ssDNA probes [CA rich, as above, or 5′-(GGTGTG)3-3′]. To normalize the ssDNA signal to total telomere DNA signal, the nylon membrane or gel was subsequently denatured and reprobed with either the CA-rich or the GT-rich ssDNA oligonucleotide.
RESULTS
Expected senescence phenotype and telomeric DNA arrangement in W303 and S288C:
Previous studies have reported that survivors could not be generated in W303 and closely related derivative strains in the absence of both RAD52 and telomerase (Le et al. 1999; Teng and Zakian 1999; Chen et al. 2001; Maringele and Lydall 2004; Larrivee and Wellinger 2006; Wen et al. 2006; Zubko and Lydall 2006). We chose a different strain background, S288C [BY4741 and isogenic strain BY4742, for which genomewide, individual ORF deletions are available for all nonessential genes (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html)], to conduct a high-throughput, liquid-based genetic screen to isolate gene deletions, like exoIΔ, that promote survival in the absence of RAD52 and telomerase (E. Rosonina, L. Maringele, D. Lydall and L. A. Harrington, unpublished results) (Cook et al. 2008). We discovered that S288C est2Δ rad52Δ or S288C tlc1Δ rad52Δ cells could survive regardless of the third gene deletion, suggesting that the genetic requirements for survival in the absence of telomerase might differ between S288C and W303.
To compare the senescence of S288C and W303 strains side by side, we constructed de novo mutant heterozygous diploids, verified their genotypes, and isolated haploids. As expected, est2Δ or tlc1Δ W303 haploids underwent senescence at approximately passage 2 (40–60 generations), and survivors were observed only when RAD52 was present (Figure 1 and data not shown). In most cases, a similar result was obtained for the S288C strain, except that senescence was marginally delayed relative to the W303 strain (Figure 1), perhaps due to the slightly longer average initial telomere length in S288C (Figure 1J). In rad52Δ S288C colonies, TRFs were slightly longer than in wild-type strains, as previously reported (Chang et al. 2007) (Figure 1J). We also examined the vitality of W303 and S288C colonies in liquid media by serial dilution of cells to 105 cells/ml every 24 hr (Figure 1, K and L). While wild-type and rad52Δ cells from each strain reached a density >108 cells/ml every 24 hr, est2Δ rad52Δ cells from both strains exhibited decreased growth potential early during the experiment and failed to recover. Consistent with the growth on plates, S288C est2Δ or tlc1Δ (with or without RAD52) reached a growth crisis later than their W303 counterparts; however, in the presence of RAD52, recovery from crisis progressed with no observable decrease in growth rate (Figure 1, K and L).
Rare escape from senescence in S288C est2Δ rad52Δ or tlc1Δ rad52Δ strains:
Consistent with our observations from the genetic screen, rare S288C est2Δ rad52Δ and tlc1Δ rad52Δ colonies escaped senescence. Of 50 individual S288C est2Δ rad52Δ or tlc1Δ rad52Δ colonies (derived from heterozygous diploids) propagated on plates every 4 days, 2–5 colonies consistently sustained growth after passage 6 (Figure 2, B–D), while the majority of colonies became senescent at approximately passage 3. Four to 10% of colonies survived regardless of the telomerase gene deleted (i.e., est2Δ rad52Δ or tlc1Δ rad52Δ) and the result was reproducible over many experiments (total n > 250 colonies). The escape from senescence was not due to a reversion of the rad52 locus (supporting information, Figure S1) and was not observed in W303 est2Δ rad52Δ or tlc1Δ rad52Δ strains (total n > 250 colonies). Thus, in contrast to strains lacking telomerase that depend on RAD52 for survival (e.g., type I or type II survivors), S288C telomerase-deficient cells could survive independently of RAD52.
S288C est2Δ rad52Δ or tlc1Δ rad52Δ survivors arise after extensive telomere loss:
Once established, RAD52-independent survivors could be propagated for many generations (Figure 2, B–D). Prior to senescence, telomeres in the est2Δ rad52Δ or tlc1Δ rad52Δ haploids were shorter in length than telomeres in the preceding heterozygous diploid and were also shorter than in wild-type, est2Δ, or rad52Δ haploid colonies (Figure 2E, lanes 1 and 2, and data not shown). Further, telomeres shortened with every passage until the cell population reached a growth crisis (Figure 2B, passage 3), which resulted in a low yield of genomic DNA at these particular passages [Figure 2, E (lane 4) and F (lanes 3 and 10)]. When the population regained growth potential [Figure 2, B (passage 4), E (lanes 5–11), and F (lanes 4–7 and 11–14)], the telomere pattern was similar to that of RAD52-dependent, type II survivors, i.e., multiple discrete fragments from 1 to 6 kbp representing telomeres containing a variable number of TG1–3 repeats (Teng et al. 2000). Little or no amplification of the Y′ element, which is typical of type I survivors, was observed (Figure 2E and data not shown). The telomeric DNA recombination in RAD52-independent survivors was further confirmed by digestion of genomic DNA with a mixture of restriction endonucleases that recognize 4-bp sequences, to which long, nonpalindromic TG1–3 tracts would be resistant (Wen et al. 2006). Indeed, genomic DNA from RAD52-independent survivors yielded a telomeric pattern characteristic of long TG1–3 tracts (Figure 2F, lanes 11–14). We extracted genomic DNA after growth of RAD52-independent survivors in liquid media for <24 hr and also detected a TRF pattern indicative only of telomeric DNA amplification. It has been previously noted that RAD52-dependent type II survivors (telomeric DNA amplification) possess a growth advantage over type I survivors (Y′ amplification) in liquid culture (Teng and Zakian 1999).
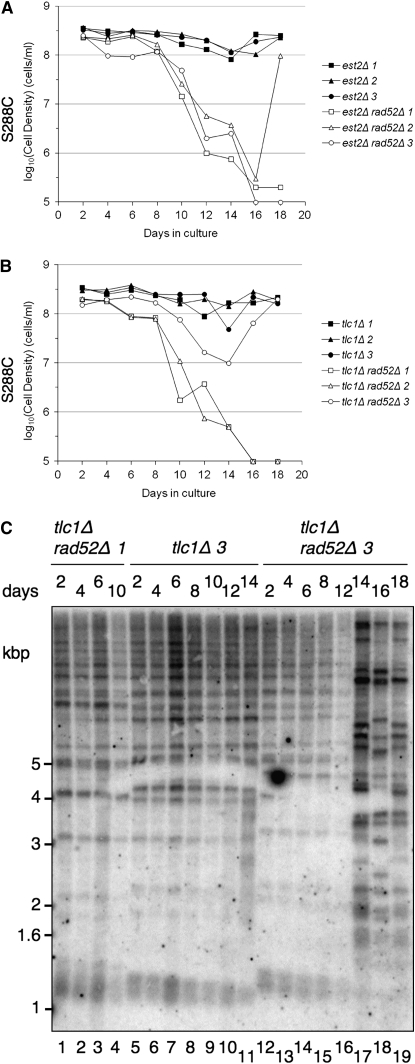
Generation of RAD52-independent survivors in S288C est2Δ rad52Δ or tlc1Δ rad52Δ in serial dilution growth assays:
Mindful of the possible bias of enrichment of type II, RAD52-dependent survivors in liquid culture, we extended the growth period between serial dilutions from 24 (Figure 1, K and L) to 48 hr (Figure 3, A and B). We examined the viability of three individual haploid colonies that originated from the same heterozygous diploid (Figure 3, A and B, and data not shown). Dilution every 48 hr nearly abrogated the transient decrease in population doubling time for est2Δ and tlc1Δ colonies (Figure 3, A and B). In this particular experiment, one of three S288C tlc1Δ rad52Δ colonies regained growth potential at day 14, concomitant with a TRF pattern reminiscent of recombination between telomeric DNA [Figure 3, B (open circles) and C (lanes 17–19)]. In addition, one of the three S288C est2Δ rad52Δ colonies regained growth potential at day 18 and also exhibited a similar TRF pattern (Figure 3A, open triangles and data not shown). In contrast, W303 est2Δ rad52Δ colonies failed to escape senescence when diluted every 48 hr (data not shown). Thus, the propagation of S288C est2Δ rad52Δ or tlc1Δ rad52Δ (but not W303 est2Δ rad52Δ or tlc1Δ rad52Δ) cells every 48 hr in liquid culture allowed the emergence of RAD52-independent survivors with a TRF pattern suggestive of telomeric DNA recombination (e.g., type II).
Figure 3.—
A subset of S288C est2Δ rad52Δ or tlc1Δ rad52Δ cells escapes senescence in liquid culture and exhibits telomeric tract recombination. (A and B) Cells were isolated from a fresh dissection plate (isolates 1–3), inoculated into YPD media, grown to saturation (1–2 × 108 cells/ml), and diluted to 105 cells/ml every 48 hr. At each serial dilution, genomic DNA was extracted for telomere analysis. (C) Telomere Southern blot of S288C haploids with the indicated genotype, as shown in B, during serial propagation in liquid culture every 2 days. Genomic DNA was isolated from cells and digested with XhoI. The membrane was hybridized to a telomeric probe. Marker sizes are indicated at left in kilobase pairs.
The similar TRF pattern between S288C RAD52-independent survivors and type II survivors in other strain backgrounds prompted us to test whether the propagation of S288C est2Δ cells in liquid culture followed by deletion of RAD52 (see materials and methods) would affect the incidence of RAD52-independent survival. We noted that S288C est2Δ rad52Δ colonies survived at an increased frequency if S288C est2Δ cells had been propagated in culture to generate type II survivors prior to deletion of RAD52, compared with removal of both genes simultaneously (p<0.01). This result is in accord with a recent finding by Grandin and Charbonneau (2009) that the generation of type II survivors permits survival upon the subsequent deletion of RAD52 (see Note added in revision).
Telomere length and escape from senescence:
We hypothesized that the slightly longer telomeres present in S288C compared to W303 (Figure 1J) might facilitate the emergence of RAD52-independent survivors. To test this prediction, we elongated telomeres in both S288C and W303 strains by transforming cells with a plasmid encoding a Cdc13-Est2 fusion protein [pVL1107; see materials and methods (Evans and Lundblad 1999)]. Strains were propagated on selective media to ensure plasmid retention, and the TRF pattern of each strain was analyzed over several passages (Figure 4A). To compare strains with similar characteristics, (i.e., transformed with pVL1107 and nearly equivalent average telomere length), we selected S288C passage 2 (Figure 4A, lane 1) and W303 passage 15 cells (Figure 4A, lane 8) for further study. Note that each strain possessed longer telomeres than wild-type W303 or S288C strains. The strains containing elongated (“EL”) telomeres were propagated in rich liquid media for 12 hr and then plated on YPD media to allow loss of the plasmid (“−pVL1107”), which was confirmed by lack of growth on media lacking leucine. Fifty colonies containing elongated telomeres (W303EL est2Δ rad52Δ−pVL1107 and S288CEL est2Δ rad52Δ−pVL1107) were serially propagated on plates every 4 days. Telomere elongation delayed the onset of senescence in both strains (Figure 4, C–J). In S288CEL est2Δ rad52Δ cells, a higher percentage of colonies sustained growth than prior to telomere elongation (Figure 4, C–F, and Figure S2). Notably, in contrast to W303 without elongated telomeres, telomere elongation in W303 est2Δ rad52Δ now allowed some cells to sustain growth for >15 passages (Figure 4, G–J, and Figure S2). In addition, the TRF pattern in both strains showed evidence of TG1–3 signal amplification and resistance to restriction endonuclease digestion (Figure 4, K–M, and Figure S2). The ability of W303 est2Δ rad52Δ cells with elongated telomeres to sustain growth after many passages is consistent with a RAD52-independent telomere maintenance mechanism. These data suggest that telomere length itself may promote survival in S288C and W303.
Single-strand G-rich extensions in S288C RAD52-independent survivors:
The occurrence of TRF patterns reminiscent of telomeric DNA recombination (type II survival) suggested that RAD52-independent survivors might similarly possess tracts of single-stranded, G-rich telomere DNA. Native in-gel analysis of RAD52-independent survivors revealed the presence of ssDNA capable of hybridization to a 32P-labeled CA-rich oligonucleotide that was not observed in the same strains prior to escape from senescence (Figure 5A, compare lanes 1 and 2, 3 and 4, etc.) or when incubated with a 32P-labeled GT-rich oligonucleotide (e.g., to detect C-rich ssDNA; data not shown). As controls, S288C est2Δ type I survivors (exhibiting Y′ amplification) and ku70Δ cells exhibited distinct G-rich ssDNA patterns typical for these strain backgrounds (Figure 5A and data not shown) (Gravel et al. 1998; Polotnianka et al. 1998). However, unlike ku70Δ cells, whose ssDNA signal is sensitive to Escherichia coli Exonuclease I (ExoI) and thus specific to the telomere 3′ terminus (Wellinger et al. 1993), the ssDNA signal observed in late passage tlc1Δ rad52Δ or est2Δ rad52Δ cells was ExoI-resistant (Figure S3). The presence of DNA in all lanes (including those lacking an overhang signal) was confirmed by Southern blotting of the corresponding denatured samples (Figure 5B and Figure S3; note that A and B represent different gels). Thus, S288C RAD52-independent survivors possessed a G-rich, telomeric ssDNA signal that differed in appearance and nature from the ssDNA signal observed in other types of telomerase-independent survivors and ku70Δ cells.
Figure 5.—
RAD52-independent survivors exhibit an increased G-rich, single-stranded telomere signal. (A) Native in-gel analysis of genomic DNA isolated from cells of the indicated genotype and passage (pass.), grown either on plates or in liquid media (liq.). DNA was digested with XhoI and the membrane was hybridized to a radiolabeled C-rich oligonucleotide (5′-CACACCCACACCCACACC-3′). (B) Denaturing Southern blot of the same samples as in A (but not the same gel, as the gel in A was resolved in the presence of radiolabeled probe). DNA was digested with XhoI and the membrane was hybridized to the radiolabeled C-rich oligonucleotide. Marker sizes are indicated at left in kilobase pairs. Bottom panel: blot was stripped and rehybridized to a CDC15 probe as a loading control. Note that the genomic DNA in lane 7 is slightly underrepresented relative to other samples.
The RAD52-independent survivor phenotype shows complex penetrance in diploids:
Makovets et al. (2008) recently demonstrated that RAD52-dependent (type I or II) survival in diploids created by mating two telomerase-deficient haploids exhibited dominance over senescence, and the ability of haploid progeny to survive could be inherited in a non-Mendelian (i.e., multigenic) manner. To examine the viability of various diploids created by mating RAD52-independent survivor haploids, we created three different S288C diploid strains: (1) two S288C est2Δ rad52Δ survivors mated together (survivor × survivor), (2) an est2Δ rad52Δ survivor mated to a presenescent est2Δ rad52Δ colony (freshly isolated from a heterozygous diploid) (survivor × presenescent), and (3) two presenescent est2Δ rad52Δ colonies mated together (presenescent × presenescent). We examined the survival of each diploid strain upon serial propagation of 50 isolated colonies. If the ability to generate RAD52-independent survivors were the result of a single dominant, extragenic suppressor mutation, then all diploids of a “survivor × presenescent” cross would be expected to survive. If the suppressor were recessive, then no survivor × presenescent strains should survive. None of the diploids obtained by mating two presenescent est2Δ rad52Δ colonies (by analyzing 50 independent diploids on plates or by inoculating 30,000 diploid cells in liquid culture) emerged as RAD52-independent survivors (Figure 6B, panels 7 and 8, and Figure S4). However, diploids obtained by mating a RAD52-independent survivor with a presenescent est2Δ rad52Δ colony exhibited an ∼50% incidence of prolonged survival, with some colonies undergoing senescence (Figure 6B, panel 4) and others sustaining growth (Figure 6B, panels 5 and 6, and Figure S4). Finally, all diploids generated by mating two RAD52-independent haploid survivors continued to grow beyond 11 passages (Figure 6B, panels 1–3, and Figure S4). These RAD52-independent survivors also exhibited a TRF pattern indicative of telomeric tract recombination (Figure 6, C and D). Therefore, unlike Makovets et al., who observed dominance of type I or type II survival upon mating a RAD52-independent survivor to a presenescent haploid, the viability of diploids created by mating a RAD52-independent survivor to a presenescent haploid was neither dominant nor recessive. The RAD52-independent survivor phenotype thus suggests complex multigenic mechanisms that may share similarities with other survivor phenotypes (Zubko and Lydall 2006; Makovets et al. 2008).
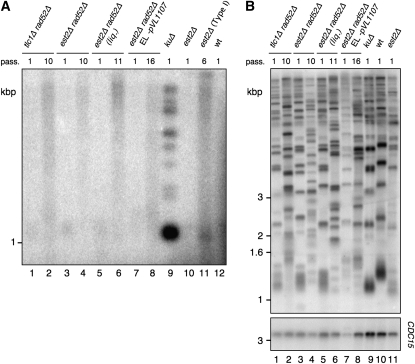
Possible genes or pathways involved in the generation of RAD52-independent survivors:
Genes involved in the generation of RAD52-dependent survivors include RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, SGS1, XRS2, and MRE11 (Le et al. 1999; Teng and Zakian 1999; Chen et al. 2001). To examine the possibility that RAD52-independent survivors might nonetheless depend on the function of one of these genes, we constructed triple-mutant haploid strains (est2Δ rad52Δ xxxΔ). Deletion of any of these genes did not affect the incidence of RAD52-independent survivors in W303 est2Δ rad52Δ or S288C est2Δ rad52Δ backgrounds (n = 50 for each strain) (Table 1). RAD5, which is nonfunctional in W303 (Fan et al. 1996) and regulates strain-specific responses to DNA damage (Demogines et al. 2008), has been implicated in tolerance to DNA damage and replication fork reversal (Blastyak et al. 2007; Klein 2007). Thus, we examined the influence of RAD5 upon the generation and maintenance of RAD52-independent survivors in W303 compared to S288C. Deletion of RAD5 in S288C, either before or after the establishment of RAD52-independent survivors, had no influence on the frequency of viable est2Δ rad52Δ colonies (P > 0.05, data not shown). In addition, the presence of wild-type RAD5 in W303 did not permit the generation of RAD52-independent survivors (L. Maringele, unpublished data).
TABLE 1.
The effect of a third gene deletion on the percentage of colonies that escape senescence in est2Δ rad52Δ strains
| S288C (% survival)
|
W303 (% survival)
|
||||||
|---|---|---|---|---|---|---|---|
| est2Δ rad52Δ | Recombination: est2Δ rad52Δ xxxΔ | Chromatin/replication: est2Δ rad52Δ xxxΔ | est2Δ rad52Δ | Recombination: est2Δ rad52Δ xxxΔ | |||
| 4–10 | 6 | rad50 | 2 | dnl4 | 0 | 0 | rad50 |
| 4 | rad51 | 4 | gcn5 | 0 | rad51 | ||
| 6 | rad54 | 4 | hta1 | 0 | rad54 | ||
| 4 | rad57 | 4 | hta2 | 0 | rad57 | ||
| 4 | rad59 | a | pol32 | 0 | rad59 | ||
| 4 | sgs1 | 4 | sir2 | 0 | sgs1 | ||
| 4 | xrs2 | 0 | xrs2 | ||||
The percentage of RAD52-independent survivors recovered from each genotype is indicated. Diploids of the appropriate genotype were generated by deletion of the locus into diploid cells or mating of appropriate haploids, followed by sporulation, microdissection, and selection of the indicated haploids (est2Δ rad52Δ xxxΔ). The majority of S288C est2Δ rad52Δ and est2Δ rad52Δ xxxΔ colonies ceased proliferation after ∼60 generations.
The S288C est2Δ rad52Δ pol32Δ haploids that could not be propagated from the microdissection plate (20 generations). The same genes were deleted in W303 est2∷NAT/EST2 rad52∷URA3/RAD52 diploids. No W303 est2Δ rad52Δ or est2Δ rad52Δ xxxΔ survivors emerged, and cell populations entered senescence at 40–60 generations.
We also examined candidate genes involved in the regulation of chromatin structure or DNA replication (DNL4, GCN5, HTA1, HTA2, POL32, and SIR2) for a potential role in the generation of RAD52-independent survivors and analyzed all eight possible haploid genotypes from the appropriate triple-heterozygous diploids (Table 1 and Figure 7B). With the exception of POL32, deletion of these genes did not alter the viability or incidence of RAD52-independent survivors in est2Δ rad52Δ xxxΔ strains (n = 50 for each strain).
The polymerase-δ subunit POL32 has been implicated in break-induced replication and telomerase-independent telomere maintenance (Lydeard et al. 2007), and pol32Δ strains exhibit TRFs slightly longer than in wild-type strains (Askree et al. 2004; Gatbonton et al. 2006). POL32 exhibited a synthetic genetic interaction with RAD52 since both rad52Δ pol32Δ and est2Δ rad52Δ pol32Δ colonies remained small after microdissection (data not shown). However, when serially passaged on plates, rad52Δ pol32Δ colonies gained growth potential and became viable to a similar extent as other colonies, whereas est2Δ rad52Δ pol32Δ microcolonies were unable to sustain growth (Figure 7B).
Since this outcome did not allow us to assess a role for POL32 in the emergence of RAD52-independent survivors, we examined whether POL32 is required for the continued viability of RAD52-independent survivors. Diploids created by crossing an established RAD52-independent haploid survivor (tlc1Δ rad52Δ, passage 9) with a pol32Δ single-mutant haploid were sporulated and tested for viability. The resulting tlc1Δ rad52Δ pol32Δ haploids exhibited heterogeneous colony sizes (data not shown). Serial propagation was possible in tlc1Δ rad52Δ pol32Δ cells arising from a colony with an initial size comparable to a tlc1Δ rad52Δ colony, whereas tlc1Δ rad52Δ pol32Δ cells arising from a microcolony failed to sustain growth (Figure 7C and data not shown). TRF analysis of viable tlc1Δ rad52Δ pol32Δ cells revealed a telomeric DNA pattern reminiscent of telomere tract recombination and “type II” survivors (Figure 7D). We were unable to obtain viable est2Δ rad52Δ pol32Δ haploids using the same mating procedure (0/100 colonies; data not shown). Taken together, these data suggest that POL32 is not strictly required for the maintenance of the RAD52-independent survivor phenotype. The synthetic lethal interaction of RAD52, POL32, and EST2 or TLC1 did not allow us to determine whether POL32 may be important for the emergence of RAD52-independent survivors.
DISCUSSION
The generation of survivors in telomerase-negative yeast has been well described in the literature. In all cases, the requisite pathways were RAD52 dependent with the exception of PAL survivors, which can be generated only in cells lacking EXO1 or SGS1 (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Teng et al. 2000; Chen et al. 2001; Maringele and Lydall 2004; Lee et al. 2008). In this study, we describe an ability of S. cerevisiae to escape senescence in the absence of both telomerase and RAD52. We speculate that the phenomenon had not been characterized fully until now because of its rarity and because most studies have been carried out in W303 or other strain backgrounds in which the survivors would normally be RAD52 dependent. RAD52-independent survival in diploids behaved as a multigenic trait, similar to other RAD52-dependent survival phenotypes (Zubko and Lydall 2006; Makovets et al. 2008); however, targeted deletion of genes required for survival in the absence of telomerase and the presence of RAD52 had no effect on RAD52-independent survival. Together with previous studies, which suggest that RAD52-dependent survivors do not absolutely require Cdc13 (Larrivee and Wellinger 2006; Petreaca et al. 2006; Zubko and Lydall 2006), our study bolsters the emerging evidence that alternate strategies exist to cap telomeres and permit cell viability even in the absence of telomerase and RAD52.
RAD52-independent survivors exhibited a telomere DNA pattern indicative of amplification of telomeric repeats (i.e., type II). We did not observe TRF properties consistent with amplification of subtelomeric DNA (Y′) or formation of terminal palindromes (Le et al. 1999; Chen et al. 2001; Maringele and Lydall 2004). It is possible that the TRF pattern in RAD52-independent survivors might comprise subtle mixtures of other TRF types or may represent a novel type of telomeric DNA amplification. One distinction between the TRF patterns of RAD52-independent and RAD52-dependent survivors was the apparent 3′–5′ exonuclease insensitivity of the G-rich ssDNA in the former, indicating that the G-rich signal is not a free 3′ overhang (Figure S3). This ssDNA signal is specific to the G strand and is detected only after the emergence of survivors. Further analysis is required to determine the precise nature of the ssDNA; one possibility could be the presence of telomeric DNA circles (Larrivee and Wellinger 2006) or ssDNA regions within the telomeric DNA that might promote signal-strand annealing and recombination (reviewed in Lyndaker and Alani 2009).
Repetitive noncoding DNA, specialized proteins, and capping structures are important features of telomere integrity and maintenance in all species. Changes in the structure of telomeric DNA (facilitated by increased length) might be important for the generation of RAD52-independent survivors. A longer telomeric “seed” sequence, especially during early generations after telomerase loss, may facilitate survival through the creation of telomeric circles via intratelomeric recombination. The generation of telomeric circles, particularly in cells possessing long telomeres, has been documented in both yeast and humans (Bucholc et al. 2001; Wang et al. 2004; Lin et al. 2005; Muntoni and Reddel 2005; Williams et al. 2005; Pickett et al. 2009). For example, in Kluyveromyces lactis the roll-and-spread mechanism of DNA synthesis in telomerase-deficient and telomerase template-mutated strains does not absolutely require RAD52 (McEachern and Blackburn 1996). It is possible that the G-rich telomeric ssDNA in RAD52-independent survivors (Figure 5, Figure S3) could be excised as a telomeric DNA circle, which could be integrated at a shorter telomere or extend a telomere via a similar rolling-circle mechanism. Elongated telomeres could then be used as a template for intertelomeric BIR to lengthen other telomeres (Natarajan and McEachern 2002; McEachern and Haber 2006). Similar events might explain the abrupt emergence of RAD52-dependent survivors harboring very long telomeric tracts in S. cerevisiae (Teng et al. 2000). Relatively few long terminal telomeric extensions might be sufficient to initiate this sequence of events.
If RAD52-independent survival were reliant on a roll-and-spread mechanism, telomere maintenance would presumably require the DNA replication machinery. Payen et al. (2008) suggest the involvement of Pol32, a nonessential subunit of DNA polymerase-δ, in segmental duplication promoting genomic instability through a RAD52-independent mechanism of template switching between microsatellites or microhomologous sequences. This new mechanism, named microhomology/microsatellite-induced replication (MMIR), differs from the known DNA double-strand repair pathways and occurs in the absence of homologous recombination and nonhomologous end-joining machineries (Payen et al. 2008). Although the synthetic lethality of the est2Δ rad52Δ pol32Δ and tlc1Δ rad52Δ pol32Δ gene deletions in S288C is notable, it did not allow us to test the requirement of POL32 for the generation of RAD52-independent survivors. Our results nonetheless indicate that the Pol32 protein is not required for viability once a RAD52-independent survivor has been established. We have not ruled out that a subset of the population may require Pol32 when telomeres become critically short. Indeed, it was suggested that in the absence of mismatches between repeated sequences, not all segmental duplications require Pol32 (Payen et al. 2008). These Pol32-independent segmental duplications likely result from unequal crossing over between repeated sequences, as would be possible at yeast telomeres. Lundblad and colleagues also found that deletion of genes important for mismatch repair (MSH2, MLH1, PMS1) promotes the generation of survivors in the absence of telomerase (Rizki and Lundblad 2001).
The incidence of survival in diploids created by mating RAD52-independent survivors suggests a multigenic and potentially epigenetic pattern of inheritance. For example, the nearly 50% incidence of escape from senescence in diploids created by mating a RAD52-independent survivor with a presenescent est2Δ rad52Δ population argues against a simple recessive or dominant extragenic suppressor mutation. Interestingly, the incidence of escape from senescence in diploids created by mating two est2Δ rad52Δ haploid strains is less frequent than in an est2Δ rad52Δ haploid (Figure 6 and data not shown). Diploidy also reduces the incidence of RAD52-dependent (type II) survivors (Liti and Louis 2003).
When RAD52-independent survivors were propagated continuously on plates, not all colonies survived indefinitely. These observations suggest that the mechanism leading to telomere maintenance and escape from senescence is the exception and not the rule. Over time, the population is able to maintain telomeres and bypass senescence even without telomerase and RAD52. Like many cellular processes, there is an overriding selection for cell survival by whatever means possible. The fact that a means to survive exists in the absence of RAD52 and telomerase suggests that multiple, redundant pathways have evolved to ensure telomere homeostasis even under the most extreme conditions. Uncovering the genetic pathways that allow telomerase- and homologous recombination-independent mechanisms of telomere maintenance should further our understanding of how some human tumors are able to bypass the reacquisition of telomerase activity during tumorigenesis (Muntoni and Reddel 2005; Muntoni et al. 2009).
Acknowledgments
We are grateful to the laboratories of M. Tyers, D. Durocher, V. Lundblad, and R.J. Wellinger for strains, plasmids, and reagents; to B.-J. Breitkreitz, L. Boucher, M. Cook, D. Edmonds, C.-Y. Ho, M. Spitzer, and M. Tyers for advice in comparative genomic hybridization analysis and genomewide screens; and to the Harrington lab, the Lingner lab, D. Blake, H. Pickersgill, E. Louis, T. Weinert, and R. J. Wellinger for discussion and comments. The reviewers are thanked for constructive suggestions. D.L. acknowledges the Wellcome Trust (WT 075294), L.M. acknowledges a Wellcome Trust Career Development Award (WT 081164), and L.A.H. acknowledges the Campbell Family Institute for Cancer Research and grants from the National Institute on Aging (National Institutes of Health R01 AG02398), the Howard Hughes Medical Institute International Scholar Award Program (HHMI 55005945), the Medical Research Council United Kingdom (G0800081-85694), and the Wellcome Trust (WT 084637).
Note added in revision: During the revision of this manuscript, Grandin and Charbonneau (N. Grandin and M. Charbonneau, 2009, Telomerase- and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol. Cell. Biol. 29: 965–985) showed that telomerase-deficient cells with long telomeres (e.g., type II survivors) could survive the subsequent deletion of RAD52. They employed a different strain background (BF264a-15D), which could not survive the simultaneous deletion of telomerase and RAD52. These cell populations, referred to as interlengthening of telomeres (ILT), also exhibited amplification of telomeric repeats, similar to type II survivors. Dissimilar to our findings, ILT survivors relied on MRE11 and RAD50 for survival, and lengthening of telomeres by a different means (rif2Δ or introduction of a Cdc13-Est1 fusion protein) failed to promote survival. Thus, multiple mechanisms exist for RAD52-independent survival in the absence of telomerase, whose gene dependence may reflect the context in which longer telomeres are introduced.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102939/DC1.
References
- Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk et al., 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak, A., L. Pinter, I. Unk, L. Prakash, S. Prakash et al., 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Bucholc, M., Y. Park and A. J. Lustig, 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakhparonian, M., and R. J. Wellinger, 2003. Telomere maintenance and DNA replication: How closely are these two connected? Trends Genet. 19 439–446. [DOI] [PubMed] [Google Scholar]
- Chan, C. S., and B. K. Tye, 1983. a A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J. Mol. Biol. 168 505–523. [DOI] [PubMed] [Google Scholar]
- Chan, C. S., and B. K. Tye, 1983. b Organization of DNA sequences and replication origins at yeast telomeres. Cell 33 563–573. [DOI] [PubMed] [Google Scholar]
- Chang, M., M. Arneric and J. Lingner, 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., A. Ijpma and C. W. Greider, 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, M. A., C. K. Chan, P. Jorgensen, T. Ketela, D. So et al., 2008. Systematic validation and atomic force microscopy of non-covalent short oligonucleotide barcode microarrays. PLoS ONE 3 e1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., 2008. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8 512–522. [DOI] [PubMed] [Google Scholar]
- Demogines, A., E. Smith, L. Kruglyak and E. Alani, 2008. Identification and dissection of a complex DNA repair sensitivity phenotype in Baker's yeast. PLoS Genet. 4 e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S. K., and V. Lundblad, 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286 117–120. [DOI] [PubMed] [Google Scholar]
- Fan, H. Y., K. K. Cheng and H. L. Klein, 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics 142 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S. S., M. K. Zubko, S. Guillard and D. Lydall, 2006. MRX protects telomeric DNA at uncapped telomeres of budding yeast cdc13–1 mutants. DNA Repair 5 840–851. [DOI] [PubMed] [Google Scholar]
- Gatbonton, T., M. Imbesi, M. Nelson, J. M. Akey, D. M. Ruderfer et al., 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2 e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2006. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 313 107–120. [DOI] [PubMed] [Google Scholar]
- Grandin, N., and M. Charbonneau, 2009. Telomerase- and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol. Cell. Biol. 29 965–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, S., M. Larrivee, P. Labrecque and R. J. Wellinger, 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 741–744. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink (Editors), 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts et al., 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11 125–129. [DOI] [PubMed] [Google Scholar]
- Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn et al., 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, H. L., 2007. Reversal of fortune: Rad5 to the rescue. Mol. Cell 28 181–183. [DOI] [PubMed] [Google Scholar]
- Larrivee, M., and R. J. Wellinger, 2006. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 8 741–747. [DOI] [PubMed] [Google Scholar]
- Larrivee, M., C. LeBel and R. J. Wellinger, 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S., J. K. Moore, J. E. Haber and C. W. Greider, 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBel, C., M. Larrivee, A. Bah, N. Laterreur, N. Lvesque et al., 2006. Assessing telomeric phenotypes. Methods Mol. Biol. 313 265–316. [DOI] [PubMed] [Google Scholar]
- Lee, J. Y., J. L. Mogen, A. Chavez and F. B. Johnson, 2008. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J. Biol. Chem. 283 29847–29858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian and V. Lundblad, 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. Y., H. H. Chang, K. J. Wu, S. F. Tseng, C. C. Lin et al., 2005. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell 4 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti, G., and E. J. Louis, 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11 1373–1378. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and E. H. Blackburn, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73 347–360. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 633–643. [DOI] [PubMed] [Google Scholar]
- Lydeard, J. R., S. Jain, M. Yamaguchi and J. E. Haber, 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448 820–823. [DOI] [PubMed] [Google Scholar]
- Lyndaker, A. M., and E. Alani, 2009. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. BioEssays 31 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets, S., T. L. Williams and E. H. Blackburn, 2008. The telotype defines the telomere state in Saccharomyces cerevisiae and is inherited as a dominant non-Mendelian characteristic in cells lacking telomerase. Genetics 178 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele, L., and D. Lydall, 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18 2663–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10 1822–1834. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., and J. E. Haber, 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75 111–135. [DOI] [PubMed] [Google Scholar]
- Muntoni, A., and R. R. Reddel, 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14(Spec. No. 2): R191–R196. [DOI] [PubMed] [Google Scholar]
- Muntoni, A., A. A. Neumann, M. Hills and R. R. Reddel, 2009. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum. Mol. Genet. 18 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan, S., and M. J. McEachern, 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov, A. M., 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41 181–190. [DOI] [PubMed] [Google Scholar]
- Payen, C., R. Koszul, B. Dujon and G. Fischer, 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4 e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreaca, R. C., H. C. Chiu, H. A. Eckelhoefer, C. Chuang, L. Xu et al., 2006. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 8 748–755. [DOI] [PubMed] [Google Scholar]
- Pickett, H. A., A. J. Cesare, R. L. Johnston, A. A. Neumann, and R. R. Reddel, 2009. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 8 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka, R. M., J. Li and A. J. Lustig, 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8 831–834. [DOI] [PubMed] [Google Scholar]
- Rizki, A., and V. Lundblad, 2001. Defects in mismatch repair promote telomerase-independent proliferation. Nature 411 713–716. [DOI] [PubMed] [Google Scholar]
- Teng, S. C., and V. A. Zakian, 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., J. Chang, B. McCowan and V. A. Zakian, 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6 947–952. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., A. K. Taggart and V. A. Zakian, 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11 1328–1335. [DOI] [PubMed] [Google Scholar]
- Walmsley, R. W., C. S. Chan, B. K. Tye and T. D. Petes, 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310 157–160. [DOI] [PubMed] [Google Scholar]
- Wang, R. C., A. Smogorzewska and T. de Lange, 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119 355–368. [DOI] [PubMed] [Google Scholar]
- Watson, J. D., 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239 197–201. [DOI] [PubMed] [Google Scholar]
- Wellinger, R. J., A. J. Wolf and V. A. Zakian, 1993. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72 51–60. [DOI] [PubMed] [Google Scholar]
- Wen, W. Y., H. J. Tsai, C. C. Lin, S. F. Tseng, C. W. Wong et al., 2006. Telomere configuration influences the choice of telomere maintenance pathways. Biochem. Biophys. Res. Commun. 343 459–466. [DOI] [PubMed] [Google Scholar]
- Williams, B., M. K. Bhattacharyya and A. J. Lustig, 2005. Mre 11 p nuclease activity is dispensable for telomeric rapid deletion. DNA Repair 4 994–1005. [DOI] [PubMed] [Google Scholar]
- Zubko, M. K., and D. Lydall, 2006. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 8 734–740. [DOI] [PubMed] [Google Scholar]