Abstract
Telomeric regions are known to be transcribed in several organisms. Although originally reported to be transcribed from all chromosomes with enrichment near the inactive X of female cells, we show that telomeric RNAs in fact are enriched on both sex chromosomes of the mouse in a developmentally specific manner. In female stem cells, both active Xs are marked by the RNAs. In male stem cells, both the X and the Y accumulate telomeric RNA. Distribution of telomeric RNAs changes during cell differentiation, after which they associate only with the heterochromatic sex chromosomes of each sex. FISH mapping suggests that accumulated telomeric RNAs localize at the distal telomeric end. Interestingly, telomeric expression changes in cancer and during cellular stress. Furthermore, RNA accumulation increases in Dicer-deficient stem cells, suggesting direct or indirect links to RNAi. We propose that telomeric RNAs are tied to cell differentiation and may be used to mark pluripotency and disease.
TRANSCRIPTOME analyses of recent years have revealed that 60–80% of the mammalian genome is transcribed (Birney et al. 2007; Gingeras 2007; Pheasant and Mattick 2007). Distinctly different from small RNAs of the RNA-interference pathway (Zamore and Haley 2005), “macroRNAs” have largely remained mysterious in function. Their potential in epigenomic regulation can be seen, for example, in fission yeast and plants, where silencing of centric heterochromatin depends on expressed noncoding RNA (ncRNA) within retrotransposable elements (Grewal and Elgin 2007). In the fruitfly, dosage compensation of the X chromosome requires the action of roX1 and roX2, two macroRNAs associated with the male-specific lethal (MSL) complex that directs hypertranscription of the male X chromosome (Kelley and Kuroda 2000). In mammals, macroRNA function is exemplified by Xist (Borsani et al. 1991; Brockdorff et al. 1991; Brown et al. 1991) and its antisense partner, Tsix (Lee and Lu 1999)—two genes that control the initiation of “X chromosome inactivation” (XCI). XCI is induced by Xist RNA, a 17-kb ncRNA that accumulates in cis and recruits silencing factors to the X destined to be inactivated (Wutz 2003). Tsix opposes Xist through cotranscriptional recruitment of repressive chromatin to Xist (Navarro et al. 2005; Sado et al. 2005; Sun et al. 2006).
Although the biological function remains unknown, noncoding RNAs have also been reported at the telomeric ends of several organisms, including birds (Solovei et al. 1994), trypanosomes (Rudenko and Van Der Ploeg 1989), and mammals (Azzalin et al. 2007; Schoeftner and Blasco 2008). Interestingly, one group reported that telomeric RNAs are enriched near the inactive X in mammals (Schoeftner and Blasco 2008). Here, by analyzing the heterogeneous patterns of Cot-1 RNA in mouse cells, we have independently found transcription originating from mammalian telomeres. In addition to what was published recently by others (Schoeftner and Blasco 2008), we show that telomeric RNAs associate not only with the inactive X in somatic cells, but generally with both sex chromosomes in stem cells. In female stem cells, both active Xs are in fact marked by the RNAs. In male stem cells, both the X and the Y also accumulate telomeric RNA. The pattern of expression changes dynamically during normal cell differentiation and in diseased cells. In aggregate, our data suggest that chromosome-specific patterns of telomeric RNA expression mark pluripotency and disease.
MATERIALS AND METHODS
Cell lines:
The following ES cell lines have been described elsewhere: Wild-type female ES cell line, 16.7, and male ES line, J1 (Lee and Lu 1999); transgenic cell lines, π2.5.5, π1.4.1, and 116.6 (Lee and Jaenisch 1997); conditional knockout ES line, XaXiΔXist (Zhang et al. 2007); and Dcr+/− and Dcr−/− ES cells (Ogawa et al. 2008). MEFs were isolated from d13.5 embryos. Transformed fibroblast lines were immortalized by SV-40 T-antigen. The colon cancer line (HCC1937), ovarian cancer line (TOV12G), cervical cancer line (HeLa), and primary human fibroblast line (WI-38) have all been purchased from ATCC. HESC lines were maintained as described (Silva et al. 2008).
DNA-, RNA-, and immuno-FISH:
For cytologic analysis, cells were either grown directly on slides (MEFs) or cytospun onto glass slides (ES cells) using Shandon Cytospin3 cytocentrifuge. Cells were then permeabilized with CSK buffer containing 0.5% Triton X-100 at 4° (30 sec for undifferentiated ES cells, 3 min for MEF), fixed in 4% paraformaldehyde at room temperature for 10 min, and stored in 70% ethanol at 4°. Before use, slides were dehydrated with ethanol series and dried in room temperature for 3 min before probes were added onto the slides. Cot-1 DNA (Invitrogen) was labeled with Cy3-12-dUTP (Amersham Biosciences) by Prime-It Fluor fluorescence labeling kit (Stratagene). One microliter of Cot-1 DNA (1 μg/μl) was mixed with 10 μl random 9-mer primers and 27 μl water. Solution was heated at 95° for 5 min to denature DNA, incubated on ice 5 min for probe annealing. Added into the reaction then were 9.2 μl 5× nucleotide buffer, 0.8 μl Cy3-12-dUTP, and 2 μl Klenow. The reaction was carried out at 37° for 30 min. Labeled DNA was ethanol precipitated and resuspended in 50 μl of hybridization buffer (probe concentration is labeled as 20 ng/μl). DNA Oligo probes for telomeric RNA FISH were ordered from Integrated DNA Technologies: Telo1, (TAACCC)7-Alexa488-3′; Telo2, (TTAGGG)6-Alexa488-3′. Oligo probes were dissolved at 1 pmol/μl in hybridization buffer for RNA FISH. For combinational RNA FISH of Cot-1 and telomeric RNA, Cot-1 probe and telomeric DNA oligo probe were mixed at the final concentration of 20 ng/μl and 0.5 pmol/μl, respectively. LINE L1, SINE B1, and SINE B2 probes (S. H. Namekawa, unpublished results) were Cy3 labeled and each used at a final concentration of 4 ng/μl. Antibodies for immuno-FISH were as follows: 1:50 monoclonal mouse anti-RNA Pol-II (H5) (Covance) and 1:200 monoclonal mouse anti-RNA pol-II (CTD, clone8WG16) (Upstate), used in combination with secondary Cy3-labeled goat anti-mouse IgG antibodies. FISH hybridization was carried out at 42° in a dark and humid oven. After 3 hr (RNA FISH) or overnight (DNA FISH) hybridization, slides were washed at 45° in a shaker waterbath. Fluorescent images were collected on a Zeiss Axioplan2 microscope (Carl Zeiss) using Openlab software (Improvision). Pictures of RNA and DNA FISH were collected in z-series (0.5 μm z-interval, 3-4 μm in total). Pictures from single focal plain are stacked and merged pictures carrying all the detectable signals within nuclei are shown. Additional details for RNA-, DNA-, and immuno-FISH have been described elsewhere (Zhang et al. 2007).
Northern analyses:
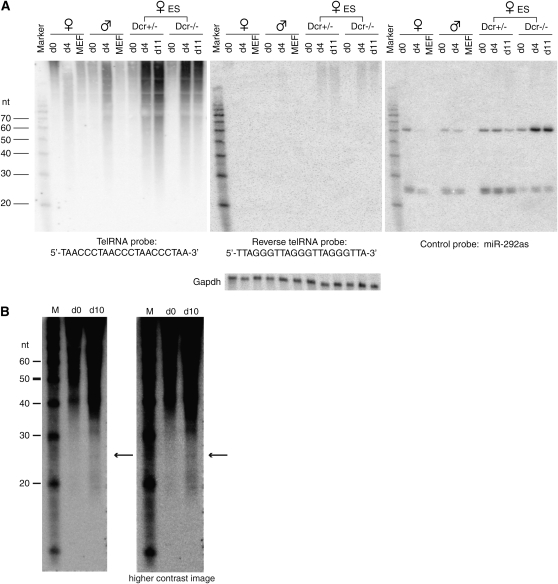
Northern blot protocol for small RNA detection has been described (Lau et al. 2001) with the following modifications. Probes used for Northern blots of Figure 8A were as follows: Telo1-21nt, TAACCCTAACCCTAACCCTAA; Telo2-21nt, TTAGGGTTAGGGTTAGGGTTA; GAPDHprobe, GTAGACTCCACGACATACTCAGCACCGGCCTCACCCCATT; and miR-292-as, ACACTCAAAACCTGGCGGCACTT. For the small RNA Northerns of Figure 8B, 20 μg of small RNAs extracted by mirVana miRNA isolation kit (Ambion) were used instead of total RNA and a 30mer oligo was used as probe to increase detection sensitivity (Telo1-30nt, TAACCCTAACCCTAACCCTAACCCTAACCC). All oligo probes were end labeled using T4 polynucleotide kinase. Hybridization was carried out at either 50° (Figure 8A) or 42° (Figure 8B) with ULTRAhyb-Oligo hybridization buffer (Ambion).
Figure 8.—
Northern analysis reveals a heterogeneous telomeric RNA population and possible small RNA species. (A) Northern analysis of telomeric RNA in d0, d4, and d11 ES cells and in primary mouse embryonic fibroblasts (MEF). Remaining levels of miR-292 in Dcr−/− cells are due to ≪5% residual Dcr activity in this line (Ogawa et al. 2008). (B) Northern analysis using increased starting material and blots that are trimmed to eliminate the highest molecular weight RNA species to visualize smaller species more effectively. The two images shown are different contrasts of the same blot. Arrows, 25-nt species.
RESULTS
X- and Y-linked Cot-1 appendages:
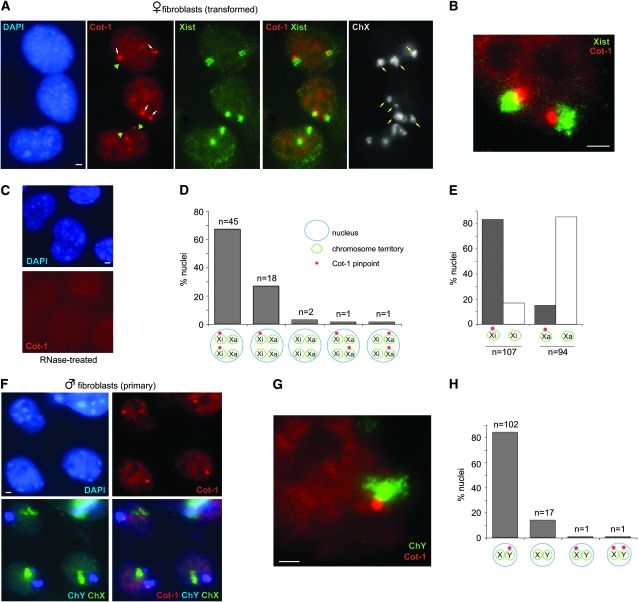
Transcriptional activity can be assessed by RNA fluorescence in situ hybridization (FISH) using Cot-1 probes on undenatured nuclei (Hall et al. 2002; Huynh and Lee 2003). “Cot-1” refers to the DNA fraction that reanneals first after genomic DNA is denatured. Because the Cot-1 fraction contains intergenic repetitive elements and those that are present in introns of nascent mRNA, Cot-1 probes broadly identify nuclear regions with ongoing transcription. We incidentally noted that, while the inactive X (Xi) generally excluded Cot-1 hybridization, the Xi in mouse fibroblasts showed frequent association with an intense Cot-1 body (Figure 1A). The Cot-1 bodies were smaller than Xist RNA clouds and were attached to the RNA cloud, which covers the majority of the Xi chromosome territory (Figure 1B). RNaseA treatment abolished the signals, indicating that the Cot-1 structure is RNA in nature (Figure 1C). The Cot-1 bodies could be seen in almost all cells, with 83% (n = 135) attached specifically to Xi. In female cells, 82% of all Xi had one (very occasionally two) Cot-1 appendages (Figure 1, D and E). Only 15% of all Xa showed association with the Cot-1 body. The association with Xi was observed in both transformed (shown) and primary (not shown) female fibroblasts (mouse embryonic fibroblasts, MEFs). Thus, while the Xi generally lacks Cot-1 expression due to a dearth of nascent transcription, a structure of intense Cot-1 activity lies immediately adjacent to Xi. Male fibroblasts also expressed Cot-1 foci (Figure 1F). Interestingly, 83% of Cot-1 structures (n = 124) were attached to the Y. Conversely, 85% of all Ys (n = 121) displayed a Cot-1 appendage next to the chromosome (Figure 1, G and H). Thus, the Cot-1 body is neither specific to females nor to the X.
Figure 1.—
A Cot-1 RNA attachment to sex heterochromatin in the mouse. (A) Sequential RNA/DNA FISH in transformed female fibroblasts. Note: SV40-transformed fibroblasts are often tetraploid. RNA FISH to detect Cot-1 pinpoints (Cy3, white arrow) and Xist RNA (FITC) was followed by slide denaturation and DNA FISH using an X chromosome paint (FITC) to detect all Xs (yellow arrows). Examples of Cot-1 holes indicated by green arrowheads. Images are merged z-stacks. Scale bar, 2 μm. (B) Magnification of a nucleus to demonstrate the spatial relationship between the Xi and the Cot-1 body. (C) RNAseA treatment prior to FISH abolishes the Cot-1 signals. (D) Patterns of Cot-1 expression in transformed female fibroblasts. n = 67. (E) Frequency of Cot-1 pinpoint attachment to Xi vs. Xa. (F) Sequential RNA/DNA FISH in primary male fibroblasts. RNA FISH to detect Cot-1 pinpoints (Cy3) was followed by DNA FISH using an X (FITC) and Y (Cy3, pseudocolored blue) paint. (G) Magnification of a nucleus to demonstrate spatial relationship between the Cot-1 pinpoint and the Y. (H) Patterns of Cot-1 pinpoint expression in primary male fibroblasts. n = 121.
Relationship to XCI:
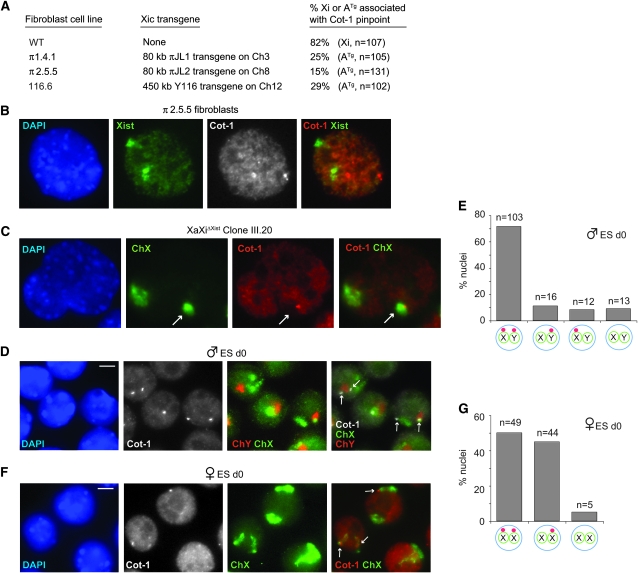
Association with the two heterochromatic sex chromosomes (Xi and Y) was curious. To determine if there is a connection to XCI, we asked whether the Cot-1 appendage occurred in XCI-mutant cell lines by testing three independent male fibroblast lines carrying Xist sequences on an autosomal transgene. Previous analysis showed that Xist RNA induces ectopic silencing of the host autosome in all three cell lines (Lee et al. 1999). Here, we saw that only 15–30% of Cot-1 bodies were attached to Xist-coated autosomes (Figure 2, A and B), implying that autosomal Xist and inactivation do not necessarily result in Cot-1 RNA recruitment. We next examined female fibroblast clones in which Xist has been conditionally deleted from the Xi [XaXiΔXist (Zhang et al. 2007)]. Previous analysis showed that the Xist deletion resulted in loss of Xi heterochromatin and partial reactivation of the Xi. Despite those changes, however, the XiΔXist chromosome retained an association with the Cot-1 pinpoint at a similar frequency (Figure 2C; Xi distinguished from Xa by its associated Cot-1 hole). Collectively, these experiments indicated that expression of the Cot-1 body is not immediately impacted by changes in Xist expression and chromosome inactivation.
Figure 2.—
Relationship between Cot-1 bodies and X inactivation. (A) Summary of Cot-1 localization in male fibroblast cell lines carrying Xic transgenes on an autosome (ATg). Right column indicates the percentage of the Xist-expressing chromosome (either X or autosomal) that is associated with a Cot-1 RNA focus. (B) RNA FISH of a representative nucleus (π2.5.5 clone) combining a Cot-1 (Cy3) and Xist (FITC) probes. Cot-1 foci and autosomal Xist RNA do not colocalize. Note: The cell lines are tetraploid due to SV-40 transformation. (C) RNA FISH detecting Xist (FITC) and Cot-1 RNA (Cy3), followed by slide denaturation and X painting (FITC) in an XaXiΔXist clone. Arrows point to XiΔXist lying within a Cot-1 hole. (D) RNA FISH detecting Cot-1 RNA (Cy3, grayscale), followed by slide denaturation and DNA FISH with X (FITC) and Y (Cy3) painting probes. Representative cells from d0 male ES cells. Arrows, Cot-1 foci. Scale bar, 5 μm. (E) Patterns of Cot-1 pinpoint in d0 XY ES cells. n = 121. (F) RNA FISH detecting Cot-1 RNA (Cy3), followed by slide denaturation and DNA FISH with X painting probes (FITC). Representative cells from d0 female ES cells. Arrows, Cot-1 foci. (G) Pattern of Cot-1 RNA expression in d0 XX ES cells. n = 98.
We then examined mouse embryonic stem (ES) cells to determine if expression of the Cot-1 body changes during differentiation. Female ES cells carry two Xa, are uncommitted to XCI, but recapitulate XCI upon cell differentiation ex vivo. Intriguingly, undifferentiated XX and XY ES cells displayed not one but two Cot-1 structures. XY cells showed predominantly two Cot-1 pinpoints per nucleus, one attached to the X and the other to the Y (Figure 2, D and E). XX cells showed two subpopulations, with ∼50% of nuclei exhibiting two pinpoints per nucleus and ∼45% exhibiting only one (Figure 2, F and G). In both female subpopulations, the Cot-1 structures were nearly always attached to the Xs. Thus, although Xist expression does not affect the Cot-1 structure, the structure shows specific sex chromosome dynamics.
Asymmetric telomeric origin:
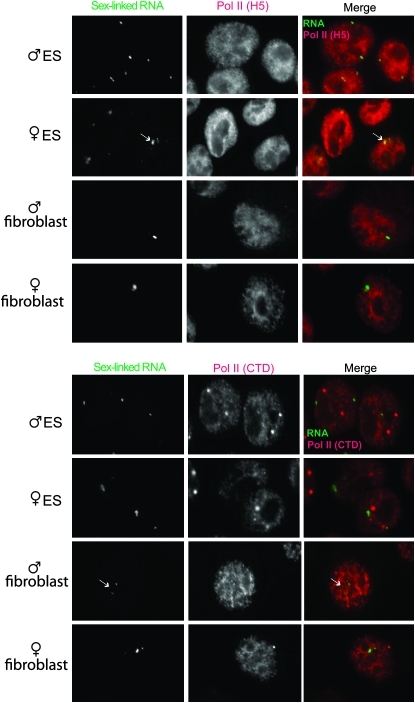
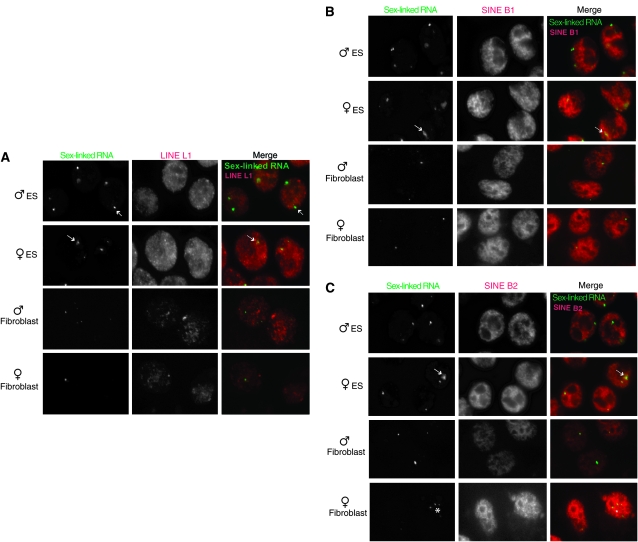
We next investigated the nature of the Cot-1 foci. To determine if the sex-linked RNA foci merely represented regions of Pol-II enrichment, we immunostained ES and primary MEFs with antibodies against Pol-II (C-terminal domain, CTD) and its elongating isoform (H5) (Figure 3). None of the RNA foci showed significantly elevated Pol-II concentration, suggesting that the Cot-1 structures represented true RNA accumulation rather than increased transcription. Because repetitive elements comprise Cot-1, we tested consensus probes for the family of LINEs (Figure 4A), SINEs B1 and B2 (Figure 4, B and C), and centromeric repeats (data not shown), but found none that specifically colocalized with sex-linked RNA foci.
Figure 3.—
Pol-II activity is not highly enriched at Cot-1 foci. RNA immuno-FISH of indicated cell lines to examine the sex-linked RNA (Cot-1, telomeric fraction, see Figure 5) and its relationship to Pol-II. Pol II (H5), elongating isoform; Pol II (CTD), C-terminal domain of Pol-II. Arrows indicate diffuse RNA clusters; asterisks denote speckled pattern. Day 0 ES cells are shown.
Figure 4.—
The sex-linked RNA foci are not of LINE and SINE origin. Two-color RNA FISH combines detection of the sex-linked RNA (Cot-1, telomeric fraction, see Figure 5) and repetitive elements of LINE L1 (A), SINE B1 (B), or SINE B2 (C) origin. Day 0 ES cells are shown. No overlap is seen.
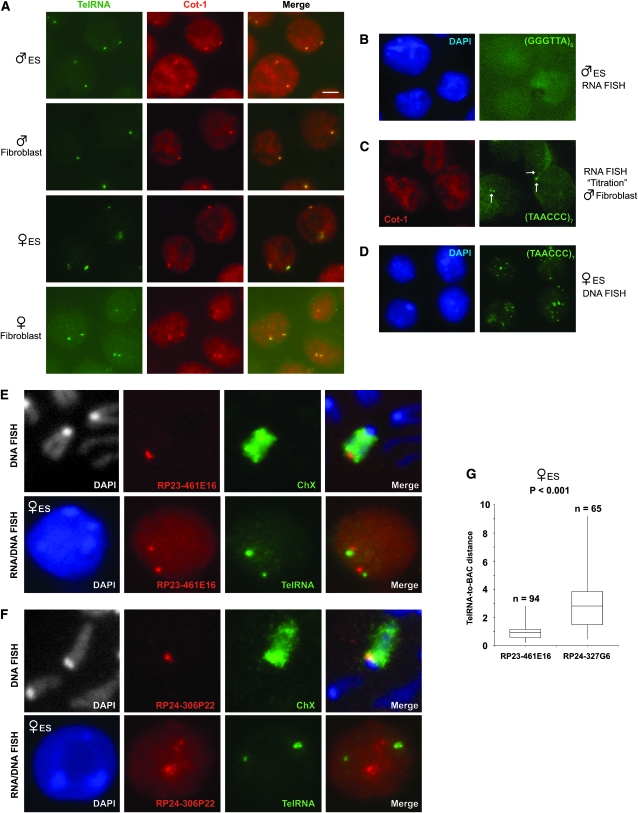
The Cot-1 fraction also contains telomeric repeats, a tandem array of TTAGGG ending in a single-stranded 3′ G-rich strand (Makarov et al. 1997; McEachern et al. 2000; De Lange 2004). Intriguingly, with a (TAACCC)7 probe, we observed pinpoint RNA FISH signals that colocalized perfectly with Cot-1 foci (Figure 5A). While the heterogeneous Cot-1 probe diffusely stained the nucleus as well as the X and Y appendages, the telomeric probe gave highly specific pinpoint signals with little to no staining of other regions. The complementary probe, (GGGTTA)6, gave no signal in any cell type (Figure 5B). Significantly, mixing the (TAACCC)7 probe at high concentration (12.5 ng/μl or 10 pmol/μl) outcompeted the heterogeneous Cot-1 probe (20 ng/μl) and specifically abolished the Cot-1 foci without affecting staining of other nuclear regions (Figure 5C). Thus, the Cot-1 fraction responsible for the sex-linked pinpoints is telomeric in origin.
Figure 5.—
The Cot-1 RNA is of telomeric origin. (A) Dual color RNA FISH on undenatured nuclei using a Cot-1 probe (Cy3) in combination with the telomeric RNA probe, (TAACCC)7-Alexa-488 (green). Scale bar, 5 μm. ES cells, d0. TelRNA, telomeric RNA. (B) RNA FISH with the complementary probe, (GGGTTA)6-Alexa-488. (C) The telomeric probe titrates away the Cot-1 pinpoint signal. Arrows, telomeric RNA foci. (D) DNA FISH on denatured nuclei with the (TAACCC)7-Alexa-488 probe shows the distribution of all telomeres. (E) Top: DNA FISH with distal telomeric probe (BAC RP23-461E16) and X paint on metaphase chromosome spread. Bottom: RNA FISH with telomeric RNA probe and BAC probe. The results show that the RNA foci are at the distal end of the Xs in d0 female ES cells. (F) Top: DNA FISH with proximal telomeric probe (BAC RP24-306P22) and X paint. Bottom: RNA FISH with telomeric RNA probe and BAC probe. The results show that the RNA foci are not at the proximal end of the Xs in d0 female ES cells. (G) Range of distances from telomeric RNA to indicated BAC markers. P < 0.001 calculated using the Student's t-test. Distances indicated in micrometers.
These data suggest that telomeres are transcribed in the mouse and transcription occurs unidirectionally in the outward orientation. In principle, telomeric transcription could be restricted to the sex chromosomes, or it could occur on multiple chromosomes but cluster in only one or two regions. To investigate, we denatured cells and performed DNA FISH using the (TAACCC)7 probe and found a multifocal staining pattern (Figure 5D). This observation ruled out telomeric clustering and implied that telomeric RNAs localize only at the sex chromosomes. To determine which telomere of the X is associated with telomeric RNA, we performed DNA FISH using two subtelomeric BAC probes: RP23-461E16, which identified the distal telomere (Figure 5E), and RP24-306P22, which identified the proximal telomere (Figure 5F). Curiously, only the distal end associated with telomeric RNA (Figure 5, E–G). We believe the distal-specific association of telomeric RNA also occurs in the male cells on Y chromosome but could not test this, as BACs are not available for this region. On the basis of these results, several possibilities could be entertained. First, telomeric RNA may be produced only from the distal telomere. Alternatively, telomeric RNA may be produced from both telomeres but be asymmetrically stabilized on the distal end of the X. Finally, telomeric RNA could be synthesized from all chromosomes but cluster at the distal end of X. We cannot distinguish among these possibilities due to a lack of chromosome-specific polymorphisms for the repeat.
Transitional states during cell differentiation:
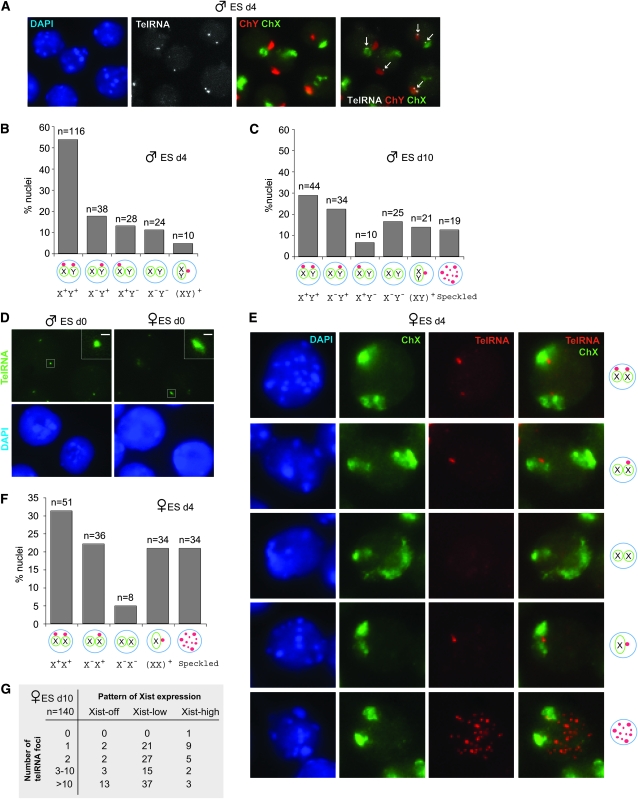
Expression of telomeric RNA changes dynamically during ES cell differentiation. While >70% of undifferentiated male ES cells [day 0 (d0)] displayed two telomeric RNA foci (Figure 2, D and E), cell differentiation (d4, d10) led to a progressive decline in nuclei with two foci (X+Y+) and increase in those with one (Figure 6, A–C). The single pinpoint was almost always attached to the X (X+Y−) or the Y (X−Y+). On d4, other patterns included those with no foci (X−Y−) and those with a single pinpoint located between adjacent X and Y chromosomes (XY)+ (Figure 6B). By d10, the cumulative number of nuclei with a single pinpoint [X−Y+, X+Y−, (XY)+] exceeded those with two (X+Y+) and the X−Y+ pattern became dominant. At this time point, multifocal speckled staining became visible in ∼12% of cells. Given that >80% of XY cells showed the X−Y+ pattern in the fully differentiated fibroblast cells (Figure 1), we believe that the patterns seen on d4 and d10 represent “transitional states,” the X+Y+ pattern identifies the original undifferentiated state, and the X−Y+ pattern signifies the differentiated state.
Figure 6.—
Transitional states of telomeric RNA in differentiating ES cells. (A) RNA FISH detecting telomeric RNA (Alexa-488, shown in grayscale) followed by DNA FISH using X (FITC) and Y (Cy3) chromosome paints in d4 male ES cells. Arrows, telomeric RNA. (B) Patterns of telomeric RNA expression in d4 male ES cells. (C) Patterns of telomeric RNA expression in d10 male ES cells. (D) A size difference in the telomeric RNA foci of male vs. female ES cells. Inset shows magnifications of the boxed signal. Scale bar, 1 μm. (E) RNA FISH detecting telomeric RNA followed by DNA FISH using X painting probes in d4 female ES cells. (F) Patterns of telomeric RNA expression in d4 female ES cells. (G) Patterns of telomeric RNA expression relative to Xist expression in d10 female ES cells.
Female ES cells also showed dynamic patterns during differentiation. Interestingly, telomeric RNA foci in d0 female cells were consistently larger than in male cells (Figure 6D). So distinct was the difference that telomeric RNA size could be used reliably to identify ES cell sex without genotyping. Differentiation also led to a progressive decrease in the percentage of nuclei with two foci (X+X+) and appearance of transitional patterns (Figure 6, E–G). Whereas ∼50% of d0 female cells showed the X+X+ pattern (Figure 2, F and G), only 32% of d4 cells remained X+X+. At the same time, there was a relative increase in the total number of nuclei with a single RNA focus [X+X−, (XX)+]. On d4, speckling became evident in female cells and ∼22% of nuclei showed juxtaposed Xs with a single telomeric RNA focus (Figure 6, E and F), consistent with homologous X-chromosome pairing during differentiation (Bacher et al. 2006; Xu et al. 2006). The data showed that female differentiation is also marked by transitional states, with the X+X+ pattern representing the undifferentiated state and the X+X− pattern representing the differentiated state. The fact that only 50% of d0 XX cells actually showed the X+X+ pattern may reflect difficulty of establishing and maintaining pluripotent female ES cells in vitro (a well-known phenomenon).
Although mutational analysis did not reveal an obvious relationship to Xist (Figure 2, A–C), XCI and changes to telomeric RNA expression occur contemporaneously (Figure 6). To test the relationship further, we performed combined Xist and telomeric RNA FISH on d10 female cells. Previous work showed that Xist has three expression states (Panning et al. 1997; Sheardown et al. 1997; Sun et al. 2006): “Low” in pre-XCI undifferentiated ES cells, “high” in post-XCI differentiated female cells when Xist is transactivated, and “off” on the Xa of differentiated male and female cells. Among Xisthigh cells—the fraction represents cells with successful XCI—we found that a single telomeric focus was the most frequent pattern (Figure 6G), consistent with the pattern in fibroblasts (Figure 1).
Among Xistlow cells (presumably still undifferentiated), 1- and 2-pinpoint patterns were common, consistent with the pattern seen in undifferentiated XX cells (Figure 2). However, a common pattern among the Xistlow subpopulation was actually the speckled pattern (52%, n = 100). Speckling was even more common in Xistoff cells (80%, n = 20). In the d10 population, the Xistoff and Xistlow states would presumably represent failed dosage compensation. Speckling was not frequent in d10 male cells (12%). These results suggested that speckling may be a sign of stress caused, in this case, by genotoxicity associated with unrealized dosage compensation. Thus, differentiation in both XX and XY cells leads to dynamic changes in telomeric RNA expression, with the overall effect of reducing two RNA foci in undifferentiated cells to a single RNA focus in mature differentiated cells.
Effects of a Dicer deficiency on telomeric RNA:
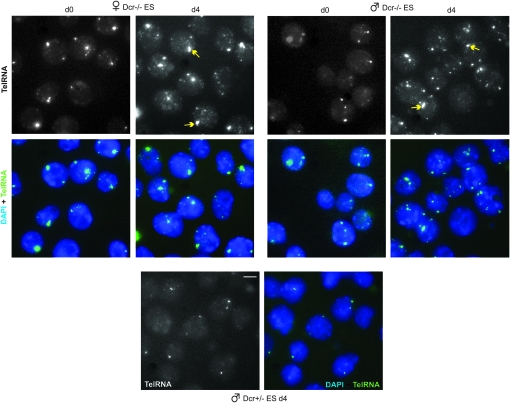
In several ways, chromatin structure at the telomere is reminiscent of pericentric heterochromatin (Grewal and Elgin 2007) in that genes inserted within or near both structures are subject to position-effect silencing (Gottschling et al. 1990; Baur et al. 2001; Tham and Zakian 2002; Vega et al. 2003). Given that gene regulation in the pericentric region is known to involve Dicer (Dcr) and the RNAi pathway (Grewal and Elgin 2007), we next asked whether telomeric RNA is impacted by Dcr. Using a Dcr-deficient ES model [Dcr−/− cells carrying a Dcr transgene expressing Dcr at <5% of normal levels (Ogawa et al. 2008)], we observed a modest increase in both number and size of telomeric RNA foci in d0 Dcr−/− cells as compared to wild-type cells of both sexes (Figure 7). Cell differentiation resulted in further amplification in number and intensity of telomeric RNA foci (d4, Figure 7, and data not shown). Interestingly, female Dcr-mutant cells showed a higher level of telomeric RNA accumulation than mutant male cells. Thus, Dcr activity affects telomeric RNA expression and telomeric RNA can accumulate elsewhere in the genome when Dcr activity is deficient.
Figure 7.—
Dcr deficiency leads to telomeric RNA upregulation. RNA FISH for telomeric RNA in d0 and d4 Dcr−/− and control Dcr+/− ES cells. Arrows, presumptive sex-linked telomeric RNA. Scale bar, 5 μm.
Because Dcr activity is often associated with processing to small RNAs (Zamore and Haley 2005; Grewal and Elgin 2007), we first performed Northern analyses using total RNA. Although control miR-292as RNA could be detected on the same blot, initial analysis revealed no discrete small RNA bands for telomeric RNA but did reveal a smear of signals, consistent with variable repeat lengths and potential usage of multiple transcription initiation sites along the repeats (Figure 8A). Hybridization using the complementary probe yielded no signals, as expected.
Because the considerably greater quantities of longer telomeric RNAs could potentially squelch hybridization to smaller RNAs, we repeated the Northern analysis using small RNA fractions from d0 and d10 female ES cells rather than total RNA, began with greater starting material, and increased the probe length to improve small RNA detection. Intriguingly, such analysis revealed a faint signal at ∼25 nt and possibly additional bands at ∼20 nt (Figure 8B). These small bands were visible in differentiated cells (d10) but were below detection levels or absent in d0 cells. The results suggested that the RNAs may be processed to small RNAs in a developmentally specific manner and raised the possibility that Dcr may regulate telomeric RNA turnover. This is consistent with the observation that Dcr-deficient cells exhibited greater telomeric RNA accumulation not only at the sex chromosomes but generally throughout the genome, as demonstrated by RNA FISH (Figure 7). Northern analysis of Dcr-deficient cells confirmed increased steady state levels of telomeric RNA (Figure 8A). Interestingly, even Dcr+/− cells [which express Dcr at ∼50% of normal levels (Ogawa et al. 2008)] showed greater telomeric RNA accumulation, suggesting that regulation of telomeric RNA levels is very sensitive to Dcr activity. The effect of Dcr on telomeric RNA levels may be direct or indirect.
Telomeric expression in normal cell growth and disease:
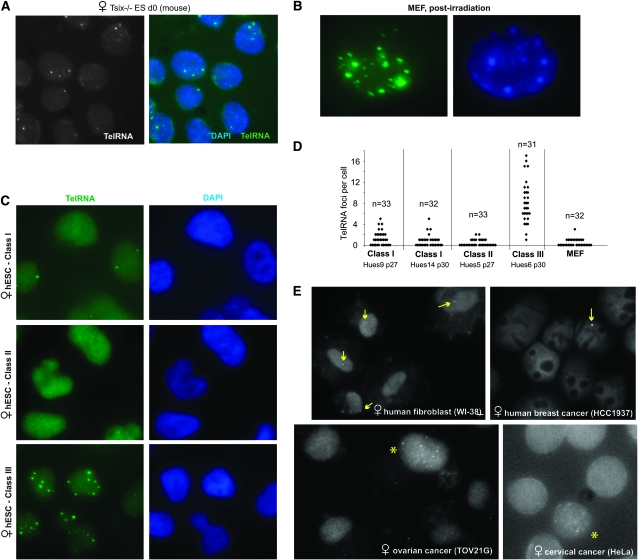
We next investigated whether telomeric expression changes under other physiological conditions such as genotoxic or general cellular stress. In Tsix−/− mouse cells, deletion of Xist's antisense regulator abolishes counting/choice and results in the appearance of cells with abnormal Xi number and cellular loss (Lee 2002, 2005). Here, we found that 64% (n = 84) of Tsix−/− cells in culture showed telomeric RNA speckling (Figure 9A). Gamma irradiation also resulted in telomeric RNA upregulation in all MEF cells of the culture (Figure 9B). Thus, widely different stimuli such as Dcr deficiency, radiation, and aberrant chromosome counting and inactivation are linked to changes in telomeric RNA expression in mice.
Figure 9.—
Altered telomeric RNA patterns in physiologically aberrant states. (A) Telomeric RNA FISH in d0 Tsix−/− mouse ES cells. (B) Irradiation induces telomeric RNA upregulation in MEF cells. (C) Aberrant classes of hESC exhibit abnormal telomeric RNA expression. (D) Expression patterns of telomeric RNA in class I, II, and III hESC lines. (E) RNA FISH in the wild-type and human cancer cell lines indicated. Arrows, cells with single telomeric RNA pinpoint. Asterisks, cells with speckling. Unmarked cells have no detectable RNA foci.
We also tested human cells. Like mouse ES cells, human embryonic stem cells (hESC) are uncommitted to XCI in the undifferentiated state but can inactivate one X during cell differentiation (Dhara and Benvenisty 2004; Hall et al. 2008; Shen et al. 2008; Silva et al. 2008). Recent work shows that many hESC are epigenetically abnormal with respect to XCI and can be grouped into three classes on the basis of XCI patterns (Enver et al. 2005; Hoffman et al. 2005; Adewumi et al. 2007; Hall et al. 2008; Shen et al. 2008; Silva et al. 2008). Here, we found that class I female cells, which undergo XCI properly, exhibited either one or two telomeric RNA foci (Figure 9, C and D), consistent with those observed in mouse ES cells. Human telomeric RNA signals, however, are much smaller and less intense than the mouse signals, perhaps correlating with shorter human telomere lengths (Harley et al. 1990; Kipling and Cooke 1990). Class II cells, which spontaneously underwent XCI (Xist+), generally displayed no RNA foci. At least one class III line (HUES6)—which lost the ability to express Xist following premature XCI—showed multifocal staining reminiscent of that seen in Tsix−/−, irradiated, and Dcr−/− cells. These data showed that aberrant dosage compensation in hESC is also associated with altered telomeric RNA expression.
In primary human female fibroblasts (WI-38), telomeric RNA expression was similar to that observed in mouse fibroblasts, with only one detectable telomeric RNA focus per nucleus (Figure 9E). Among cancer cells of various origins, telomeric RNA patterns were distinctly different from those seen in normal cells. A majority of cells of a breast cancer line (HCC1937), an ovarian cancer line (TOV21G), and a cervical cancer line (HeLa) showed no detectable telomeric RNA foci at all, consistent with the general shortening of telomeres in cancer cells (Blackburn et al. 2006; Feldser and Greider 2007). In all cancer lines examined, some degree of speckling could be observed in a small subset of cells. In HCC1937, a minority of cells showed a single large focus—much brighter and larger than was seen in control WI-38 cells.
Thus, in general, telomeric RNA foci are smaller in human cells than in mouse cells, possibly reflecting shorter telomeric lengths in humans. Although telomeric foci are relatively small in humans, the pattern of expression in healthy cells clearly differs from that in stressed or diseased cells. We emphasize the fact that changes to telomeric expression in stress and disease can involve either increases or decreases in RNA foci number and size.
DISCUSSION
Telomeric RNA dynamically associates with both X and Y chromosomes:
We have shown that Cot-1 RNA does not accumulate homogeneously in the mammalian genome and that Cot-1's telomeric fraction is enriched near sex chromosomes. The finding that telomeric repeats are transcribed agrees with previous studies (Rudenko and Van Der Ploeg 1989; Solovei et al. 1994; Azzalin et al. 2007; Schoeftner and Blasco 2008). However, we find that telomeric RNAs associate not only with the inactive X in somatic cells (Schoeftner and Blasco 2008), but generally associate with both sex chromosomes in undifferentiated stem cells. In female stem cells, both active Xs are marked by the RNAs. Furthermore, in male stem cells, the Y also accumulates telomeric RNA. During differentiation of both male and female ES cells, telomeric RNA is downregulated from the active sex chromosome and continues to associate only with the heterochromatic sex chromosome (Xi, Y). Our observation suggests that sex chromosome differentiation occurs not only for two Xs in female ES cells but also for the X and Y in male ES cells. Telomeric RNA accumulates more in XX than in XY cells. The expression dynamics suggest a “sex chromosome differentiation process” phenomenologically similar to XCI. The RNA also accumulates asymmetrically on each sex chromosome, associating preferentially with the distal telomeric end where the pseudoautosomal region is located.
Telomeric RNA as markers for pluripotency and cell health:
Given the association with sex chromosomes and apparent sex differences, we initially suspected functional linkage to dosage compensation and chromosome expression states, but the data so far do not support such a role (Figure 2). However, because deleting Xist does not cause instant X reactivation (Zhang et al. 2007), it is possible that loss of telomeric expression would reveal gross anomalies only after long-term culture. The low percentage of telomeric RNA accumulation on Xist-bearing autosomes may account for why autosomal silencing by Xist is generally not as robust (Lee and Jaenisch 1997; Heard et al. 1999; Lee et al. 1999). Regardless, while the purpose of telomeric transcription is currently not known, our current work suggests an intimate link with cell differentiation.
Its dynamic changes during normal cell differentiation and in disease states suggest that telomeric RNA could be used for diagnostic purposes. We propose that its RNA pattern can be used to identify the pluripotent ES cells—an especially useful application, given that ES cells [and presumably their induced pluripotent stem (iPS) cell counterparts] are prone to spontaneous differentiation in culture (Osafune et al. 2008; Silva et al. 2008). The two-foci pattern could help identify isolates and passages that remain undifferentiated and pluripotent. Those displaying only one focus may be partially differentiated. We also propose that telomeric RNA can be used to identify diseased and stressed cells. To the extent that we have tested them here, aberrant cell physiology seems to be generally associated with anomalous telomeric expression. In some aberrant states, telomeric RNA accumulation increases (e.g., irradiation, absence of dosage compensation, cellular transformation); in other disease states, RNA accumulation decreases (e.g., some malignancies, altered stem cell fates).
Regulation of telomeric RNA expression:
Our work leaves open the origin and regulation of telomeric RNA. Two intriguing questions are why telomeric RNA accumulates near the sex chromosomes instead of autosomes and why the heterochromatic sex chromosome is preferred in differentiated cells. One idea is that the RNA is produced only by the sex chromosomes, initially by both sex chromosomes and then only by the heterochromatic sex chromosome upon cell differentiation. Another is that the RNA is synthesized by all telomeres but concentrates only near the sex chromosomes. For example, all chromosomes may produce equal quantities of telomeric RNA, but the RNA may be stable only when made from the X or the Y. Alternatively, telomeric RNA produced off autosomes may relocalize to a specific nuclear compartment that just happens to be near the X and the Y.
In considering the regulation of telomeric RNA, we offer two potentially related observations. First, under stress and in some disease states, autosomes appear to accumulate telomeric RNA as well. (However, we cannot rule out the alternative possibility that the speckling represents relocalization of RNAs produced elsewhere.) In such states, a profound “speckling” of the nucleus becomes apparent, but accumulation elsewhere does not usually occur to the same extent as that seen on the Xi or the Y. These observations argue that autosomes can also express and localize telomeric RNA nearby, in agreement with previous findings (Rudenko and Van Der Ploeg 1989; Solovei et al. 1994; Azzalin et al. 2007; Schoeftner and Blasco 2008). Therefore, association of telomeric RNA with the X and Y in normal cells may reflect greater transcription or accumulation on sex chromosomes, rather than relocalization of autosomal RNAs.
Second, although we have not provided direct evidence, one might consider the possibility that the RNAi pathway impacts telomeric expression. In Dcr-deficient cells, telomeric RNA also becomes significantly upregulated, not only on the X and Y but also elsewhere in the nucleus. This effect could be direct or indirect. The Dcr-deficient state qualifies as a high-stress situation, as Dcr knockout cells are known to grow and differentiate poorly (Kanellopoulou et al. 2005; Murchison et al. 2005). This situation could result in upregulation of telomeric expression as we observed under other stressful conditions. An alternative, perhaps more intriguing scenario is the possibility of telomeric regulation by post-transcriptional gene silencing (PTGS), a mechanism already known to regulate pericentric heterochromatin (Grewal and Elgin 2007). For instance, autosomal telomeric RNA could be subject to PTGS via a Dcr-dependent pathway and be cleaved to small RNAs, with the consequence that the RNA accumulates near sex chromosomes. Consistent with this idea, we observe small RNAs of apparent telomeric origin (Figure 8). Furthermore, knocking down Dcr in both male and female ES cells leads to significant accumulation of telomeric RNA throughout the nucleus (Figures 7 and 8). These findings may suggest that telomeric RNA are transcribed from all chromosomes but are stabilized and accumulate only on the sex chromosomes.
Relevant to these ideas are several differences between our findings and those in previous reports. One difference concerns the change in telomeric expression in Dcr-deficient ES cells. Whereas Schoeftner and Blasco observed a slight reduction in total telomeric RNA (Schoeftner and Blasco 2008), we detect significantly more robust telomeric foci in Dcr-deficient cells. It is possible that our Dcr-deficient cells are more stressed than the control ES cells and also more stressed than those of Schoeftner and Blasco (2008), which may therefore explain the increased telomeric expression. The discrepancy may also stem from different methods of detection and quantitation (dot blot vs. Northern blot/RNA FISH methods). We emphasize that, in our hands, the two independent methods of assessment showed increased RNA accumulation when Dcr is deficient (Figures 7 and 8).
At face value, another difference relates to the number of visible telomeric RNA foci. Previous studies reported that RNA occurs on all telomeres (Rudenko and Van Der Ploeg 1989; Solovei et al. 1994; Azzalin et al. 2007; Schoeftner and Blasco 2008), whereas our data suggest that visible accumulation occurs predominantly on sex chromosomes. One difference may be that we used oligo probes targeted specifically at the tandem repeats, whereas previous experiments used larger probes (BAC) that may contain sequences in addition to the TAACCC repeat (Azzalin et al. 2007; Schoeftner and Blasco 2008). A more significant difference may be in usage of primary vs. transformed/stressed cell lines. The multifoci pattern reported by Azzalin et al. (2007) and by Schoeftner and Blasco (2008) occurred in immortalized or transformed cell lines (stressed cells), whereas our sex-linked patterns occurred predominantly in undifferentiated ES cells and in primary cell lines. We observe the multifocal pattern primarily in stressed cells after irradiation and in Dcr- and XCI-deficient states.
Collectively, our data sets lend support to the hypothesis that, under normal physiological conditions, telomeric RNA may be transcribed from all telomeres but be stably and visibly associated primarily with the X and the Y. We suggest that, under conditions of stress and disease, the RNA becomes more visible elsewhere in the genome. In summary, our work adds telomeric RNA to a growing list of large noncoding RNAs with potential function in regulating the epigenome.
Acknowledgments
We thank Jerome DeJardin, Yesu Jeon, Eda Yildirim, and Bernhard Payer for critical reading of the manuscript and all members of the lab for stimulating discussion. This work was funded by an award from the Howard Hughes Medical Institute (to J.T.L.).
References
- Adewumi, O., B. Aflatoonian, L. Ahrlund-Richter, M. Amit, P. W. Andrews et al., 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25 803–816. [DOI] [PubMed] [Google Scholar]
- Azzalin, C. M., P. Reichenback, L. Khoriauli, E. Giulotto and J. Lingner, 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318 798–801. [DOI] [PubMed] [Google Scholar]
- Bacher, C. P., M. Guggiari, B. Brors, S. Augui, P. Clerc et al., 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8 293–299. [DOI] [PubMed] [Google Scholar]
- Baur, J. A., Y. Zou, J. W. Shay and W. E. Wright, 2001. Telomere position effect in human cells. Science 292 2075–2077. [DOI] [PubMed] [Google Scholar]
- Birney, E., J. A. Stamatoyannopoulos, A. Dutta, R. Guigo, T. R. Gingeras et al., 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E. H., C. W. Greider and J. W. Szostak, 2006. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12 1133–1138. [DOI] [PubMed] [Google Scholar]
- Borsani, G., R. Tonlorenzi, M. C. Simmler, L. Dandolo, D. Arnaud et al., 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351 325. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N., A. Ashworth, G. F. Kay, P. Cooper, S. Smith et al., 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351 329. [DOI] [PubMed] [Google Scholar]
- Brown, C. J., A. Ballabio, J. L. Rupert, R. G. Lafreniere, M. Grompe et al., 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349 38. [DOI] [PubMed] [Google Scholar]
- de Lange, T., 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 5 323–329. [DOI] [PubMed] [Google Scholar]
- Dhara, S. K., and N. Benvenisty, 2004. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 32 3995–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver, T., S. Soneji, C. Joshi, J. Brown, F. Iborra et al., 2005. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 14 3129–3140. [DOI] [PubMed] [Google Scholar]
- Feldser, D. M., and C. W. Greider, 2007. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell 11 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras, T. R., 2007. Origin of phenotypes: genes and transcripts. Genome Res. 17 682–690. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63 751–762. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and S. C. Elgin, 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L. L., M. Byron, J. Butler, K. A. Becker, A. Nelson et al., 2008. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J. Cell. Physiol. 216 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L. L., M. Byron, K. Sakai, L. Carrel, H. F. Willard et al., 2002. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. USA 99 8677–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley, C. B., A. B. Futcher and C. W. Greider, 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345 458–460. [DOI] [PubMed] [Google Scholar]
- Heard, E., F. Mongelard, D. Arnaud, C. Chureau, C. Vourc'h et al., 1999. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96 6841–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. M., L. Hall, J. L. Batten, H. Young, D. Pardasani et al., 2005. X-inactivation status varies in human embryonic stem cell lines. Stem Cells 23 1468–1478. [DOI] [PubMed] [Google Scholar]
- Huynh, K. D., and J. T. Lee, 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426 857. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin et al., 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, R., and M. Kuroda, 2000. Noncoding RNA genes in dosage compensation and imprinting. Cell 29 9–12. [DOI] [PubMed] [Google Scholar]
- Kipling, D., and H. J. Cooke, 1990. Hypervariable ultra-long telomeres in mice. Nature 347 400–402. [DOI] [PubMed] [Google Scholar]
- Lau, N. C., L. P. Lim, E. G. Weinstein and D. P. Bartel, 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, J. T., 2002. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat. Genet. 32 195–200. [DOI] [PubMed] [Google Scholar]
- Lee, J. T., 2005. Regulation of X-chromosome counting by Tsix and Xite sequences. Science 309 768–771. [DOI] [PubMed] [Google Scholar]
- Lee, J. T., and R. Jaenisch, 1997. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386 275–279. [DOI] [PubMed] [Google Scholar]
- Lee, J. T., and N. Lu, 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99 47–57. [DOI] [PubMed] [Google Scholar]
- Lee, J. T., N. Lu and Y. Han, 1999. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc. Natl. Acad. Sci. USA 96 3836–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov, V. L., Y. Hirose and J. P. Langmore, 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88 657–666. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., A. Krauskopf and E. H. Blackburn, 2000. Telomeres and their control. Annu. Rev. Genet. 34 331–358. [DOI] [PubMed] [Google Scholar]
- Murchison, E. P., J. F. Partridge, O. H. Tam, S. Cheloufi and G. J. Hannon, 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 102 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, P., S. Pichard, C. Ciaudo, P. Avner and C. Rougeulle, 2005. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 19 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, Y., B. K. Sun and J. T. Lee, 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune, K., L. Caron, M. Borowiak, R. J. Martinez, C. S. Fitz-Gerald et al., 2008. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26 313–315. [DOI] [PubMed] [Google Scholar]
- Panning, B., J. Dausman and R. Jaenisch, 1997. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90 907–916. [DOI] [PubMed] [Google Scholar]
- Pheasant, M., and J. S. Mattick, 2007. Raising the estimate of functional human sequences. Genome Res. 17 1245–1253. [DOI] [PubMed] [Google Scholar]
- Rudenko, G., and L. H. Van der Ploeg, 1989. Transcription of telomere repeats in protozoa. EMBO J. 8 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado, T., Y. Hoki and H. Sasaki, 2005. Tsix silences Xist through modification of chromatin structure. Dev. Cell 9 159–165. [DOI] [PubMed] [Google Scholar]
- Schoeftner, S., and M. A. Blasco, 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10 228–236. [DOI] [PubMed] [Google Scholar]
- Sheardown, S. A., S. M. Duthie, C. M. Johnston, A. E. T. Newall, E. J. Formstone et al., 1997. Stabilization of Xist RNA mediate initiation of X chromosome inactivation. Cell 91 99–107. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Y. Matsuno, S. D. Fouse, N. Rao, S. Root et al., 2008. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl. Acad. Sci. USA 105 4709–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, S. S., R. K. Rowntree, S. Mekhoubad and J. T. Lee, 2008. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl. Acad. Sci. USA 105 4820–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei, I., E. R. Gaginskaya and H. C. Macgregor, 1994. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 2 460–470. [DOI] [PubMed] [Google Scholar]
- Sun, B. K., A. M. Deaton and J. T. Lee, 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21 617–628. [DOI] [PubMed] [Google Scholar]
- Tham, W. H., and V. A. Zakian, 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21 512–521. [DOI] [PubMed] [Google Scholar]
- Vega, L. R., M. K. Mateyak and V. A. Zakian, 2003. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell. Biol. 4 948–959. [DOI] [PubMed] [Google Scholar]
- Wutz, A., 2003. RNAs templating chromatin structure for dosage compensation in animals. BioEssays 25 434–442. [DOI] [PubMed] [Google Scholar]
- Xu, N., C. L. Tsai and J. T. Lee, 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 1149–1152. [DOI] [PubMed] [Google Scholar]
- Zamore, P. D., and B. Haley, 2005. Ribo-gnome: the big world of small RNAs. Science 309 1519–1524. [DOI] [PubMed] [Google Scholar]
- Zhang, L. F., K. D. Huynh and J. T. Lee, 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129 693–706. [DOI] [PubMed] [Google Scholar]