Abstract
The transcription factors Adr1 and Cat8 act in concert to regulate the expression of numerous yeast genes after the diauxic shift. Their activities are regulated by Snf1, the yeast homolog of the AMP-activated protein kinase of higher eukaryotes. Cat8 is regulated directly by Snf1, but how Snf1 regulates Adr1 is unknown. Mutations in Adr1 that alleviate glucose repression are clustered between amino acids 227 and 239. This region contains a consensus sequence for protein kinase A, RRAS230F, and Ser230 is phosphorylated in vitro by both protein kinase A and Ca++ calmodulin-dependent protein kinase. Using an antiphosphopeptide antibody, we found that the level of Adr1 phosphorylated on Ser230 was highest in glucose-grown cells and decreased in a Snf1-dependent manner when glucose was depleted. A nonphosphorylatable Ser230Ala mutant was no longer Snf1 dependent for activation of Adr1-dependent genes and could suppress Cat8 dependence at genes coregulated by Adr1 and Cat8. Contrary to expectation, neither protein kinase A (PKA) nor Ca++ calmodulin-dependent protein kinase appeared to have an important role in Ser230 phosphorylation in vivo, and a screen of 102 viable kinase deletion strains failed to identify a candidate kinase. We conclude that either Ser230 is phosphorylated by multiple protein kinases or its kinase is encoded by an essential gene. Using the Ser230Ala mutant, we explain a long-standing observation of synergy between Adr1 constitutive mutants and Snf1 activation and conclude that dephosphorylation of Ser230 via a Snf1-dependent pathway appears to be a major component of Adr1 regulation.
TRANSCRIPTION factors activate gene expression by recruiting coactivators to a promoter to create a pre-initiation complex (PIC) containing RNA polII and general factors (Featherstone 2002; Fry and Peterson 2002). Signal transduction pathways often regulate this process by acetylation or phosphorylation of transcription factors to influence their ability to recruit coactivators (Kwok et al. 1994; Papoutsopoulou and Janknecht 2000; Zhong et al. 2002; Goel and Janknecht 2003). Adr1 is a yeast transcription factor that acts in concert with Cat8 and Oaf1/Pip2 to activate numerous genes that are expressed after glucose depletion, allowing cells to use nonfermentable carbon sources such as ethanol, glycerol, and fatty acids (Schuller 2003; Young et al. 2003; Santangelo 2006). The signal transduction pathway that activates Adr1 and Cat8 is mediated by Snf1 (Rahner et al. 1999; Schuller 2003; Charbon et al. 2004; Santangelo 2006), the yeast homolog of AMP-activated protein kinase that is widely regarded as the “energy sensor of the cell” (Hardie et al. 1998). Snf1 is activated by phosphorylation on Thr210 by three upstream kinases (Hong et al. 2003). Snf1-Thr210 is dephosphorylated by the PP1-type protein phosphatase Glc7, and regulation of this event appears to be through the targeting factor Reg1 (Sanz et al. 2000; Rubenstein et al. 2008). As expected, removing the negative regulatory mechanism by deleting REG1 causes constitutive activation of Snf1, as measured by expression in glucose of Snf1-dependent genes that are normally glucose repressed (Dombek et al. 1993; Tu and Carlson 1995; McCartney and Schmidt 2001; Orlova et al. 2008). Control of Snf1 must include additional mechanisms beyond simple phosphorylation and Glc7·Reg1-dependent dephosphorylation, however, because in a reg1Δ strain, additional phosphorylation of Snf1 and further activation of some Snf1-dependent genes is achieved by glucose depletion (Dombek et al. 1993; Tachibana et al. 2007; Orlova et al. 2008). Interaction with regulatory proteins or changes in protein structure have been suggested as additional levels of Snf1 control (Rubenstein et al. 2008).
Snf1 is necessary for the activity of both Adr1 and Cat8, enhancing transcription of CAT8 through inactivation of the repressor Mig1, as well as activating Cat8 directly by phosphoryation (Hedges et al. 1995; Rahner et al. 1996, 1999; Charbon et al. 2004; reviewed in Schuller 2003). Snf1 is required for Adr1 binding (Young et al. 2002) and recruitment of coactivators to Adr1-dependent promoters (Biddick et al. 2008). Promoter binding by Adr1 is regulated in part by phosphorylation of its DNA-binding domain (Kacherovsky et al. 2008) and by acetylation of promoter nucleosomes (Verdone et al. 2002; Tachibana et al. 2007), but it is not known whether Snf1 activates Adr1 directly or indirectly or has another role at Adr1-dependent promoters, such as nucleosome modification (Lo et al. 2001).
Selection of mutants that allowed ADH2 expression to escape glucose repression identified rare semidominant ADR1c alleles (ADR1-constitutive) (Ciriacy 1979; Denis et al. 1991). ADR1c alleles allow ADH2 expression to occur in the presence of glucose and enhance derepressed transcription. Surprisingly, the level of Adr1c protein is much lower than the level of wild-type Adr1 (Taylor and Young 1990; Dombek and Young 1997), suggesting that its high transcriptional potency might be associated with rapid turnover as has been observed for some other activators (Tansey 2001). ADR1c shows strong synergism with deletion of the Snf1 regulatory factor REG1. In combination with Adr1c, partial activation of Snf1 by deleting REG1 allows much higher levels of ADH2-repressed expression than are observed with either mutation acting alone (Dombek et al. 1993). Adr1c and reg1Δ are also synergistic in activating a poised but inactive PIC (Tachibana et al. 2007). Although this suggests that Adr1c and activation of Snf1 by reg1Δ act through different pathways to cause constitutive expression of Snf1-dependent genes, the precise mechanism of the synergism is unknown.
Cloning and molecular analysis of ADR1c alleles showed mutations between amino acids 227 and 239, a region that contains a consensus sequence for a cyclic AMP-dependent PKA, Arg-Arg-Ala-Ser-Phe (Denis and Gallo 1986; Denis et al. 1992). No other mutations in the Adr1 ORF causing constitutive ADH2 expression were isolated, suggesting that the 227–239 region plays a unique role in regulating Adr1 activity. The ADR1c mutations suggest that Adr1 might be post-translationally inhibited by phosphorylation of Ser230 in the PKA consensus sequence (Denis and Gallo 1986; Cherry et al. 1989). Adr1 appears to be phosphorylated at Ser230 in vivo because subtle alterations in its SDS–PAGE electrophoretic mobility and partial sensitivity to phosphatase treatment are observed (Vallari et al. 1992; Dombek and Young 1997). Recent studies of the yeast phosphorylome also suggested Adr1 phosphorylation, although the reported site was Ser232, not Ser230 (Chi et al. 2007).
In support of phosphorylation of Adr1 by PKA, both PKA and calmodulin-dependent protein kinase (CMK) efficiently phosphorylate Ser230 in vitro (Denis et al. 1991; Hook et al. 1999). Hyperactivation of PKA by deletion of its BCY1-encoded regulatory subunit causes loss of ADH2 expression (Cherry et al. 1989), suggesting that unregulated PKA activity inhibits Adr1 by constitutively phosphorylating Ser230. However, replacing Ser230 with phosphomimetic Asp gave the same constitutively active phenotype as other alterations in the region, suggesting that either Asp does not mimic phosphorylated Ser or phosphorylation of Ser230 is not the only determinant of the ADR1c phenotype (Denis et al. 1992; Dombek and Young 1997). In addition, several of the Adr1c mutations appear to refute PKA as a candidate for the Ser230 kinase in vivo. The Ala229Pro Adr1c mutant shows greatly increased affinity for PKA binding in a peptide phosphorylation assay, which would predict greater phosphorylation and lower Adr1 activity. However, the Ala229Pro mutant is one of the most constitutive of the Adr1c mutants (Denis et al. 1992). Therefore, it is not known if phosphorylation of Ser230 affects the activity of Adr1 and what kinase is responsible for the phosphorylation in vivo (Denis et al. 1992; Cook et al. 1994; Dombek and Young 1997).
How Adr1c achieves its high transcriptional potency is unknown (Denis et al. 1992; Cook et al. 1994; Dombek and Young 1997). ADR1c alleles map far from the DNA-binding domain and Adr1-Ser230A does not have enhanced in vitro DNA-binding activity (Taylor and Young 1990). After characterizing a series of mutants in amino acids 227–239, as well as internal deletions in this region of Adr1, Cook et al. (1994) suggested that this region either binds a repressive factor that cannot interact with the mutants or interacts with and blocks the activation domain. Some evidence has suggested that these Adr1c mutants suppress the requirement for Snf1 (Ciriacy 1979; Denis 1984), which would imply that it is not affected by Snf1, but this is contradictory to the enhanced effect on ADH2 expression seen when Adr1c is combined with the Snf1-activating reg1Δ mutation. Thus, the questions of how Snf1 affects the activity of Adr1 and whether it interacts with the Adr1c allele are unresolved.
We used an antibody against a Ser230-phosphorylated peptide of Adr1 amino acids 217–234 to investigate the conditions under which Ser230 is phosphorylated (Adr1-pSer230) and the effect of Snf1 on this modification. The antibody was used to screen for a specific Adr1-Ser230 kinase. Phosphorylation was not increased by enhanced PKA activity, nor decreased by loss of PKA or CMK. These and other data suggest that Ser230 may be redundantly phosphorylated in vivo by multiple kinases or phosphorylated by an essential kinase. Using the nonphosphorylatable Adr1c, we probed the relationship between Adr1-pSer230, Snf1, and the coregulator Cat8, with the results supporting an earlier model for the function of the Ser230-containing domain.
MATERIALS AND METHODS
Yeast strains, plasmids, culture conditions, and enzyme assays:
The strains used in the study are listed in Table 1. Multiple deletion mutants of TPK and CMK genes were constructed by standard genetic techniques. Epitope tagging, gene deletion, and marker swapping were according to Guldener et al. (1996), Cross (1997), and Knop et al. (1999), respectively. Deletions and epitope tagging were confirmed by PCR and, for marker swapping, by Western blots. Yeast strains were grown in complete or synthetic media as described in Sherman (1991). Repressing medium contained 5% glucose; derepressing medium contained 0.05% glucose with or without 3% glycerol or 2% ethanol. The plasmids used were described previously (Yu et al. 1989; Dombek and Young 1997) and modified in some cases by introducing an epitope tag at the C terminus of Adr1 as described below. For gene expression studies, CEN-TRP1 plasmids expressing wild-type ADR1 (pKD16) or the S230A (pKD14), R228K (pKD27), or Δ3 (a deletion that removes Adr1 amino acids 226–233; pKD26) alleles of ADR1 from its native promoter were used. To facilitate detection of Adr1-S230 phosphorylation by Western blotting, 2μ plasmids expressing wild-type ADR1 (pKD17-HA∷kanMX (TRP1) and the ADR1-S230A allele (pKD20-HA∷kanMX (TRP1) from the ADH1 promoter and tagged with an HA epitope were employed. Alternatively, strains with four additional copies of ADR1 integrated at the leu2 locus were used (Sloan et al. 1999). ADR1 was epitope tagged with HA∷kanMX in this strain. The Adr1-dependent reporter was a UAS1-lacZ plasmid, pHDY10, containing 10 Adr1-binding sites (Yu et al. 1989). β-Galactosidase assays were performed as described in Guarente (1983). ADH enzyme activity was analyzed by separating proteins on nondenaturing polyacrylamide gels and visualized by in-gel chromogenic staining as described (Dombek and Young 1997).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| TYY201 (aka W303-1A) | MATaade2 can1-100 his3-11,15 leu2-13,112 trp1-1 ura3-1 | Yeast stock center |
| VBY20 | MATα adh3 ura3 his3 leu2∷(pRS315-ADR1)X3 ADH2∷YIpADH2/lacZ(TRP1)ADH2∷ YIpADH2/GFP (URA3) | Voronkova et al. (2006) |
| NKY85 | VBY20 ADR1:HA∷kanMX | This study |
| NKY87 | NK85 reg1Δ∷natMX | This study |
| NKY91 | NK85 snf1Δ∷natMX | This study |
| TYY204 | W303-1A adr1Δ1∷LEU2 ADH2∷YIpADH2/lacZ∷TRP1 | Young et al. (2003) |
| TYY497 | W303-1A adr1Δ1∷LEU2 ADH2∷YIpADH2/lacZ∷trp1∷HIS3 | Young et al. (2008) |
| TYY498 | W303-1A snf1Δ∷URA3 adr1Δ1∷LEU2 ADH2∷YIpADH2/lacZ∷trp1∷HIS3 | Young et al. (2008) |
| CKY13 | W303-1A adr1Δ∷kanMX | This study |
| CKY26 | W303-1A adr1Δ∷natMX snf1Δ∷kanMX | This study |
| CKY5 | W303-1A adr1Δ∷kanMX reg1Δ∷natMX | This study |
| CKY23 | W303-1A adr1Δ∷natMX cat8Δ∷kanMX | This study |
| SRY60 | W303-1A reg1Δ∷kanMX cat8Δ∷natMX adr1Δ∷HYG | This study |
| NKY108 | NKY85 cmk1Δ∷hphmx cmk2Δ∷natMX cmk3Δ∷natMX | This study |
| SGP406 | MATahis3 leu2-3,112 tpk1∷URA3 tpk2∷HIS3 tpk3∷TRP1 trp1 ura3-52 yak1∷LEU2 | Garrett and Broach (1989) |
| NKY111 | NKY85 bcy1Δ∷natMX | This study |
| CMY323 | MATaade2-101° can1 his3Δ200 lys2-801 trp1Δ1 ura3-52 | Carl Mann |
| CMY328 | MATaade2-101°adr1Δ2∷URA3 can1 his3Δ200 lys2-801atrp1Δ1 ura3-52 | Dombek and Young (1997) |
| CMY468 | MATaade2-101° his3Δ leu2Δ1 lys2-801 tpk1∷URA3 tpk2∷HIS3tpk3ts-1 trp1Δ1 ura3-52 | Carl Mann |
| SGP400 | MATahis3 leu2-3,112 trp1 ura3-52 yak1∷LEU2 | Garrett and Broach (1989) |
Real-time quantitative PCR:
For expression analysis, RNA was isolated by hot phenol extraction (Collart and Oliveiro 1993) and converted to cDNA with a SuperScript III kit (Invitrogen) according to the manufacturer's directions. cDNA was quantified by real-time quantitative PCR (RT–qPCR) (Tachibana et al. 2007) with an MJ Research Chromo4 system, using ABI or Quantace SYBR Master Mix, according to the manufacturer's instructions. Primer sequences are available on request.
Immunoblotting:
Whole-cell extracts were analyzed on 3–8% or 6% polyacrylamide gels (NuPage system, Invitrogen) and transferred to PVDF membranes after electrophoresis. Western blots were probed with α-pSer230 followed by a rabbit-specific Licor secondary Ab (λ680). A second identical blot was probed with a monoclonal α-HA antibody and a mouse-specific Licor secondary antibody (λ800). Probing a single blot with both antibodies resulted in bleed-through from the more abundant HA-epitope-derived signal so either two blots were done or a single blot was probed sequentially—first, with α-pSer230 and an appropriate Licor secondary antibody and, second, with the α-HA antibody and an appropriate Licor secondary antibody. Cell extracts were prepared as described previously (Dombek and Young 1997) or by a boiling procedure with the addition of agitation with glass beads for 30 sec before the boiling step (Invitrogen) (Hahn 2008). An Odyssey Infrared imaging system (Licor Biosciences) was used for quantitation. α-HA (Y-11, Santa Cruz Biochemicals), α-Adr1 (Dombek et al. 1993), or antibodies against an Adr1-derived synthetic peptide were used at 1:1000 dilution. The peptides, representing Adr1 amino acids 217–234, VKRKYLKKLTRRA(pS)FSAQ, and its nonphosphorylated version were synthesized and used to generate and affinity purify the α-pSer230 antibody by Bethyl Laboratories (Montgomery, TX). Secondary IR-dye-conjugated antibodies used at 1:1000–1:3000 were goat α-mouse Alexa 680 (Molecular Probes) or IRdye800-conjugated α-rabbit IgG (Rockland Immunochemicals).
RESULTS
Adr1-Ser230 phosphorylation in vivo is glucose dependent and dephosphorylation is Snf1 dependent:
Antibodies against a phosphorylated peptide representing Adr1 amino acids 217–234 (VKRKYLKKLTRRA(pS)FSAQ) were generated to assess the level of Adr1 phosphorylated on Ser230 in vivo (designated Adr1−pSer230). ELISA assays indicated that the antiphosphopeptide antibody (α-pSer230) recognized the phosphorylated peptide 5000× better than the nonphosphorylated peptide (data not shown). Figure 1A shows that α-pSer230 recognized a protein the size of Adr1 in a strain expressing Adr1-HA from the strong ADH1 promoter on a multicopy plasmid, but not in a strain lacking Adr1. Adr1 containing a Ser230Ala mutation (hereafter referred to as Adr1c unless other ADR1c alleles are specified) was recognized poorly by α-pSer230, demonstrating the specificity of the antibody for Adr1 with phosphorylated Ser230. Competition with the phosphorylated and nonphosphorylated peptides demonstrated the specificity of α-pSer230 (Figure 1B). These data confirmed that α-pSer230 recognized Adr1-pSer230 and cross-reacted weakly, if at all, with Adr1 lacking phosphate on Ser230.
Figure 1.—
Antiphosphopeptide antibody recognizes Adr1 phosphorylated on Ser230. (A) α-pSer230 recognizes Adr1 in whole-cell extracts. Wild type (wt) and S230A refer to TYY497 (adr1Δ1) with pKD17-HA and pKD20-HA, expressing HA-epitope-tagged versions of wild-type ADR1 or ADR1-Ser230, respectively; “Δ” is the strain TYY497 with a vector lacking ADR1. The amount of protein loaded on the gel is shown above each lane. The cells were grown in trp− synthetic medium in repressing conditions (5% glucose). (B) Peptide competition. Ten ng/ml of nonphosphorylated or Ser230-phosphorylated peptide (materials and methods) representing Adr1 amino acids 217–234 was present during Western blotting with the primary antibody. Cell extracts were made from strain TYY497 carrying pKD17-HA (wt multicopy ADR1) grown in repressing conditions in trp− synthetic medium. The amount of protein loaded on the gel is shown above each lane. A very low amount of cell extract (nanograms) was used to allow effective competition with the peptide.
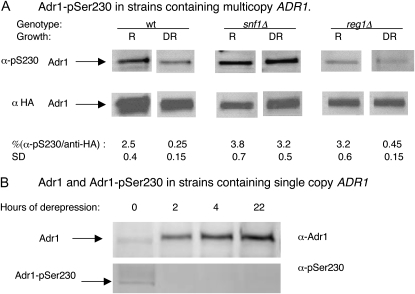
A strain containing multiple integrated copies of ADR1 was employed to facilitate quantitation of Adr1-pSer230 in different growth conditions. At least one copy of Adr1 in this strain was tagged with an HA epitope to provide a measure of the total amount of Adr1 in the extract. Adr1 levels in the multicopy strain grown in repressing conditions are similar to Adr1 levels in wild-type cells grown in derepressing conditions, and ADH2 regulation appears unperturbed by the modest increase in Adr1 levels (Sloan et al. 1999; Voronkova et al. 2006).
Western blotting for both HA and Adr1-pSer230 revealed that the amount of pSer230 relative to total Adr1 was highest in glucose-grown cells and decreased ∼10-fold when cells were grown in low glucose plus ethanol (Figure 2A, wt lanes). The decrease in pSer230 in the absence of glucose was not due to the elevated Adr1 level in the multicopy strain because a decrease was also observed in a strain with a single copy of ADR1 (Figure 2B). Although the low level of Adr1-pSer230 in the single-copy strain precluded quantification of the change in phosphorylation, Adr1 increased at least 10-fold after 22 hr of growth in the absence of glucose, and the level of pSer230 decreased to an undetectable level after 2 hr.
Figure 2.—
Phosphorylation of Adr1-pSer230 is glucose and Snf1 regulated. Preparation of cell extracts, electrophoresis, and Western blotting were carried out as described in materials and methods. (A) Multicopy ADR1 strains. Wild type (wt) is NKY85; snf1Δ is NKY91, and reg1Δ is NKY87; R, repressing growth conditions, 5% glucose; derepression (DR) was for 6 hr in 0.05% glucose–2% ethanol. Cell extract (25 μg) was analyzed. The signal intensity was measured as described in materials and methods for several biological replicates. The values shown are the means ± one standard deviation (SD). (B) A single copy ADR1 strain (TYY201) was analyzed in a similar manner using 100 μg of protein extracted from repressed cells (0 hr derepression) and from cells derepressed for 2, 4, and 22 hr in 0.05% glucose–2% ethanol.
A strain lacking SNF1 is unable to activate ADH2 expression presumably due to inactive Adr1. Consistent with the hypothesis that phosphorylation of Ser230 inactivates Adr1, in a snf1Δ strain, the amount of Adr1-pSer230 relative to total Adr1 appeared to be elevated in the presence of glucose and did not decrease significantly after its removal (Figure 2A, snf1Δ lanes). Since Snf1 is a kinase, any regulation of phosphorylated Ser230 must be indirect, for example, through control of another kinase or a phosphatase. Surprisingly, although dephosphorylation of Ser230 appeared to be Snf1 dependent, deleting REG1 or inactivating GLC7 with a temperature-sensitive mutation to activate Snf1 in repressing conditions did not affect the relative level of Adr1-pSer230 in either repressing or derepressing conditions (Figure 2A, reg1Δ; unpublished data for Glc7-127). The experiment was repeated multiple times with similar results. Snf1 control of Adr1-pSer230 apparently requires additional levels of Snf1 regulation, such as control over access to the Snf1 T210 phosphorylation site, that work with dephosphorylation by Reg1·Glc7 to mediate Snf1 activity (Orlova et al. 2008; Rubenstein et al. 2008). Alternatively, Ser230 dephosphorylation in glucose may be prevented by a mechanism that is independent of Reg1·Glc7. In summary, inactivation of Snf1 prevented the decrease in Ser230 phosphorylation that normally accompanies derepression and activation of Adr1-dependent genes, which is consistent with phosphorylation playing a negative role in regulating the activity of Adr1.
The stoichiometry of phosphorylation on Ser230 was assessed by immunoprecipitation of Adr1 in whole-cell extracts derived from the multicopy ADR1 strain grown in repressing conditions. Adr1 was immunoprecipitated with α-Adr1 and α-pSer230 antisera, and the precipitated and supernatant fractions were analyzed by SDS–PAGE. Western blotting using α-Adr1 antisera, which recognizes both phosphorylated and nonphosphorylated Adr1, demonstrated that α- and α-pSer230 antisera immunoprecipitated 65% and 45%, respectively, of the total Adr1 (supporting information, Figure S1). Dephosphorylation of pSer230 during immunoprecipitation may contribute to the lower level of Adr1 immunoprecipitated by the α-pSer230 antisera compared to the α-Adr1 antisera, or there may be a fraction of Adr1 that is not phosphorylated on Ser230 even in repressing growth conditions. The high level of Adr1 precipitated by the α-pSer230 antisera indicates that the majority of Adr1 is phosphorylated in vivo in repressing conditions.
Adr1c activation is Snf1 independent and synergism with reg1Δ is from Snf1 activation of Cat8:
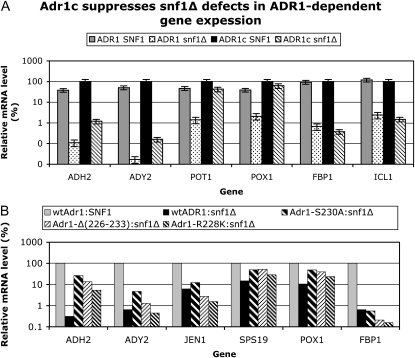
Since the decrease of Adr1-Ser230 phosphorylation did not occur in a snf1Δ strain, we hypothesized that, if this decrease were important for the activity of Adr1, Adr1c, which has the phosphorylatable serine mutated to alanine, would be insensitive to snf1Δ. As predicted, both ADH2/lacZ reporter assays and in-gel assays of endogenous ADHII activity showed that Adr1c was highly efficient at activating ADH2, even in the absence of Snf1. ADH2 derepression by Adr1c was reduced only fourfold in the snf1 mutant, compared to an almost complete lack of expression with wild-type Adr1 (Figure S2). Snf1 independence of Adr1c was also seen when mRNA from ADH2 was measured directly by RT–qPCR. With this assay, we found that other Adr1-dependent genes, including POX1 and SPS19, were also activated in a Snf1-independent manner by Adr1c. Adr1c did not change the expression of FBP1, which is not Adr1 dependent (Figure 3A). Other Adr1c variants of the Ser230 region were used to show that the Snf1 independence was a general effect, because two other alleles, Adr1-Δ3 [Δ(226-223)] and Adr1-R228K, also activated the Adr1-dependent genes without Snf1. The CAT8-dependent FBP1 gene was not derepressed in snf1Δ strains by Adr1c alleles (Figure 3B). ADR1c could not overcome the Snf1 requirement for growth on the nonfermentable carbon sources ethanol, glycerol, lactate, or acetate, however (data not shown). This is not surprising because Cat8 is essential for growth on these carbon sources and expression of Cat8-dependent genes such as FBP1 were unaffected by Adr1c. In addition, there are other Snf1-dependent transcription factors such as Mig1 that are involved in gene expression for these metabolic pathways.
Figure 3.—
Snf1 dependence of Adr1c-dependent gene expression. (A) Derepressed transcript levels in SNF1 or snf1Δ, with wild-type (wt) ADR1 or ADR1c (ADR1-S230A). Analysis of mRNA isolated from cells derepressed in 0.05% glucose–3% glycerol for 4 hr. RT–qPCR was performed and analyzed as described in materials and methods. Strains were TYY497 (adr1ΔSNF1) and TYY498 (adr1Δsnf1Δ) carrying either wild-type ADR1 (pKD16) or ADR1-S230A (pKD14). The mRNA values were normalized to ACT1 mRNA and are expressed as a percentage of mRNA in the wild-type SNF1 strain with wild-type Adr1 as the activator. The values represent the mean ± a standard deviation of two biological replicates assayed in duplicate. (B) Relative transcript levels in snf1Δ with various Adr1c alleles. Transcript levels were quantified and displayed as in A. The strains were CKY13 (adr1Δ) and CKY26 (adr1Δ snf1Δ). The plasmids were as in section A and additionally pKD26 [Adr1-Δ(226-233)] and pKD27 (Adr1-R228K).
The lack of Snf1 control over Adr1c alleles raised a question about earlier results showing synergism between Adr1c and reg1Δ (Dombek et al. 1993; Tachibana et al. 2007), in which Adr1c-containing strains show additional activation of ADH2 when Snf1 is activated in glucose by reg1Δ. Adr1c was Snf1 independent, so it should not be influenced by reg1Δ. A clue to the mechanism of the synergism came from the high Snf1 dependence seen for the Cat8-dependent FBP1 gene (Figure 3, A and B) and the lack of synergism seen at Cat8-independent genes such as POX1 (data not shown). We hypothesized that the additional activation of ADH2, seen when REG1 was deleted in an Adr1c-containing strain, is from Snf1 activation of Cat8. If this hypothesis is correct, reducing the Cat8 effect should reduce the synergism. We measured ADH2 activation independently of other activators using a reporter that lacked binding sites for Cat8 and had only the Adr1-binding site (UAS1) of the ADH2 promoter (Yu et al. 1989). Snf1 dependence and synergism with reg1Δ was determined in strains carrying the reporter and either wild-type Adr1 or Adr1c. The activity of the reporter in the REG1 strain with wild-type Adr1 was very low in repressing conditions and increased 260-fold in derepressing conditions (Table 2). The Adr1c allele showed the expected constitutive activity and hyperactivation. In the reg1Δ mutant, wild-type Adr1 showed constitutive β-galactosidase activity to ∼10% of the 8-hr derepressed value in the REG1 strain. The same degree of constitutive activity was seen when Adr1c was the activator in the reg1Δ strain, instead of the synergistic effects seen previously in assays that measured expression from the entire ADH2 promoter (Dombek et al. 1993; Tachibana et al. 2007). In the reg1Δ strain, both the wild-type and Adr1c alleles showed a slight increase in constitutive activity over the REG1 strain, possibly due to the 2- to 3-fold elevated ADR1 transcription that is observed in a reg1Δ mutant (Dombek et al. 1993).
TABLE 2.
Expression from an ADR1-dependent reporter lacking Cat8-binding sites
| β-Galactosidase activitya
|
|||
|---|---|---|---|
| Genotypeb | Activatorc | Rd | DR (8 hr) |
| REG1 | Adr1 | 1.2 (0.7) | 260 (98) |
| REG1 | Adr1-S230A | 21 (10) | 450 (180) |
| reg1Δ | Adr1 | 30 (3.8) | NMe |
| reg1Δ | Adr1-S230A | 36 (7.3) | NM |
R, repressing growth conditions. DR, derepressing growth conditions.
UAS1 reporter activity (pHDY10). Data show average values in Miller units for at least three transformants with standard deviations in parentheses.
The strains are REG1: CKY13; reg1Δ: CKY5; both are adr1Δ∷kanmx.
Wild-type Adr1 is carried on pKD16; Adr1c is on pKD14 (S230A).
Growth conditions are described in materials and methods.
NM, not measured.
We additionally tested the reliance of Adr1c and reg1Δ synergism on Cat8 by deleting CAT8 in an adr1Δ reg1Δ mutant strain and by introducing wild-type Adr1 or Adr1c on plasmids. ADH2 expression was measured by isolating mRNA and performing RT–qPCR. The results indicated that Cat8 was required for the high, glucose-resistant ADH2 expression in the reg1Δ strain when either wild-type Adr1 or Adr1c was the activator (Table 3). Efficient derepression by Adr1c was also abolished in the double cat8Δreg1Δ mutant.
TABLE 3.
Synergistic activation of ADH2 by Adr1creg1Δ is Cat8 dependent
| Genotypea
|
ADH2 mRNA level/ACT1b
|
|||
|---|---|---|---|---|
| ADR1 | CAT8 | REG1 | R | DR |
| Wild type | Wild type | Wild type | 0.0025(0.005) | 2.3 (0.5) |
| Wild type | Wild type | Δ | 0.54 (0.12) | 1.3 (0.3) |
| Wild type | Δ | Wild type | 0.0016 (0.001) | 0.26 (0.02) |
| Wild type | Δ | Δ | 0.004 | 0.0025 |
| Adr1c | Wild type | Wild type | 0.14 (0.03) | 11 (2.5) |
| Adr1c | Wild type | Δ | 1.4 (0.16) | 5.6 (0.2) |
| Adr1c | Δ | Δ | 0.001 (0.0005) | 0.13 (0.02) |
| Adr1c | Δ | Wild type | 0.1 (0.02) | 7.2 (0.38) |
R, repressing growth conditions. DR, derepressing growth conditions.
Strains used are adr1Δ strains CKY13 (CAT8 REG1), CKY23 (cat8Δ REG1), and SRY60 (cat8Δ reg1Δ) with plasmid pKD16 (ADR1 wild type) or pKD14 (Adr1c).
mRNA levels were measured by RT–qPCR as described in materials in methods. Data are the mean of two or three replicates (standard deviation).
Adr1c partially suppresses Cat8 dependence at genes activated by both Adr1- and Cat8:
In addition to showing that Cat8 was required for the constitutivity conferred by reg1Δ, the results with the cat8Δ strains indicated that ADH2 expression mediated by Adr1c in the presence of Reg1 was largely independent of Cat8 (Table 3: compare Adr1c in the Cat8 wild type and Cat8Δ rows). This was in contrast to the strong Cat8 dependence when wild-type Adr1 was the activator. Thus, Adr1c acts independently of, and even rescues, the nonactivating ADH2 defect of a cat8Δ mutation. In a previous study, we found that, at some promoters regulated by both Adr1 and Cat8 such as ADH2, overexpression of Adr1 partially compensates for the loss of Cat8 (Biddick et al. 2008), suggesting somewhat redundant functions for Adr1 and Cat8 at coregulated promoters. We tested whether Adr1c in single copy could suppress the requirement for Cat8 as overexpressed wild-type Adr1 does. Expression of Adr1- and Cat8-dependent genes was measured by RT–qPCR of RNA from isogenic adr1Δ CAT8 and adr1Δ cat8Δ strains with either wild-type Adr1 or Adr1c on plasmids and grown in repressing and derepressing conditions.
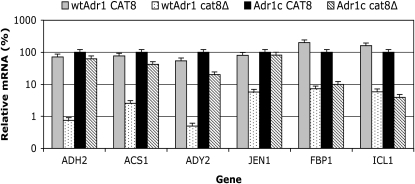
The mRNA levels of several Adr1- and Cat8-dependent genes in derepressing conditions are shown in Figure 4. Table S1 has data from both repressed and derepressed conditions for Adr1- and Cat8-dependent genes, which is expressed in a manner that allows comparison of mRNA levels between genes. The dependence on Cat8 was markedly reduced for ADH2, ACS1, ADY2, and JEN1 when Adr1c was the activator compared to the strong Cat8 dependence of expression of these genes when wild-type Adr1 was the activator. These results were confirmed for ADH2 by reporter assays (Figure S3A) and by in-gel staining of ADH activity in cell extracts (Figure S3B). Genes dependent mainly on Cat8, such as FBP1 and ICL1, were unaffected by Adr1c (Figure 4). In summary, Adr1c efficiently suppressed the Cat8 requirement for numerous genes whose derepression is normally codependent on Adr1 and Cat8. These results are consistent with those in the previous section that documented the partial Snf1 independence of Adr1c.
Figure 4.—
Adr1c suppresses cat8Δ-associated defects in Adr1-dependent gene expression. Cell growth is described in materials and methods. RT–qPCR was performed and analyzed as described in materials and methods. Strains were wild type (wt) CKY13 (adr1Δ) and cat8Δ CKY23 (adr1Δ cat8Δ), each carrying either wild-type ADR1 (pKD16) or ADR1-S230A (Adr1c, pKD14). Derepression was for 4 hr in 0.05% glucose–3% glycerol. The mRNA values were normalized to ACT1 mRNA and are expressed as the percentage of the mRNA level in the wild-type CAT8 strain with Adr1c as the activator. The values represent the mean ± one standard deviation of three biological replicates assayed in duplicate.
Protein synthesis is required to regulate Adr1-pSer230 levels:
The importance of pSer230 and the Ser230 domain was demonstrated by the activity of Adr1c, even in low glucose and even without its coregulator Cat8. To address the mechanism by which Ser230 phosphorylation levels are regulated, we measured phosphorylation levels of Adr1-Ser230 in the absence of protein synthesis. The amount of total and Ser230-phosphorylated Adr1 was determined by Western blotting. In the presence or absence of glucose after the addition of cycloheximide (Cyh), the Adr1-pSer230 and total Adr1 decreased at the same rate, suggesting that Ser230-phosphorylated Adr1 and total Adr1 turn over at the same rate in both repressing and derepressing growth conditions in the absence of protein synthesis (data not shown).
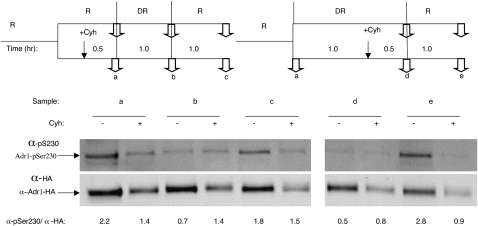
In a more complex experiment diagrammed in Figure 5, the inhibitor was added to either repressed (lanes a–c, +Cyh) or derepressed cultures (samples in lanes d and e, +Cyh), and samples were removed at various times to assay for total Adr1 and Adr1-pSer230. Control cultures lacking the inhibitor were sampled at the same time. In the absence of Cyh, the level of phosphorylation decreased 3- and 4.5-fold after 1 and 1.5 hr of derepression, respectively (compare samples in lanes a, b, and d, no Cyh). Adding glucose back into the control derepressed cultures resulted in a return to the repressed level of phosphorylation (samples in lanes c and e, −Cyh). However, if protein synthesis was inhibited in repressing conditions (left panels) before the culture was shifted to derepressing conditions, the relative level of Adr1-pSer230 did not decrease (compare samples in lanes a and b, +Cyh). Moreover, in these conditions, the relative level of Adr1-pSer230 did not change when glucose was added back (compare samples in lanes a and c, +Cyh). Similarly, if protein synthesis was inhibited in derepressing conditions (right panels) and then the culture was shifted to repressing conditions, the relative level of Adr1-pSer230 did not increase (compare samples in lanes d and e, +Cyh). These results suggest that synthesis of a phosphatase, a kinase, or new Adr1 is needed to modulate the ratio of Adr1-pSer230 to total Adr1 when cells encounter different glucose concentrations.
Figure 5.—
Stability of Adr1-HA and Adr-pSer230 in the absence of protein synthesis. Strain NKY85 was grown in complete medium containing glucose at a final concentration of 5% (R, repressing; sample lanes a–c) At the time indicated by the vertical bars (top), a portion of the culture was shifted to derepressing (DR) conditions (sample lanes d and e) by centrifugation and resuspension in low-glucose medium. In the cases indicated, glucose was added to the derepressed cultures at the times indicated by the vertical bars to reinstate repressive conditions. Cyh was added to a final concentration of 100 μg/ml at the times indicated by a vertical arrow to a portion of the culture to inhibit protein synthesis. Samples were removed for electrophoresis and Western blotting at the points indicated by two arrows at a–e. The signal intensity was measured as described in materials and methods for several biological replicates. The values shown are the mean ± one standard deviation.
Screening for the Adr1-Ser230 kinase:
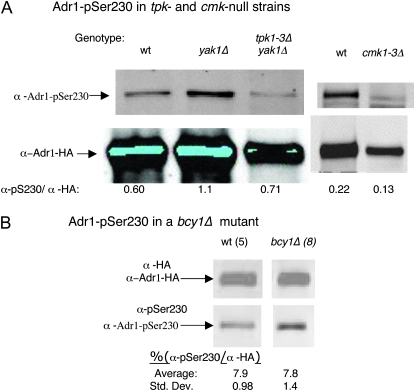
A peptide representing the Ser230 region of Adr1 is an efficient substrate in vitro for both PKA (Denis et al. 1992) and CMK (Hook et al. 1999). Early studies suggested that Adr1 was inactivated in vivo by phosphorylation of Ser230 by PKA (Cherry et al. 1989), but genetic analysis was less supportive of this model (Denis et al. 1992; Dombek and Young 1997). To determine whether Ser230 is phosphorylated in vivo by these protein kinases, we used yeast strains lacking all PKA activity (tpk1Δtpk2Δtpk3Δyak1Δ and tpk1Δtpk2Δtpk3-ts) or all CMK activity (cmk1Δcmk2Δcmk3Δ). The tpk-null strain was kept viable by deleting YAK1, encoding another protein kinase (Garrett and Broach 1989). This strain showed normal glucose regulation of ADH2 (data not shown), consistent with previous studies with a tpk1Δtpk2Δtpk3-ts mutant (Vallari et al. 1992; Dombek and Young 1997). The triple cmk-null strain was viable but grew slowly, and its ADH2 regulation was also normal (data not shown). We expected pSer230 to be absent if the mutations eliminated the Ser230 kinase, but phosphorylated Adr1 was clearly detectable in strains lacking either PKA or CMK. The relative level of Adr1-pSer230 compared to total Adr1 was measured by Western blotting in strains containing overexpressed Adr1 and was found to be altered by a factor of less than two for both PKA and CMK mutant strains (Figure 6A). The 30% reduction in the cmk1-3Δ strain may be a consequence of the partial degradation of Adr1 that occurred in this strain. A similar result was obtained in a tpk1Δtpk2Δtpk3-ts mutant (N. Kacherovsky, unpublished results).
Figure 6.—
Adr1-pSer230 phosphorylation is not significantly altered in tpkΔ and cmkΔ mutants or a bcy1Δ mutant. Preparation of cell extracts, electrophoresis, and Western blotting were carried as described in materials and methods and in the legend to Figure 1. (A) Wild type (wt) is CMY323, yak1Δ is SGP400, tpk1Δ tpk2Δ tpk3Δ yak1Δ is SGP406, cmk1Δ cmk2Δ cmk3Δ is NKY108. All three strains were transformed with plasmid pNKA1, which has HA-tagged Adr1 expressed from the ADH1 promoter on a multicopy TRP1 plasmid. Cells were grown in trp− synthetic medium with 5% glucose (repressing conditions). Fifty micrograms of protein extract was analyzed. (B) Wild type is NKY85 and bcy1Δ is NKY111, both of which are multicopy ADR1. Protein extracts (25 μg wild type, 100 μg bcy1Δ) were analyzed as described above. The numbers in parentheses indicate the number of replicates analyzed and represent both repressed (complete medium with 5% glucose) and derepressed (2, 4, and 22 hr in complete medium with 0.05% glucose–2% ethanol) cultures.
The presence of Adr1 phosphorylated on Ser230 in the tpk-null strain was not due to overexpression. Adr1-pSer230 was also detected in the tpk1-3ΔyakΔ strain with single-copy Adr1 after immunoprecipitation. The immune precipitates from wild type and the quadruple mutant were analyzed by SDS–PAGE and Western blotting for Adr1. The Licor secondary antibody revealed a similar amount of Adr1 in both immune precipitates (Figure S4). The low concentration of Adr1-pSer230 in the supernatant fraction precluded its detection.
As a second method of testing for in vivo phosphorylation of pSer230 by PKA, we measured the relative level of Adr1-pSer230 in a strain lacking Bcy1, the regulatory subunit of PKA. The bcy1Δ mutation was introduced into the same multicopy strain used to study pSer230 in the mutants described in Figures 2 and 5. The total amount of Adr1, as determined by blotting for the HA epitope on Adr1, was lower in the bcy1Δ strain than in the wild-type parent, as expected, because constitutive activation of Tpk reduces expression from the ADR1 promoter (Dombek 1997). We observed a slow rate of ADH2 derepression, measured by both in situ staining of ADH activity and by reporter assays (Figure S5), most likely because of the lower levels of Adr1. If PKA phosphorylates Ser230 in vivo, we expected to see a significant increase in Adr1-pSer230 in this strain. However, the relative level of Adr1-pSer230 was unaffected in the bcy1Δ mutant (Figure 6B).
We considered the possibility that Adr1-pSer230 might already be fully phosphorylated in repressing conditions (Figure S1). If so, the effect of the constitutive activation of PKA in the bcy1Δ strain might be observed only in derepressing conditions, when Adr1-pSer230 decreases in the BCY1 parent strain (Figure 2). However, loss of BCY1 did not increase the relative level of pSer230 in either repressing or derepressing conditions (Figure 6B and data not shown). This result is unlikely to be a consequence of the reduced level of Adr1 in the bcy1Δ mutant because the level of Adr1 in the multicopy bcy1Δ strain is similar to the level of Adr1 in a single-copy ADR1 BCY1 wild-type strain.
In combination with the results from the PKA triple-mutant strains, the results of two different approaches suggested that PKA is not the main kinase of Adr1 Ser230 in vivo. Adr1-Ser230 was clearly phosphorylated in a CMK mutant, so we concluded that neither PKA nor CMK is the main Adr1-Ser230 kinase. PKA or CMK might have partial or nonspecific effects on Adr1 phosphorylation.
Because we were unable to demonstrate a major reduction in Adr1-pSer230 in mutants lacking PKA and CMK, we screened 102 viable kinase deletion mutants (as designated by the Saccharomyces Genome Database) for their ability to phosphorylate Adr1-Ser230. Each mutant was transformed with a high-copy plasmid overexpressing wild-type Adr1 to facilitate the screen. Immunoprecipitation with anti-Adr1 antisera, followed by Western blotting with α-pSer230, was used to detect the phosphorylated protein. No mutants that were defective in phosphorylating Adr1-Ser230 were identified (data not shown).
DISCUSSION
Although Ser230 is part of a canonical consensus sequence for PKA and is phosphorylated by PKA in vitro (Denis et al. 1992), two different methods failed to confirm PKA as the Ser230-specific kinase. In addition, a mutant lacking three yeast genes encoding Cmk homologs had only a minor effect on Ser230 phosphorylation. A screen of 102 viable yeast protein kinase mutants failed to identify a candidate kinase for Ser230. In the experiments to test Ser230 kinase candidates, overexpression of Adr1 was necessary to quantify the level of Adr1-pSer230. Although we cannot rule out the possibility that nonspecific phosphorylation of the overexpressed protein masked identification of the important kinase, the wild-type control with the same plasmid did not show excessive or nonspecific phosphorylation, making this explanation unlikely. Moreover, in a strain expressing normal low levels of Adr1, Ser230 phosphorylation appeared to be present at the same level in wild-type and tpk-null strains. In vivo phosphorylation of Ser230 might involve both Cmk and Tpk, but unregulated Tpk activity did not significantly increase the level of Adr1-pSer230, making this possibility less likely. Therefore, we tentatively conclude that either Ser230 can be phosphorylated by multiple protein kinases that may include PKA and CMK or its specific kinase is essential or absent from our deletion collection for another reason. Protein synthesis appeared to be necessary to change the relative level of Adr1-pSer230. This could be interpreted to indicate that an Adr1-specific protein kinase must be synthesized as cells return to repressing growth conditions or that a protein phosphatase must be synthesized de novo when cells are shifted from repressing to derepressing conditions.
Our results are consistent with the original hypothesis that phosphorylation of Adr1 on Ser230 plays an important role in Adr1 function by altering its ability to regulate downstream genes in a glucose-dependent manner (Cherry et al. 1989). Further investigations are required to address this issue and the role of Snf1 in modulating the level of Ser230 phosphorylation. Regardless of the mechanism, regulating the level of Adr1 phosphorylated on Ser230 appears to be a major function of Snf1 in controlling the expression of Adr1-dependent genes because a nonphosphorylatable Adr1c can activate both Adr1-dependent genes and a UAS1-containing reporter without Snf1. In contrast, wild-type Adr1 was unable to activate the same genes in the absence of Snf1. This is consistent with our interpretation that pSer230 is a major, albeit indirect, target by which Snf1 regulates Adr1 activity. The remaining Snf1 dependence when Adr1c is the activator of genes uniquely dependent on Adr1 could be a function of other Snf1-dependent pathways that are important for activation by Adr1 (Voronkova et al. 2006) or could reflect the requirement for Snf1 in the dephosphorylation of pSer98. This site is adjacent to the DNA-binding domain of Adr1, and Ser98 phosphorylation appears to play a repressive role in Adr1 activity by reducing Adr1 promoter binding (Kacherovsky et al. 2008). Like phosphorylation of Ser230, phosphorylation of Ser98 is modulated by Snf1 activity and is reduced in derepressing growth conditions. Whether the two modifications are causally or functionally related is under investigation.
We previously suggested that Adr1c and Snf1 act on ADH2 expression through separate pathways (Dombek et al. 1993) on the basis of the strong synergism between ADR1c and the deletion of REG1. The data presented here indicate that the synergism is due to Cat8. Thus the primary effect of Snf1 on ADH2 expression in a strain carrying ADR1c is due to Cat8 whose expression and activity are Snf1 dependent. The enhanced activity of Adr1c may be related to the observation that it activated genes normally codependent on Adr1 and Cat8 in the absence of Cat8. Eukaryotic promoters generally respond to different environmental signals through the action of multiple transcription factors, leading to combinatorial control of gene expression. Many Adr1-dependent genes are activated by other transcription factors as well. For example, the promoters of genes encoding peroxisomal proteins and the enzymes of β-oxidation bind both Adr1 and the heterodimeric, oleate-responsive transcription factors Oaf1 and Pip2 (Simon et al. 1991, 1992; Gurvitz et al. 2000, 2001; Young et al. 2003). We are investigating whether Adr1c can compensate for the loss of Oaf1/Pip2 in the expression of peroxisomal genes, as it does for the loss of Cat8 at Adr1- and Cat8-dependent promoters.
The finding that Adr1c can suppress the requirement for Cat8 suggests that, in spite of its low protein levels, Adr1c has characteristics similar to overexpressed wild-type Adr1, which also escapes Snf1 regulation (Denis 1987) and also compensates for loss of Cat8 at coregulated promoters (Biddick et al. 2008). In vitro, by electrophoretic mobility shift assays, Adr1c does not have exceptional DNA-binding activity (Taylor and Young 1990); however, it may be able to bind promoters efficiently in vivo, where factors such as chromatin structure and stabilization by coactivator interaction are proposed to influence Adr1 binding (Tachibana et al. 2007; Young et al. 2008). The wide variety of constitutive Adr1 mutations, such as a complete deletion of amino acids 220–263 and 262–330 (Cook et al. 1994), suggests that not only the Ser230 phosphorylation site, but also the entire region is important for Adr1 regulation. Our results are consistent with a model that mutations in this area affect protein structure or alter interaction with other factors (Cook et al. 1994). Although no Adr1-specific repressor has been found in spite of several genetic screens, these findings about Adr1c suggest a model that can drive further investigation of the novel interaction between Adr1 and Snf1.
Acknowledgments
We thank other members of our lab for advice, encouragement, and reagents; Ella Chang for performing some of the quantitative RT–qPCR analyses; Catherine Kehl and Kenneth Dombek for strain construction; Carl Mann for strains; and Chris Tachibana for valuable comments on the manuscript. This work was supported by a grant (no. GM26079) from the National Institutes of Health to E.T.Y.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103432/DC1.
References
- Biddick, R. K., G. L. Law and E. T. Young, 2008. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS ONE 3 e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon, G., K. D. Breunig, R. Wattiez, J. Vandenhaute and I. Noel-Georis, 2004. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 24 4083–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. R., T. R. Johnson, C. Dollard, J. R. Shuster and C. L. Denis, 1989. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 56 409–419. [DOI] [PubMed] [Google Scholar]
- Chi, A., C. Huttenhower, L. Y. Geer, J. J. Coon, J. E. Syka et al., 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 104 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy, M., 1979. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol. Gen. Genet. 176 427–431. [DOI] [PubMed] [Google Scholar]
- Collart, M., and S. Oliveiro (Editors), 1993. Current Protocols in Molecular Biology. Greene/Wiley Interscience, New York.
- Cook, W. J., D. Chase, D. C. Audino and C. L. Denis, 1994. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol. Cell. Biol. 14 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13 647–653. [DOI] [PubMed] [Google Scholar]
- Denis, C. L., 1984. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 108 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, C. L., 1987. The effects of ADR1 and CCR1 gene dosage on the regulation of the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. Mol. Gen. Genet. 208 101–106. [DOI] [PubMed] [Google Scholar]
- Denis, C. L., and C. Gallo, 1986. Constitutive RNA synthesis for the yeast activator ADR1 and identification of the ADR1–5c mutation: implications in posttranslational control of ADR1. Mol. Cell. Biol. 6 4026–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, C. L., B. E. Kemp and M. J. Zoller, 1991. Substrate specificities for yeast and mammalian cAMP-dependent protein kinases are similar but not identical. J. Biol. Chem. 266 17932–17935. [PubMed] [Google Scholar]
- Denis, C. L., S. C. Fontaine, D. Chase, B. E. Kemp and L. T. Bemis, 1992. ADR1c mutations enhance the ability of ADR1 to activate transcription by a mechanism that is independent of effects on cyclic AMP-dependent protein kinase phosphorylation of Ser-230. Mol. Cell. Biol. 12 1507–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek, K. M., and E. T. Young, 1997. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol. Cell. Biol. 17 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek, K. M., S. Camier and E. T. Young, 1993. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol. Cell. Biol. 13 4391–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone, M., 2002. Coactivators in transcription initiation: here are your orders. Curr. Opin. Genet. Dev. 12 149–155. [DOI] [PubMed] [Google Scholar]
- Fry, C. J., and C. L. Peterson, 2002. Transcription. Unlocking the gates to gene expression. Science 295 1847–1848. [DOI] [PubMed] [Google Scholar]
- Garrett, S., and J. Broach, 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3 1336–1348. [DOI] [PubMed] [Google Scholar]
- Goel, A., and R. Janknecht, 2003. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol. Cell. Biol. 23 6243–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L., 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101 181–191. [DOI] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fiedler, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz, A., L. Wabnegger, H. Rottensteiner, I. W. Dawes, A. Hartig et al., 2000. Adr1p-dependent regulation of the oleic acid-inducible yeast gene SPS19 encoding the peroxisomal beta-oxidation auxiliary enzyme 2,4-dienoyl-CoA reductase. Mol. Cell Biol. Res. Commun. 4 81–89. [DOI] [PubMed] [Google Scholar]
- Gurvitz, A., J. K. Hiltunen, R. Erdmann, B. Hamilton, A. Hartig et al., 2001. Saccharomyces cerevisiae Adr1p governs fatty acid beta-oxidation and peroxisome proliferation by regulating POX1 and PEX11. J. Biol. Chem. 276 31825–31830. [DOI] [PubMed] [Google Scholar]
- Hahn, S., 2008. Rapid yeast protein prep for SDS–PAGE and Western. http://www.fhcrc.org/science/labs/hahn/methods/biochem_meth/yeast_prot_SDS.html).
- Hardie, D. G., D. Carling and M. Carlson, 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855. [DOI] [PubMed] [Google Scholar]
- Hedges, D., M. Proft and K. D. Entian, 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15 1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. P., F. C. Leiper, A. Woods, D. Carling and M. Carlson, 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100 8839–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook, S. S., B. E. Kemp and A. R. Means, 1999. Peptide specificity determinants at P-7 and P-6 enhance the catalytic efficiency of Ca2+/calmodulin-dependent protein kinase I in the absence of activation loop phosphorylation. J. Biol. Chem. 274 20215–20222. [DOI] [PubMed] [Google Scholar]
- Kacherovsky, N., C. Tachibana, E. Amos, D. Fox, III and E. T. Young, 2008. Promoter binding by the Adr1 transcriptional activator may be regulated by phosphorylation in the DNA-binding region. PLoS ONE 3 e3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 963–972. [DOI] [PubMed] [Google Scholar]
- Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger et al., 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370 223–226. [DOI] [PubMed] [Google Scholar]
- Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane et al., 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293 1142–1146. [DOI] [PubMed] [Google Scholar]
- McCartney, R. R., and M. C. Schmidt, 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276 36460–36466. [DOI] [PubMed] [Google Scholar]
- Orlova, M., L. Barrett and S. Kuchin, 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25 745–754. [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou, S., and R. Janknecht, 2000. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol. Cell. Biol. 20 7300–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahner, A., A. Scholer, E. Martens, B. Gollwitzer and H. J. Schuller, 1996. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 24 2331–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahner, A., M. Hiesinger and H. J. Schuller, 1999. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34 146–156. [DOI] [PubMed] [Google Scholar]
- Rubenstein, E. M., R. R. McCartney, C. Zhang, K. M. Shokat, M. K. Shirra et al., 2008. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 283 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, P., G. R. Alms, T. A. Haystead and M. Carlson, 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller, H. J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43 139–160. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194 3–21. [DOI] [PubMed] [Google Scholar]
- Simon, M., G. Adam, W. Rapatz, W. Spevak and H. Ruis, 1991. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol. Cell. Biol. 11 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M., M. Binder, G. Adam, A. Hartig and H. Ruis, 1992. Control of peroxisome proliferation in Saccharomyces cerevisiae by ADR1, SNF1 (CAT1, CCR1) and SNF4 (CAT3). Yeast 8 303–309. [DOI] [PubMed] [Google Scholar]
- Sloan, J. S., K. M. Dombek and E. T. Young, 1999. Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J. Biol. Chem. 274 37575–37582. [DOI] [PubMed] [Google Scholar]
- Tachibana, C., R. Biddick, G. L. Law and E. T. Young, 2007. A poised initiation complex is activated by SNF1. J. Biol. Chem. 282 37308–37315. [DOI] [PubMed] [Google Scholar]
- Tansey, W., 2001. Transcriptional activation: risky business. Genes Dev. 15 1045–1050. [DOI] [PubMed] [Google Scholar]
- Taylor, W. E., and E. T. Young, 1990. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc. Natl. Acad. Sci. USA 87 4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, J., and M. Carlson, 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallari, R. C., W. J. Cook, D. C. Audino, M. J. Morgan, D. E. Jensen et al., 1992. Glucose repression of the yeast ADH2 gene occurs through multiple mechanisms, including control of the protein synthesis of its transcriptional activator, ADR1. Mol. Cell. Biol. 12 1663–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone, L., J. Wu, K. van Riper, N. Kacherovsky, M. Vogelauer et al., 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronkova, V., N. Kacherovsky, C. Tachibana, D. Yu and E. T. Young, 2006. Snf1-dependent and Snf1-independent pathways of constitutive ADH2 expression in Saccharomyces cerevisiae. Genetics 172 2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, E. T., N. Kacherovsky and K. van Riper, 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcriptional activation. J. Biol. Chem. 277 38095–38103. [DOI] [PubMed] [Google Scholar]
- Young, E. T., K. M. Dombek, C. Tachibana and T. Ideker, 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278 26146–26158. [DOI] [PubMed] [Google Scholar]
- Young, E. T., C. Tachibana, H. W. Chang, K. M. Dombek, E. M. Arms et al., 2008. Artificial recruitment of mediator by the DNA-binding domain of Adr1 overcomes glucose repression of ADH2 expression. Mol. Cell. Biol. 28 2509–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., M. S. Donoviel and E. T. Young, 1989. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 9 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, H., M. J. May, E. Jimi and S. Ghosh, 2002. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell 9 625–636. [DOI] [PubMed] [Google Scholar]