Abstract
Epigenetic regulation of transcriptional silencing is essential for normal development. Despite its importance, in vivo systems for examining gene silencing at cellular resolution have been lacking in developing vertebrates. We describe a transgenic approach that allows monitoring of an epigenetically regulated fluorescent reporter in developing zebrafish and their progeny. Using a self-reporting Gal4-VP16 gene/enhancer trap vector, we isolated tissue-specific drivers that regulate expression of the green fluorescent protein (GFP) gene through a multicopy, upstream activator sequence (UAS). Transgenic larvae initially exhibit robust fluorescence (GFPhigh); however, in subsequent generations, gfp expression is mosaic (GFPlow) or entirely absent (GFPoff), despite continued Gal4-VP16 activity. We find that transcriptional repression is heritable and correlated with methylation of the multicopy UAS. Silenced transgenes can be reactivated by increasing Gal4-VP16 levels or in DNA methyltransferase-1 (dnmt1) mutants. Strikingly, in dnmt1 homozygous mutants, reactivation of gfp expression occurs in a reproducible subset of cells, raising the possibility of different sensitivities or alternative silencing mechanisms in discrete cell populations. The results demonstrate the power of the zebrafish system for in vivo monitoring of epigenetic processes using a genetic approach.
THE ability to constrain transcription is fundamental to genome maintenance. Underscoring its importance, several conserved mechanisms maintain repressive chromatin at transposons, repetitive sequences, and gene promoters (Hsieh and Fire 2000; Bernstein et al. 2007; Girard and Hannon 2008). Histone modifications, small RNAs, and chromosome organization within the nucleus contribute to transcriptional silencing (Strahl and Allis 2000; Heard and Bickmore 2007; Girard and Hannon 2008). Although basic components of the silencing machinery are conserved, evolution of transposons has driven organisms to evolve specialized responses. For example, DNA methylation is required for silencing in plants, vertebrates, and some fungi, but is absent from other species, including many invertebrates (Goll and Bestor 2005).
Epigenetic regulation of transcription is essential for normal development. Mouse embryos lacking DNA methyltransferase or histone-modifying enzymes are abnormal (Li et al. 1992; Peters et al. 2001; Tachibana et al. 2002). Loss of methylation leads to H2AX phosphorylation, ATM activation, and initiation of the G2/M checkpoint in cell culture (Chen et al. 2007). Similarly, p53-dependent apoptosis is observed in embryos lacking DNA methyltransferase-1 (Dnmt1) (Jackson-Grusby et al. 2001; Stancheva et al. 2001). Although these observations implicate programmed cell death as an underlying cause of lethality in dnmt1 mutants, some evidence suggests that not all tissues are similarly affected by the loss of DNA methylation. For example, in zebrafish, methylation of DNA and histone H3K9 appear to be required for terminal differentiation of the intestine, exocrine pancreas, and retina, but not of the endocrine pancreas (Rai et al. 2006). Similarly, hypomethylation-induced cell death was observed only in some Xenopus tissues at early stages of development (Stancheva et al. 2001). Tissue-specific differences in expression of chromatin-associated factors have been reported in mice and zebrafish, suggesting that gene requirements for silencing may not be equivalent in all cells (Jarvis et al. 1996; Bourc'his et al. 2001; Rai et al. 2006; Sun et al. 2008).
Transcriptional silencing and methylation of transgenes have been studied in many organisms (for example: Sapienza et al. 1989; Stuart et al. 1990; Weber et al. 1990; Kilby et al. 1992; Matzke et al. 1994; and references in Dorer 1997). Transgenes containing the LacZ reporter allow transcriptional silencing to be visualized in dissected tissues and fixed embryos (Allen et al. 1990; Sutherland et al. 2000; Chevalier-Mariette et al. 2003; Strathdee et al. 2008). In the mouse, a dominant screen for new genes involved in epigenetic regulation relied on fluorescence-activated cell sorting of blood cells expressing a green fluorescent protein (GFP) transgene (Blewitt et al. 2005; Ashe et al. 2008). While such approaches have yielded useful information, transgenic tools to monitor silencing in live developing embryos have been lacking.
The zebrafish is highly amenable to such an in vivo strategy. The transparency of the embryo permits visualization of fluorescent reporters at the level of individual cells throughout development and over multiple generations. In addition, a short generation time and large clutches of embryos facilitate unbiased genetic screens.
A gene/enhancer trap system was previously developed that integrates a self-reporting Gal4-VP16; 14× upstream activator sequence (UAS):GFP vector into the zebrafish genome by Tol2 transposition (Kawakami 2004; Davison et al. 2007). Although recovered transgenic lines initially exhibit robust tissue-specific patterns of GFP labeling, mosaic expression is observed in subsequent generations.
In this report, we systematically characterize the onset and progression of GFP silencing using a newly identified gene trap insertion that shows brain-specific expression. Through the generation of independent epiallelic lines with high (GFPhigh), reduced (GFPlow), or no GFP (GFPoff) labeling, we demonstrate that the 14× UAS is subject to DNA methylation, which correlates with the heritable loss of gfp expression. We examine the impact of Gal4-VP16 activity on methylation and demonstrate that mutation of the dnmt1 gene overrides transcriptional repression. Reactivation of gfp expression in dnmt1 mutants occurs only in a stereotypic region of the brain, raising the possibility that discrete cell populations possess different sensitivities to silencing mechanisms. The results highlight the value of an in vivo system for monitoring epigenetic regulation of transcription in developing tissues.
MATERIALS AND METHODS
Zebrafish strains and lines:
Zebrafish were raised under standard conditions at 27° (Westerfield 2000). The splice acceptor-Gal4-VP16, UAS:GFP (SAGVG) construct (Davison et al. 2007) and Tol2 transposase mRNA (Kawakami 2005) were injected into one-cell embryos of the AB strain (Walker 1999), and injected G0 larvae were raised to adulthood. Adults were mated with AB fish, and the resultant F1 progeny were screened for restricted patterns of GFP labeling. One F1 founder male generated larvae with GFP labeling in the forebrain, midbrain retina, and rhombomeres 5–6 (r5–6) and was designated as carrying the c269 allele. Transgenic F2 adults carrying a single-gene trap insertion were used to establish the c269 epiallelic lines. Additional Gt(Gal4-VP16;UAS:EGFP) gene/enhancer trap lines and Tg(UAS-E1b:NfsB-mCherry)c264/+ were previously described (Davison et al. 2007) and renamed according to nomenclature conventions (http://zfin.org/zf_info/nomen.html). The N-ethyl-N-nitrosourea (ENU)-induced mutation dnmt1s872 (mixed AB, TU, and TL background) was identified in a mutagenesis screen and is a G-to-A transition at nucleotide 4376 (R. Anderson and D. Stainier, unpublished results). To identify dnmt1s872 homozygotes, genomic DNA was isolated from individual larvae and polymerase chain reaction (PCR) amplified using the primers GCCTACATGCCATATGCTGA and TCTCCTGCTTCACAGGCTCT.
Assessment of transgene copy number:
Genomic DNA (2.5 μg) was isolated from fin clips of c269 F2 adults that had been sorted for GFPhigh or GFPlow labeling at larval stages and reared separately. DNA was digested with BamHI and subjected to Southern blotting (Southern 1975). Membranes were hybridized with radiolabled GFP coding sequence.
Transmission of epialleles:
Progeny from the c269 F1 founder male were scored for high, low, or no GFP labeling at 3 days post fertilization (dpf). Sorted groups of F2 larvae were raised separately to adulthood and mated to AB adults. Resultant F3 larval progeny were scored for GFP labeling. GFPoff larvae that maintained the c269 transgene insertion were identified by mating F2 GFPlow adults with 14× UAS:mCherry transgenic fish and raised to adulthood to establish the stable, nonexpressing c269 GFPoff epiallelic line.
Methylation-sensitive Southern blots:
Genomic DNA (5 μg) from pools of ∼20 GFPhigh or GFPoff larvae was digested with SacI and then divided, with one-half further digested by HpaII. Following digestion, samples were subjected to Southern blotting (Southern 1975). Membranes were probed with radiolabeled 14× UAS coding sequence amplified by PCR from SAGVG (Davison et al. 2007) using the primers CGGCCATCAAGCTTAGGCTC and TCAAAGTGAGGCGAGACGC.
Bisulfite sequencing:
Individual larval heads and bodies were dissected by cutting anterior to the yolk at 3 dpf. DNA isolation and bisulfite conversion were achieved using the EZ DNA Methylation Direct kit (Zymo Research). Bisulfite converted DNA was amplified using the primers TTTTATGTTTTAGGTTTAGGG and CCCTTACTCACCATAATAAC followed by TATGTTTTAGGTTTAGGGGGA and CCCTTACTCACCATAATAAC. The resultant fragment was purified and sequenced. Analysis and statistical comparison of bisulfite data was performed using QUMA (Kumaki et al. 2008). P-values were calculated using both Fisher's exact test and the Mann–Whitney U-test.
Gal4-VP16 overexpression:
Plasmid DNA (25 pg) encoding Gal4-VP16 under the control of the EF1α promoter was injected into one-cell embryos from GFPoff intercrosses. At 1 dpf, larvae were assessed for GFP-labeled cells under a fluorescent stereomicroscope (Leica MZ16F). DNA was collected from injected larvae that showed the highest levels of gfp reactivation or mock-injected controls and analyzed by bisulfite sequencing.
Quantification of fluorescence intensity in c269 larvae:
Larvae showing the highest number of GFP-labeled cells in r5–6 were imaged for GFP and mCherry fluorescence under a Zeiss Axio Imager.D1 with the ApoTome system using AxioVision digital imaging software. GFP- and mCherry-integrated fluorescent intensities were independently assessed in the mid/forebrain region and in r5–6 using MetaMorph software (Version 7, Molecular Devices). The ratio of fluorescence was calculated for each region by dividing the integrated intensity of GFP by the integrated intensity of mCherry.
RESULTS
gfp expression is epigenetically regulated in transgenic zebrafish lines:
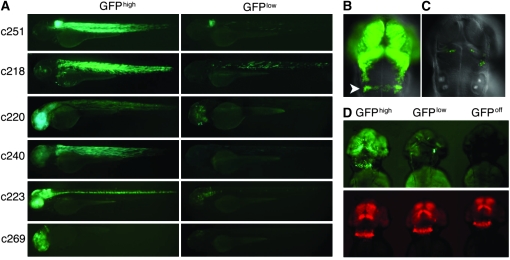
Larvae carrying Gal4-VP16; 14× UAS:GFP gene/enhancer trap insertions exhibit robust tissue-specific patterns of GFP labeling in the F1 generation, but some showed variegated expression (GFPlow) in subsequent generations (Figure 1A). To assess the origins of the variegation systematically, expression from a newly identified insertion was tracked over multiple generations. F1 larvae carrying the c269 insertion showed robust expression of gfp in the retina, forebrain, and midbrain and in a broad stripe in the hindbrain corresponding to rhombomeres (r) 5–6. This pattern was evident at 1 dpf and persisted until at least 7 dpf (data not shown and Figure 1B). In the F2 generation, two classes of transgenic larvae were detected: those with robust GFP labeling that recapitulated the F1 pattern (GFPhigh) and those showing only a small subset of GFP-positive cells spatially restricted within the F1 pattern of labeling (GFPlow) (Figure 1C). Using linker-mediated PCR, the c269 insertion was mapped within the fourth intron of the zebrafish homolog of the odd Oz/ten-m homolog 4 (odz4) gene (data not shown). Brain-specific fluorescence in GFPhigh larvae corresponded with the previously described expression pattern for the odz4 gene (Mieda et al. 1999).
Figure 1.—
Variegated expression of UAS-regulated GFP in zebrafish larvae. (A) Representative larva carrying unique insertions of the gene/enhancer trap vector designated by c numbers. For a given insertion, sibling F2 larvae express gfp either in a pattern similar to the F1 generation (GFPhigh, left panels) or in a smaller subset of cells within this pattern (GFPlow, right panels). Larvae were imaged at 3 dpf. (B) Fluorescent labeling of the forebrain, midbrain, and hindbrain (r5–6, arrowhead) of c269 GFPhigh larvae at 2 dpf. (C) GFPlow c269 sibling larvae show significantly fewer GFP-labeled cells, which are restricted to within the c269 GFPhigh pattern of expression. (D) Corresponding images of GFP (top) and mCherry (bottom) fluorescence in c269 GFPhigh, GFPlow, and GFPoff larvae carrying a newly integrated UAS:mCherry transgene at 2 dpf. Irrespective of the extent of GFP expression, mCherry labeling recapitulates the complete c269 GFPhigh expression pattern.
Differences in GFP labeling between c269 sibling larvae could result from the segregation of multiple independent insertions of the gene/enhancer trap vector, mutation within the transgene, or a decrease in the number of UAS sites due to DNA polymerase slippage. Tol2-mediated trangenesis results in dispersed single integration events rather than large concatomeric arrays (Kawakami et al. 2000). Southern blots revealed GFPhigh and GFPlow individuals that carried only a single insertion of the gene/enhancer trap in the odz4 gene (supporting information, Figure S1). Therefore, differences in transgene copy number cannot account for the mosaicism observed in GFPlow individuals. To rule out mutation of the transgene, DNA fragments encoding Gal4-VP16, the 14× UAS, or GFP were amplified by PCR from GFPlow individuals. Sequencing of the amplified products failed to reveal any mutations and confirmed the presence of 14 intact UAS copies (data not shown). Therefore, GFPhigh and GFPlow larvae have the identical transgene insertion yet are phenotypically distinct, suggesting that the transgene is regulated epigenetically.
Heritability of GFP epialleles:
In mammals, genomewide erasure and reestablishment of epigenetic marks early in development frequently leads to metastable inheritance of silenced epialleles (Rakyan et al. 2002). To test whether epialleles of the c269 transgene were stably inherited, a single c269 F1 adult was mated to wild type. The resultant F2 GFPhigh and GFPlow sibling larvae were separated and independently raised to adulthood. All F2 c269 GFPhigh adults produced both GFPhigh and GFPlow F3 progeny (Table 1). In contrast, F2 c269 GFPlow adults did not produce any GFPhigh progeny and GFPlow F3 larvae were recovered at less than the expected frequency of 50%. F2 GFPlow adults also produced F3 larvae that retained the transgene but did not show any GFP labeling (GFPoff). GFP-labeled cells were never observed in progeny (n > 5000) from GFPoff adults in the F3 or F4 generations (Table 1 and data not shown). Similar inheritance of GFP epialleles was found upon mating other transgenic lines and occurred irrespective of the sex of the parent carrying the transgene (Table 1; Table S1; Table S2). The results indicate that gfp expression is subject to silencing, which, once established, is stably inherited.
TABLE 1.
Inheritance of c269 epialleles
| Class of progeny (%)
|
No. of progeny | |||
|---|---|---|---|---|
| c269/+ × wild type | GFPhigh | GFPlow | No GFP | |
| F1 GFPhigh male | 29 | 23 | 48 | 225 |
| F2 GFPhigh female 1 | 34 | 14 | 52 | 380 |
| F2 GFPhigh male 1 | 45 | 4 | 51 | 84 |
| F2 GFPhigh male 2 | 5 | 5 | 90 | 400 |
| F2 GFPlow female 1 | 0 | 11 | 89 | 163 |
| F2 GFPlow male 1 | 0 | 10 | 90 | 122 |
| F2 GFPlow male 2 | 0 | 1 | 99 | 108 |
| F3 GFPoff male 1 | 0 | 0 | 100 | 215 |
| F3 GFPoff female 1 | 0 | 0 | 100 | 192 |
Strain-specific modifiers that affect transgene expression have been previously reported (Sapienza et al. 1989; Allen et al. 1990; Martin and McGowan 1995). Although the c269 transgene was introduced and maintained in the AB background, this background is unlikely to be homogeneous. To assess whether genetic variation affected expression of the transgene, GFPoff adults (AB) were mated to adults of the WIK, Tubingen (TU), and mixed AB, TU, and Tupfel long fin (TL) genetic backgrounds. Progeny expressing gfp were never observed (data not shown), indicating that genetic background differences do not account for transcriptional silencing.
All transgenic lines express functional Gal4-VP16:
The bipartite nature of the gene trap construct meant that loss of gfp expression could result from the failure to produce functional Gal4-VP16 or of Gal4-VP16 to activate UAS sites. To distinguish between the two possibilities, c269 GFPhigh, GFPlow, and GFPoff adults were mated to adults carrying a newly integrated 14× UAS-regulated mCherry reporter. Expression of mCherry depends on Gal4-VP16 from the trapped transgene. Therefore, if Gal4-VP16 is absent in GFPoff cells, mCherry will not be expressed. Alternatively, if activation of the 14× UAS:GFP is compromised, Gal4-VP16 should still transactivate UAS:mCherry in GFP-negative cells. Labeling of mCherry was observed in GFPhigh, GFPlow, and GFPoff c269 larvae in a pattern characteristic of endogenous odz4 expression (Figure 1D). Robust mCherry fluorescence was also observed in larvae from other transgenic lines that exhibited variegated expression, irrespective of the degree of GFP labeling (Figure S2). Thus, Gal4-VP16 is produced in GFPoff cells at levels sufficient for activation of a second UAS-regulated transgene. These results also suggest that epigenetic modification of the multicopy UAS regulating GFP prevents activation by Gal4-VP16. Further implicating the 14× UAS in silencing, in later generations, some larvae carrying the 14× UAS:mCherry transgene also exhibited mosaic labeling of mCherry (data not shown).
DNA methylation correlates with silencing:
One mechanism for transcriptional silencing is DNA methylation. In vitro methylation of UAS sites was previously shown to prevent Gal4 activation of a luciferase reporter following transfection into cultured cells (Li et al. 2007). The 14× UAS (CGG-N11-CCG) may be particularly susceptible to methylation in vivo because it is a CpG-rich tandem repeat (Giniger et al. 1985). Methylation occurs predominately at CpG dinucelotides in vertebrates, and tandem repeats are frequent targets of DNA methylation (Garrick et al. 1998).
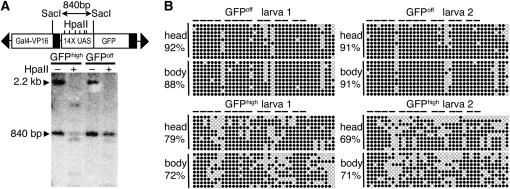
To test for DNA methylation, genomic DNA was collected from pools of c269 GFPhigh or GFPoff heterozygous larvae and digested with SacI to release an 840-bp fragment corresponding to the 14× UAS upstream of gfp. One-half of each sample was further digested with HpaII, which cleaves only at unmethylated CCGG sites. To visualize the fragment containing the UAS, Southern blots were performed on digested DNA and hybridized using a radiolabeled 14× UAS DNA fragment as probe. Full digestion at the six HpaII sites within the UAS results in small fragments that are difficult to detect. Therefore, decreased intensity of the 840-bp fragment was used as a measure of the extent of HpaII digestion. Following digestion with HpaII, the 840-bp fragment from GFPhigh larvae was significantly decreased in intensity when compared to the HpaII-digested sample from GFPoff larvae, suggesting a correlation between methylation of the 14× UAS and loss of gfp expression (Figure 2A).
Figure 2.—
Methylation of the multicopy UAS is correlated with decreased GFP labeling. (A) Genomic DNA from pooled GFPhigh or GFPoff larvae was digested with SacI to produce an 840-bp SacI fragment encompassing the 14× UAS upstream of gfp. The 840-bp SacI fragment has increased sensitivity to HpaII cleavage in GFPhigh larvae compared to GFPoff larvae. Transactivation of a 14× UAS:mCherry transgene was used to identify GFPoff larvae that inherited the c269 transgene. The 14× UAS probe also recognizes the 14× UAS regulating mCherry, although SacI cleavage releases a larger fragment (2.2 kb). This fragment, containing the 14× UAS upstream of mCherry and the mCherry coding sequence, is largely digested, indicating that it is unmethylated in GFPhigh and GFPoff samples. (B) DNA from heads and bodies of individual GFPhigh or GFPoff larvae was subjected to bisulfite sequencing. The methylation status of each CG site is reported as a percentage of total sites tested for 10 clones from the head and 10 clones from the body of each individual. Methylated sites are solid circles; horizontal bars indicate pairs of CpG dinucleotides within individual Gal4-binding sites. The final 8 CpG sites reside within the minimal promoter sequence.
To assess DNA methylation at higher resolution and to determine if differences in methylation could be detected between Gal4-VP16-expressing and -nonexpressing tissues, sodium bisulfite sequencing was performed. Genomic DNA was separately isolated from dissected heads and bodies of individual c269 GFPhigh or GFPoff heterozygous larvae. Following bisulfite treatment, all unmethylated cytosine residues were converted to thymidine while methylated cytosine residues remained unconverted. Differences in methylation were not observed when DNA from Gal4-VP16-expressing tissues of the head were compared to nonexpressing body tissues from the same individual (Figure 2B). However, consistent with the Southern blot data presented in Figure 2A, a statistically significant increase (P < 0.001) in the number of methylated CpG dinucleotides was observed when GFPoff sequences were compared to GFPhigh sequences (Figure 2B). In addition, DNA from GFPhigh larvae showed a higher number of clustered unmethylated sites, generally including at least one fully unmethylated Gal4-binding site. Fully unmethylated Gal4-binding sites were not observed in GFPoff larvae (Figure 2B). Consistent with the heritability of the GFPoff epiallele, DNA methylation patterns in sperm from GFPoff adults were very similar to those in sperm from GFPoff larvae (average of 88.5% of CpG's methylated; Figure S3). Collectively, the bisulfite sequencing data and Southern blot data indicate a correlation between methylation and silencing of gfp expression and suggest that methylation directs the stable inheritance of GFPoff.
Reactivation of gfp by exogenous Gal4-VP16:
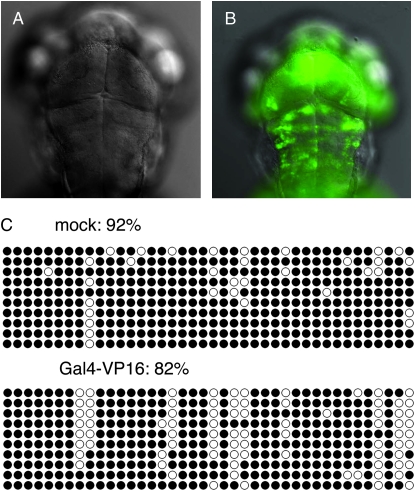
The UAS remains methylated in GFPoff larvae despite the presence of Gal4-VP16. In addition, differences in DNA methylation were not observed between Gal4-VP16-expressing and -nonexpressing tissues in GFPhigh and GFPlow individuals. However, previous reports indicate that DNA-binding proteins can trigger demethylation of recognition sites (Matsuo et al. 1998; Lin et al. 2000). To test whether higher levels of Gal4-VP16 could cause UAS demethylation and reactivation of gfp expression, GFPoff larvae were injected with plasmid DNA containing Gal4-VP16 under the control of a ubiquitous promoter. Labeled cells were not detected in mock-injected controls or in controls injected with an unrelated plasmid (Figure 3A). In contrast, GFP labeling was detected throughout the entire larva in the presence of excess Gal4-VP16 (Figure 3B). Reactivation of gfp in Gal4-VP16-injected larvae correlated with a modest decrease in DNA methylation as assessed by bisulfite sequencing (Figure 3C). Therefore, increasing the levels of Gal4-VP16 can reactivate gfp expression, likely through partial demethylation of the UAS.
Figure 3.—
Reactivation of GFP expression by exogenous Gal4-VP16. (A) Mock-injected larvae from c269 GFPoff intercrosses show no GFP labeling (n = 100). (B) GFPoff siblings injected with ∼25 pg of plasmid DNA encoding the EF1α ubiquitous promoter driving Gal4-VP16 expression are extensively labeled with GFP (98%; n = 60). (C) DNA from injected or mock-injected larvae was subjected to bisulfite sequencing, and the methylation status of each CG site was reported as a percentage of the total sites tested for 10 clones. Methylated sites are solid circles. Data are representative of two biological replicates.
Mutation of dnmt1 reactivates selective gfp expression:
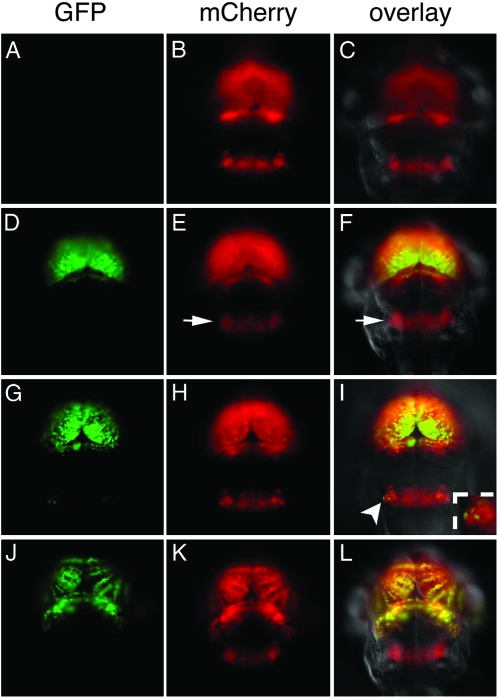
If methylation is responsible for suppression of UAS-regulated gene expression, then blocking the methylation process should lead to reactivation of gfp expression. Zebrafish homozygous for an ENU-induced G1459D amino acid change in the AdoMet-binding motif X of DNA methyltransferase-1 (dnmt1s872) show morphological abnormalities and a reduction in global methylation (Figure S4 A; R. Anderson and D. Stainier, unpublished results). To test for reactivation of gfp, the dnmt1s872 mutation was introduced into the c269 GFPoff, UAS:mCherry transgenic background. Adults heterozyogous for the mutation and the transgenes were intercrossed, and their progeny were examined for gfp expression. At 1 dpf, GFP-labeled cells were not detected despite transactivation of UAS:mCherry in the complete c269 brain-specific pattern. The presence of maternal Dnmt1 activity likely accounts for transgene silencing at this stage. However, by 2 dpf, robust GFP labeling appeared in the midbrain, forebrain, and retina in 25% of larvae showing mCherry transactivation (Figure 4, D–F). Expression persisted until 7 dpf, when dnmt1 homozygous mutants are no longer viable. Genotyping confirmed that GFP-positive larvae were homozygous for the dnmt1s872 mutation (n = 10), while those without GFP-labeled cells had one or no mutant alleles (n = 6; Figure 4, A–C). Methylation-sensitive Southern blots revealed significant demethylation of the UAS in gfp-reactivated larvae compared to their GFPoff siblings (Figure S4). These results indicate that Dnmt1-mediated methylation is important for silencing of the UAS-regulated transgene.
Figure 4.—
Distinct regions of the brain show differential silencing and reactivation. Corresponding images of GFP fluorescence (A, D, G, and J), mCherry fluorescence (B, E, H, and K), and an overlay of mCherry, GFP, and differential interference contrast (C, F, I, and L) from 3 dpf larvae. (A–C) c269 GFPoff larvae that are wild type or heterozygous for the dnmt1s872 mutation do not show GFP labeling. (D–F) c269 GFPoff larvae that are homozygous dnmt1s872 mutants show GFP labeling in the midbrain and forebrain. Despite transactivation of mCherry, GFP labeling is not observed in r5–6 (arrow in E and F). (G–I) The homozygous dnmt1s872 mutant with gfp reactivation in a small number of bilateral cells in r5–6. The insert in I is an enlargement of reactivated cells indicated by the arrowhead. (J–L) Larvae from a c269 GFPhigh adult mated to wild type. Labeling of GFP is present in the forebrain and midbrain, but not in r5–6 mCherry-positive cells.
While reactivation of gfp was reproducibly observed in the midbrain, forebrain, and retina, cells of the r5–6 hindbrain region were refractory to reactivation. Transactivation of UAS:mCherry confirmed that cells in r5–6 express Gal4-VP16 in homozygous mutant larvae. However, GFP-labeled cells were not detected in r5–6 in 65% of larvae (n = 135) that showed reactivation elsewhere in the brain (Figure 4, D–F). In the remaining 35%, two small groups of GFP-expressing cells were found at the lateral edges of the r5–6 stripe (Figure 4, G–I, arrowhead in I). The difference in reactivation between the midbrain/forebrain and the hindbrain stripe was quantified by determining the integrated fluorescent intensity of GFP and mCherry for each brain region using larvae that showed the greatest number of GFP-labeled cells in r5–6. The average ratio of GFP to mCherry fluorescence was 0.68 ± 0.13 in the midbrain/forebrain region and 0.05 ± 0.07 in r5–6 (n = 5), indicating at least a 13-fold difference in labeling. Cells in the hindbrain are therefore more refractory to reactivation upon loss of Dnmt1 activity.
Curiously, cells in r5–6 are often the first to be silenced in the c269 transgenic background, even when the forebrain and midbrain show high levels of GFP labeling (Figure 4, J–L). Among 112 GFPhigh F2 progeny with robust gfp expression in the forebrain, midbrain, and retina, only 16 had any labeled cells in r5–6. Cells in the r5–6 stripe are therefore more susceptible to silencing than those in the midbrain and forebrain regions.
DISCUSSION
In this study, we used a multicopy UAS and fluorescent reporter genes to monitor transcriptional silencing in transgenic zebrafish. Independent GFPhigh, GFPlow, and GFPoff epiallelic lines were established, which allowed silencing of gfp expression to be followed in developing tissues and over multiple generations. Complete heritable silencing of single-copy GFPoff transgenes was observed and correlated with increased methylation of the tandemly repeated UAS. Reactivation of GFPoff transgenes was demonstrated by increasing Gal4-VP16 levels and by mutation of DNA methyltransferase-1. In addition, we observed distinct profiles of gfp silencing and reactivation in different cell populations, underscoring the value of an in vivo system for monitoring epigenetic regulation of transcription. Collectively, these results demonstrate a powerful system for studying transcriptional silencing during vertebrate development using genetic approaches.
Although prior work in zebrafish provided evidence for transgene silencing, integrated DNA existed in concatemeric arrays and lacked trackable fluorescent reporters (Gibbs et al. 1994; Martin and McGowan 1995). Silencing of a single-copy GFP transgene was described in one report, but only in adult tissues (Thummel et al. 2006). Epiallelic transgenic lines were not previously established, and inheritance of a fully silenced transgene had not been followed over generations. In this study, transgenesis was mediated by Tol2 transposition, resulting in random, dispersed integrations rather than complex concatemers (Kawakami 2005). Single insertions were isolated, allowing epigenetic modifications to be compared at a short defined repeat, the multicopy UAS. Silencing may be triggered by the repetitive nature of the 14× UAS. UAS-regulated transgenes are being used more widely in zebrafish research (Halpern et al. 2008); however, variegated expression could be detrimental in experiments where uniform transgene expression is required. Optimization of Gal4/UAS constructs may be necessary to increase their utility, and decreasing UAS copy number could potentially mitigate silencing.
Silencing appears to be a stochastic event because only some progeny from GFPhigh adults showed variegated expression, and only a subset of transgenic progeny from GFPlow individuals completely lacked GFP labeling. The progression from GFPhigh to GFPlow to GFPoff also suggests that more than one generation is required for extinction of gfp expression. Widespread methylation of sperm from GFPoff adults suggests that, once extensive methylation of the transgene is achieved, it is propagated through the gametes. The heritable complete silencing observed at the UAS is in contrast to mouse transgenes, which frequently show metastable inheritance of silencing whereby restablishment of the epigenetic state in the next generation is a probabilistic event (Rakyan et al. 2002).
Heritable silencing in GFPoff larvae provides a tool to explore the mechanisms of silencing. Previous studies have shown that DNA-binding proteins can cause demethylation of their recognition sites (Matsuo et al. 1998; Hsieh 2000; Lin et al. 2000). It has been suggested that protein binding during replication may protect sites from methylation by Dnmt1, thereby demethylating genes designated for activation (Matsuo et al. 1998; Hsieh 2000). We similarly found that Gal4-VP16 could cause demethylation of the UAS, leading to reactivation of gfp. However, reactivation was achieved using very high levels of Gal4-VP16. In contrast, methylation and silencing persist in GFPoff lines that express lower levels of Gal4-VP16, and differences in methylation were not observed between Gal4-VP16-expressing and -nonexpressing tissues. These data imply that physiological concentrations of DNA-binding proteins may not generally be sufficient to drive demethylation as a means of activating transcription.
Mutation of dnmt1 also resulted in reactivation of gfp expression. Although transcriptional repression independent of methyltransferase activity has been reported for Dnmt1 (Milutinovic et al. 2004; Dunican et al. 2008), the dnmt1S872 allele is a point mutation expected to compromise catalytic function without disrupting N-terminal regulatory domains. Therefore, it is most likely that the methyltransferase activity of Dnmt1 is directly required for silencing at the UAS.
Reactivation of gfp expression in dnmt1S872 mutants occurs in a cell-type-specific manner. Although strong GFP labeling was observed in the forebrain, midbrain, and retina, cells in r5–6 of the hindbrain showed limited or no reactivation of gfp. There are several potential explanations for this difference. It is possible that partially methylated UAS sites may be more sensitive to Gal4-VP16 levels. Transcription from the odz4 promoter appears similar in the forebrain, midbrain, and hindbrain as assessed by whole-mount RNA in situ hybridization (Mieda et al. 1999); however, subtle differences in expression between brain regions might not be detected by this method. Slightly lower odz4 promoter activity in the r5–6 stripe would selectively reduce Gal4-VP16 activity in this region, which could account for the differences in the onset of silencing and resistance to reactivation at a partially methylated UAS.
Another explanation for differences in reactivation between brain regions could be differences in the timing of neurogenesis. Reactivation of gfp expression in dnmt1 mutants was not observed at 1 dpf despite transactivation of UAS:mCherry. The delay in reactivation likely reflects the presence of maternal Dnmt1 at this time. If neurons in r5–6 differentiate prior to depletion of maternal Dnmt1, they would retain methylation, and reactivation would not be expected. However, we do not favor this hypothesis. By 48 hours post fertilization (hpf), reactivation is observed in the fore- and midbrain, suggesting that maternal Dnmt1 activity is no longer present. At this time point, cell proliferation is observed throughout the brain, including in r5–6 (Mueller and Wullimann 2005). Moreover, another 1400 neurons are born in r4–5 after 48 hpf (Lyons et al. 2003). Finally, it is unclear how early maternal Dnmt1, which is not spatially restricted, could specifically influence gfp expression in r5–6 of GFPhigh larvae.
A third hypothesis is that different regions of the brain have different gene requirements for silencing. In addition to a single dnmt1 homolog, the zebrafish genome also contains six DNA methyltransferase-3 (dnmt3) homologs (Shimoda et al. 2005). Although tissue-specific roles for these de novo methyltransferases have not been reported, increased activity of Dnmt3 proteins might compensate for the loss of Dnmt1. Alternatively, overexpression of the histone methyltransferase suv39h1 rescues phenotypes caused by depletion of dnmt1 in zebrafish (Rai et al. 2006). Although suv39h1 expression is higher in the forebrain compared to the hindbrain (Rai et al. 2006), it is possible that other histone-modifying enzymes or as-yet-unidentified molecular pathways could influence silencing in r5–6.
Reactivation of gfp in dnmt1 mutants provides proof of principle that factors required for transcriptional silencing may be uncovered genetically by screening for reactivation of expression in GFPoff larvae. It is unlikely that the full complement of genes required for silencing in vertebrates has been identified, and cell-type-specific roles for known factors may have been overlooked. GFPoff transgenic lines provide an opportunity to probe the mechanism of silencing through candidate testing, mutagenesis, or drug screening approaches. Because such experiments are performed in vivo, they have the potential to further our understanding of developmental and tissue-specific requirements for gene silencing in a developing embryo.
Acknowledgments
We thank Michelle Macurak and Brian Hollenback for technical support and Steve Leach, Michael Parsons, and Courtney Akitake for useful discussions and reagents. This project was supported in part by the National Institute of Child Health and Human Development (grant no. R01HD058530) to M.E.H. M.G.G. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-#1945-07). A.C.S. is an investigator of the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102079/DC1.
References
- Allen, N. D., M. L. Norris and M. A. Surani, 1990. Epigenetic control of transgene expression and imprinting by genotype-specific modifiers. Cell 61 853–861. [DOI] [PubMed] [Google Scholar]
- Ashe, A., D. K. Morgan, N. C. Whitelaw, T. J. Bruxner, N. K. Vickaryous et al., 2008. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 9 R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., A. Meissner and E. S. Lander, 2007. The mammalian epigenome. Cell 128 669–681. [DOI] [PubMed] [Google Scholar]
- Blewitt, M. E., N. K. Vickaryous, S. J. Hemley, A. Ashe, T. J. Bruxner et al., 2005. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102 7629–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman and T. H. Bestor, 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294 2536–2539. [DOI] [PubMed] [Google Scholar]
- Chen, T., S. Hevi, F. Gay, N. Tsujimoto, T. He et al., 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39 391–396. [DOI] [PubMed] [Google Scholar]
- Chevalier-Mariette, C., I. Henry, L. Montfort, S. Capgras, S. Forlani et al., 2003. CpG content affects gene silencing in mice: evidence from novel transgenes. Genome Biol. 4 R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, J. M., C. M. Akitake, M. G. Goll, J. M. Rhee, N. Gosse et al., 2007. Transactivation from Gal4–VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer, D. R., 1997. Do transgene arrays form heterochromatin in vertebrates? Transgenic Res. 6 3–10. [DOI] [PubMed] [Google Scholar]
- Dunican, D. S., A. Ruzov, J. A. Hackett and R. R. Meehan, 2008. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development 135 1295–1302. [DOI] [PubMed] [Google Scholar]
- Garrick, D., S. Fiering, D. I. Martin and E. Whitelaw, 1998. Repeat-induced gene silencing in mammals. Nat. Genet. 18 56–59. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. D., A. Peek and G. Thorgaard, 1994. An in vivo screen for the luciferase transgene in zebrafish. Mol. Marine Biol. Biotechnol. 3 307–316. [PubMed] [Google Scholar]
- Giniger, E., S. M. Varnum and M. Ptashne, 1985. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40 767–774. [DOI] [PubMed] [Google Scholar]
- Girard, A., and G. J. Hannon, 2008. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 18 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll, M. G., and T. H. Bestor, 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74 481–514. [DOI] [PubMed] [Google Scholar]
- Halpern, M. E., J. Rhee, M. G. Goll, C. M. Akitake, M. Parsons et al., 2008. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard, E., and W. Bickmore, 2007. The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 19 311–316. [DOI] [PubMed] [Google Scholar]
- Hsieh, C. L., 2000. Dynamics of DNA methylation pattern. Curr. Opin. Genet. Dev. 10 224–228. [DOI] [PubMed] [Google Scholar]
- Hsieh, J., and A. Fire, 2000. Recognition and silencing of repeated DNA. Annu. Rev. Genet. 34 187–204. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough et al., 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27 31–39. [DOI] [PubMed] [Google Scholar]
- Jarvis, C. D., T. Geiman, M. P. Vila-Storm, O. Osipovich, U. Akella et al., 1996. A novel putative helicase produced in early murine lymphocytes. Gene 169 203–207. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., 2004. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77 201–222. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., 2005. Transposon tools and methods in zebrafish. Dev. Dyn. 234 244–254. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., A. Shima and N. Kawakami, 2000. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97 11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby, N. J., H. M. Leyser and I. J. Furner, 1992. Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol. Biol. 20 103–112. [DOI] [PubMed] [Google Scholar]
- Kumaki, Y., M. Oda and M. Okano, 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 36 W170–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E., T. H. Bestor and R. Jaenisch, 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 915–926. [DOI] [PubMed] [Google Scholar]
- Li, F., M. Papworth, M. Minczuk, C. Rohde, Y. Zhang et al., 2007. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 35 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, I. G., T. J. Tomzynski, Q. Ou and C. L. Hsieh, 2000. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, D. A., A. T. Guy and J. D. Clarke, 2003. Monitoring neural progenitor fate through multiple rounds of division in an intact vertebrate brain. Development 130 3427–3436. [DOI] [PubMed] [Google Scholar]
- Martin, C. C., and R. McGowan, 1995. Genotype-specific modifiers of transgene methylation and expression in the zebrafish, Danio rerio. Genet. Res. 65 21–28. [DOI] [PubMed] [Google Scholar]
- Matsuo, K., J. Silke, O. Georgiev, P. Marti, N. Giovannini et al., 1998. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 17 1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, A. J., F. Neuhuber, Y. D. Park, P. F. Ambros and M. A. Matzke, 1994. Homology-dependent gene silencing in transgenic plants: epistatic silencing loci contain multiple copies of methylated transgenes. Mol. Gen. Genet. 244 219–229. [DOI] [PubMed] [Google Scholar]
- Mieda, M., Y. Kikuchi, Y. Hirate, M. Aoki and H. Okamoto, 1999. Compartmentalized expression of zebrafish ten-m3 and ten-m4, homologues of the Drosophila ten(m)/odd Oz gene, in the central nervous system. Mech. Dev. 87 223–227. [DOI] [PubMed] [Google Scholar]
- Milutinovic, S., S. E. Brown, Q. Zhuang and M. Szyf, 2004. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J. Biol. Chem. 279 27915–27927. [DOI] [PubMed] [Google Scholar]
- Mueller, T., and M. Wullimann, 2005. Atlas of Early Brain Development. Elsevier, Amsterdam/New York.
- Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer et al., 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107 323–337. [DOI] [PubMed] [Google Scholar]
- Rai, K., L. D. Nadauld, S. Chidester, E. J. Manos, S. R. James et al., 2006. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol. Cell. Biol. 26 7077–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan, V. K., M. E. Blewitt, R. Druker, J. I. Preis and E. Whitelaw, 2002. Metastable epialleles in mammals. Trends Genet. 18 348–351. [DOI] [PubMed] [Google Scholar]
- Sapienza, C., J. Paquette, T. H. Tran and A. Peterson, 1989. Epigenetic and genetic factors affect transgene methylation imprinting. Development 107 165–168. [DOI] [PubMed] [Google Scholar]
- Shimoda, N., K. Yamakoshi, A. Miyake and H. Takeda, 2005. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev. Dyn. 233 1509–1516. [DOI] [PubMed] [Google Scholar]
- Southern, E. M., 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98 503–517. [DOI] [PubMed] [Google Scholar]
- Stancheva, I., C. Hensey and R. R. Meehan, 2001. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 20 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B. D., and C. D. Allis, 2000. The language of covalent histone modifications. Nature 403 41–45. [DOI] [PubMed] [Google Scholar]
- Strathdee, D., C. B. Whitelaw and A. J. Clark, 2008. Distal transgene insertion affects CpG island maintenance during differentiation. J. Biol. Chem. 283 11509–11515. [DOI] [PubMed] [Google Scholar]
- Stuart, G. W., J. R. Vielkind, J. V. McMurray and M. Westerfield, 1990. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development 109 577–584. [DOI] [PubMed] [Google Scholar]
- Sun, X. J., P. F. Xu, T. Zhou, M. Hu, C. T. Fu et al., 2008. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS ONE 3 e1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, H. G., M. Kearns, H. D. Morgan, A. P. Headley, C. Morris et al., 2000. Reactivation of heritably silenced gene expression in mice. Mamm. Genome 11 347–355. [DOI] [PubMed] [Google Scholar]
- Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta et al., 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel, R., S. Bai, M. P. Sarras, Jr., P. Song, J. McDermott et al., 2006. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev. Dyn. 235 336–346. [DOI] [PubMed] [Google Scholar]
- Walker, C., 1999. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 60 43–70. [DOI] [PubMed] [Google Scholar]
- Weber, H., C. Ziechmann and A. Graessmann, 1990. In vitro DNA methylation inhibits gene expression in transgenic tobacco. EMBO J. 9 4409–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield, M., 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Institute of Neuroscience, University of Oregon, Eugene, OR.