Abstract
To gain new mechanistic insight into ER homeostasis and the biogenesis of secretory proteins, we screened a genomewide collection of yeast mutants for defective intracellular retention of the ER chaperone, Kar2p. We identified 87 Kar2p-secreting strains, including a number of known components in secretory protein modification and sorting. Further characterization of the 73 nonessential Kar2p retention mutants revealed roles for a number of novel gene products in protein glycosylation, GPI-anchor attachment, ER quality control, and retrieval of escaped ER residents. A subset of these mutants, required for ER retrieval, included the GET complex and two novel proteins that likely function similarly in membrane insertion of tail-anchored proteins. Finally, the variant histone, Htz1p, and its acetylation state seem to play an important role in maintaining ER retrieval pathways, suggesting a surprising link between chromatin remodeling and ER homeostasis.
THE endoplasmic reticulum (ER) is a central organelle in the biogenesis of secretory and membrane proteins. In the model eukaryote, Saccharomyces cerevisiae, fully one-third of the proteome is delivered to the ER en route to various destinations within the cell (Ghaemmaghami et al. 2003). Newly synthesized secretory proteins enter the ER as linear polypeptide chains, which are subsequently acted on by a variety of resident enzymes that promote protein folding through chaperone action, glycosylation, and disulfide bond formation. In addition to these protein-oriented events, the ER is also an important site of lipid biosynthesis, housing a suite of enzymes that function in various steps of fatty acid elongation and desaturation, ergosterol synthesis, and attachment of glycosylphospho-inositol (GPI) lipids to a class of lipid-tethered membrane proteins. Proteins and lipids destined for downstream compartments of the secretory pathway are exported from the ER via transport vesicles that are sculpted from the donor membrane by the action of a set of cytoplasmic coat proteins, known as the COPII coat (Gurkan et al. 2006; Lee et al. 2004).
In maintaining ER function and homeostasis, a central event is the efficient homeostasis of the resident proteins that perform the essential functions of protein and lipid biogenesis. Several pathways contribute to this process. First, ER resident proteins lack the positive sorting signals that mediate efficient capture into ER-derived COPII vesicles (Barlowe 2003; Otte and Barlowe 2004), making them poor clients for forward transport. However, nonspecific export of proteins may result from stochastic sampling of the ER membrane and lumen during vesicle formation in a process known as “bulk flow” (Klumperman 2000; Lee et al. 2004). Unchecked, this nonspecific ER egress could deplete the ER of relatively abundant proteins that would be packaged into transport vesicles at their prevailing concentrations. This highlights the importance of the second mechanism of ER retention: retrieval of escaped ER residents from the Golgi apparatus via retrograde COPI-coated transport vesicles (Lee et al. 2004). This process often employs a transmembrane receptor, Erd2p, that recognizes the canonical ER retrieval motif, K/HDEL on soluble ER residents (Townsley et al. 1994). The contribution of each of these pathways can be appreciated by considering the fate of the ER chaperone, Kar2p/BiP, that lacks its HDEL retrieval signal; this protein is efficiently secreted from cells (Schuldiner et al. 2005; Semenza et al. 1990), suggesting that ER residents can access transport vesicles, presumably by nonspecific capture, but are efficiently retrieved to maintain the correct steady state localization.
In addition to these pathways that regulate the localization of ER residents, further mechanisms likely govern the export of the large cast of client proteins that transit through the ER, a population of proteins that must be carefully managed (Ellgaard and Helenius 2003; Anelli and Sitia 2008). Improperly assembled proteins are prevented from premature egress, and terminally misfolded proteins are destroyed by a process known as ER-associated degradation (ERAD). This ER quality control checkpoint ensures that aberrant proteins are not deployed within the cell, where they may interfere with normal protein function (Ron 2002). Although the mechanisms that contribute to ER retention of misfolded proteins are not well understood, the improper presentation of sorting signals and retrieval from post-ER compartments may play important roles (Caldwell et al. 2001; Vashist et al. 2001; Kincaid and Cooper 2007). Furthermore, an aberrant form of the viral protein, VSV-G, is excluded from the dedicated ER exit sites that give rise to transport vesicles, suggesting additional layers of regulation function to segregate proteins prior to ER egress (Mezzacasa and Helenius 2002). Should the ER become overburdened with misfolded proteins, a transcriptional pathway known as the unfolded protein response (UPR) is activated, which functions to upregulate both ER chaperones and the degradation machinery (Travers et al. 2000). These multiple mechanisms interface with each other to contribute to ER homeostasis; when Erd2p-mediated retrieval is crippled, the UPR can compensate to maintain viability by increasing the levels of ER chaperones (Beh and Rose 1995). However, the extent of cross-talk between these processes is unclear, and the full spectrum of components that maintain the fidelity of protein biogenesis and the integrity of ER function remain unknown.
We aimed to identify components that act in ER homeostasis using an unbiased, genomewide approach focused on protein processing and transport through the ER. We reasoned that mutants that secrete ER resident proteins to the cell surface could have defects in either retention of ER resident proteins or in the quality control process that contributes to the fidelity of protein secretion. We screened the yeast genome for mutants that secrete significant amounts of the ER chaperone, Kar2p (orthologous to mammalian BiP) to the cell surface, and identified 87 yeast mutants that secrete at least twofold more Kar2p than wild-type (WT) cells. We classified each of these mutants according to their involvement in protein folding, protein glycosylation, maturation of GPI-anchored proteins, ER quality control, and ER retrieval to define a number of novel components that function in each of these pathways.
MATERIALS AND METHODS
Yeast strains and growth:
Cultures were grown at 30° in standard rich media (YPD: 1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete media (SC: 0.67% yeast nitrogen base and 2% glucose, supplemented with amino acids appropriate for auxotrophic growth). Cells were transformed using standard lithium acetate protocols. The haploid deletion and Tet-repressible strain collections were purchased from Open Biosystems (Huntsville, AL). We employed a synthetic genetic array approach to systematically introduce Erd2-GFP and the hac1Δ allele into the MATa collection of Kar2p secretors. The parental query strain [MATα, hmrΔ0∷URA3 (C. albicans), leu2Δ0, his3Δ1, can1Δ0∷PGAL1-TDH1-MFA1pr-his5 + (S. pombe), lyp1Δ0] was constructed by transforming Y15578-1.2b (Singh et al. 2009) with PCR products to integrate GFP∷LEU2 at the ERD2 locus or replace the HAC1 locus with the NATMX selection cassette to generate LMY446 and LMY445, respectively. These query strains were then mated to the haploid MATa strains that showed significant Kar2p secretion. Diploid cells were selected on SE medium (0.17% yeast nitrogen base without amino acids or ammonium sulfate; 1 g/liter monosodium glutamate; 2% glucose) supplemented with the appropriate amino acid dropout mix (Sunrise Science Products, San Diego, CA) and the selective agents G418 (Fisher, Hanover Park, IL) and/or nourseothricin (Werner Bioagents, Jena, Germany), followed by replica plating onto sporulation medium. Haploid double mutants were selected by virtue of the mating-type regulated auxotrophy cassette allowing selection of MATa haploid double mutants following multiple rounds of replica plating onto appropriate selective media lacking histidine and arginine and containing canavanine, G418, and nourseothricin. In some cases, strains were also constructed by traditional yeast mating or PCR-mediated integration (supporting information, Table S3). Strains YM1729 (WT), YM1730 (htz1Δ), YM1530 (sir2Δ), YM1589 (hmrΔ), YM1536 (htz1Δsir2Δ), and YM1590 (htz1ΔhmrΔ) were the gifts of Hiten Madhani [University of California at San Francisco (UCSF)]. Jasper Rine [University of California, Berkeley (UC Berkeley)] kindly provided JRY7981 (htz1Δ∷HIS5, pHTZ1-3FLAG), JRY7985 (htz1Δ∷HIS5, pHTZ1-K3R-3FLAG), JRY7986 (htz1Δ∷HIS5, pHTZ1-K8R-3FLAG), JRY7987 (htz1Δ∷HIS5, pHTZ1-K10R-3FLAG), and JRY7988 (htz1Δ∷HIS5, pHTZ1-K14R-3FLAG).
Plasmids:
The UPRE-lacZ reporter plasmid was derived from pMZ11 by EcoRI/XhoI digest and subcloning into pRS423 to create pLM38. A HA-tagged form of CPY* was encoded on plasmid pCP258, originally from Peter Walter's lab (UCSF). ILM1 was expressed from a multicopy plasmid, pLM550, which was generated by PCR amplification of the ILM1 gene plus 1000 bp upstream of the open reading frame, followed by subcloning into the EcoRI/SpeI sites of pRS426. The plasmid encoding GFP-Frt1p was a gift from Traude Beilharz (Victor Chang Cardiac Research Institute, Sydney, Australia). Plasmids encoding SEC22, BET1, and BOS1 on a 2μ pRS426 plasmid were from the Schekman lab strain collection (RSB1241, RSB1247, and RSB1248, respectively). A SED5 overexpression construct was made by PCR amplification of the SED5 locus, including ∼350 bp upstream and downstream followed by subcloning into the pRS426 vector.
Kar2p secretion assays:
For genomewide screening purposes, cells were grown from glycerol stocks in 96-well trays for ∼2 days. Saturated cultures (5 μl) were spotted onto YPD plates and incubated at 30° for 8 hr, at which point colonies were overlaid with nitrocellulose membranes and incubation continued for 30 min. Nitrocellulose membranes were washed under running water to remove any adhering cells and then incubated sequentially with TBS, 5% milk powder, and anti-Kar2p antibodies (kindly provided by Randy Schekman,UC Berkeley) diluted 1/10,000 in TBS. In initial screens, secreted Kar2p was detected with HRP-conjugated goat-anti-rabbit antibodies followed by ECL detection (Pierce, Rockford, IL). To quantify the amount of Kar2p secretion, we detected Kar2p using IRDye800-conjugated anti-rabbit antibodies (Rockland Immunochemicals, Gilbertsville, PA) followed by imaging on the LiCor Odyssey fluorescent scanner and analysis using Odyssey software. The Kar2p secretion signal was normalized to that observed in a wild-type strain to yield the Kar2p secretion index listed in Table 1. Kar2p secretion was also monitored from liquid cultures according to published protocols (Belden and Barlowe 2001).
TABLE 1.
Kar2p retention mutants identified from haploid deletion collections
| Gene | ORF | Functional categorya | Kar2p secretion indexb | Synthetic sick/lethal with hac1Δc | Kar2p secretion in hac1Δd |
|---|---|---|---|---|---|
| Erd1 | YDR414C | Ambiguous | 9.0 | − | + |
| Emp24 | YGL200C | ER-Golgi traffic | 9.0 | + | ND |
| Bst1 | YFL025C | O-linked glyc./GPI | 8.8 | + | ND |
| Erv25 | YML012W | ER-Golgi traffic | 8.4 | + | ND |
| Lhs1 | YKL073W | Protein maturation | 6.8 | + | ND |
| Gup1 | YGL084C | O-linked glyc./GPI | 6.7 | − | − |
| Per1 | YCR044C | O-linked glyc./GPI | 6.1 | − | − |
| Erp1 | YAR002C-A | ER-Golgi traffic | 5.9 | − | + |
| Arv1 | YLR242C | Lipid | 5.6 | + | ND |
| Ted1 | YIL039W | ER-Golgi traffic | 5.6 | − | + |
| Slp1 | YOR154W | Ambiguous | 5.0 | − | − |
| Get1 | YGL020C | ER-Golgi traffic | 5.0 | − | + |
| Ics3 | YJL077C | Ambiguous | 4.7 | − | − |
| Get3 | YDL100C | ER-Golgi traffic | 4.6 | − | + |
| Scj1 | YMR214W | Protein maturation | 4.3 | + | ND |
| YHR078W | Unknown | 4.2 | − | + | |
| YOR164C | Unknown | 4.1 | − | + | |
| Get2 | YER083C | ER-Golgi traffic | 4.1 | − | − |
| She10 | YGL228W | ambiguous | 3.8 | − | + |
| Alg3 | YBL082C | N-linked glyc. | 3.8 | + | ND |
| Pps1 | YBR276C | Cell cycle | 3.7 | − | − |
| Mnn11 | YJL183W | N-linked glyc. | 3.7 | + | ND |
| Csf1 | YLR087C | Ambiguous | 3.7 | + | ND |
| Pho88 | YBR106W | Ambiguous | 3.7 | + | ND |
| Eos1 | YNL080C | N-linked glyc. | 3.7 | + | ND |
| Erv26 | YHR181W | ER-Golgi traffic | 3.7 | − | − |
| Sec22 | YLR268W | ER-Golgi traffic | 3.5 | + | ND |
| Sum1 | YDR310C | Chromatin | 3.4 | − | + |
| YLR065C | Unknown | 3.4 | + | ND | |
| YMR031W-A | Unknown | 3.4 | + | ND | |
| Mdy2 | YOL111C | Ambiguous | 3.3 | − | + |
| YBL083C | Unknown | 3.2 | + | ND | |
| Ccw12 | YLR110C | Ambiguous | 3.2 | − | − |
| Arp6 | YLR085C | Chromatin | 3.2 | − | + |
| Htz1 | YOL012C | Chromatin | 3.2 | − | + |
| Vps71/Swc6 | YML041C | Chromatin | 3.1 | − | + |
| Cwc21 | YDR482C | Ambiguous | 3.1 | − | + |
| YLR374C | Unknown | 3.1 | − | + | |
| Ubx2 | YML013W | ERAD | 3.1 | + | ND |
| Erp2 | YAL007C | ER-Golgi traffic | 3.1 | − | + |
| YML013C-A | Unknown | 3.0 | + | ND | |
| Mga2 | YIR033W | Lipid | 3.0 | − | − |
| Hoc1 | YJR075W | N-linked glyc. | 3.0 | − | + |
| Yaf9 | YNL107W | Chromatin | 3.0 | N/D | ND |
| Ste24 | YJR117W | Protein maturation | 3.0 | + | ND |
| YER140W | Unknown | 3.0 | − | + | |
| Pin4 | YBL051C | Cell cycle | 2.9 | − | − |
| Tcb2 | YNL087W | Ambiguous | 2.8 | − | − |
| Pro1 | YDR300C | Amino acid biosynth. | 2.8 | − | − |
| YLR111W | Unknown | 2.7 | − | − | |
| Vps72/Swc2 | YDR485C | Chromatin | 2.6 | − | + |
| Swc3 | YAL011W | Chromatin | 2.6 | − | + |
| Ost3 | YOR085W | N-linked glyc. | 2.6 | + | ND |
| Pmt1 | YDL095W | O-linked glyc./GPI | 2.6 | − | − |
| Vps74 | YDR372C | Intra-Golgi traffic | 2.5 | − | − |
| Ilm1 | YJR118C | Ambiguous | 2.5 | + | ND |
| Van1 | YML115C | N-linked glyc. | 2.5 | − | + |
| Gsg1 | YDR108W | ER-Golgi traffic | 2.4 | − | + |
| Fat1 | YBR041W | Lipid | 2.4 | − | − |
| Mnn10 | YDR245W | N-linked glyc. | 2.4 | − | − |
| Ioc3 | YFR013W | Chromatin | 2.3 | − | − |
| Rce1 | YMR274C | Protein maturation | 2.3 | − | + |
| Swr1 | YDR334W | Chromatin | 2.3 | − | + |
| Sir4 | YDR227W | Chromatin | 2.3 | ND | ND |
| Las21 | YJL062W | O-linked glyc./GPI | 2.2 | + | ND |
| Grh1 | YDR517W | ER-Golgi traffic | 2.2 | − | + |
| Mrpl16 | YBL038W | Mitochondria | 2.2 | + | ND |
| Sdc1 | YDR469W | Chromatin | 2.2 | − | + |
| YJL123C | Unknown | 2.2 | − | + | |
| Gds1 | YOR355W | Ambiguous | 2.1 | − | − |
| Mih1 | YMR036C | Cell cycle | 2.1 | − | + |
| Nem1 | YHR004C | Ambiguous | 2.1 | − | + |
| Bug1 | YDL099W | ER-Golgi traffic | 2.0 | − | − |
ND, not determined.
Functional category assigned from SGD annotations.
Kar2p secretion index was determined by quantitative immunoblotting and represents a fold increase over wild type.
Synthetic genetic interactions between individual mutations and a hac1Δ allele (+ indicates significant synthetic growth defect).
Kar2p secretion was determined in the hac1Δ/yfgΔ double mutant strains by quantitative immunoblot; a twofold increase over a hac1Δ strain was considered significant secretion (+).
Network analysis:
Relationships among Kar2p retention mutants were examined and visualized using the BioGrid online resource (http://www.thebiogrid.org) and Osprey software (Breitkreutz et al. 2003).
Detection of UPR:
Strains expressing the UPRE-lacZ reporter plasmid (pLM38) were grown to midlog phase in selective medium and UPR measured by β-galactosidase activity assays as described (Ng et al. 2000). Data were normalized to the appropriate BY4742 or BY4741 wild-type strains.
Pulse-chase analysis:
Pulse-chase analysis of CPY and Gas1p maturation was performed as described (Pagant et al. 2007). CPY and Gas1p were immunoprecipitated using antibodies kindly provided by Randy Schekman. To measure the acquisition of α-1,6-mannose, CPY* was first immunoprecipitated using anti-HA antibodies; the precipitated protein was released from the beads with 1% SDS followed by dilution with IP buffer without SDS and a second round of immunoprecipitation using antibodies against the α-1,6-mannose moiety, also provided by Randy Schekman. The degree of α-1,6-mannose modification was calculated relative to a parallel immunoprecipitation that yielded the total CPY* present at each time point.
Microscopy:
Strains expressing Erd2p-GFP were grown in YPD to midlog phase and imaged using a Nikon TE300 inverted microscope (Melville, NY) with 100× N.A. 1.4 PlanApo optics and a Hamamatsu Orca-ERG charge-coupled device camera. Images were collected with the Openlab 5.0 (Improvision, Waltham, MA) software system and analyzed using Adobe Photoshop (Adobe Systems, Mountain View, CA). Strains expressing GFP-Frt1p were grown in selective medium and shifted briefly to YPD, which exaggerated the mislocalization phenotype of Frt1p, prior to imaging as described above.
RESULTS AND DISCUSSION
To uncover novel mechanisms that contribute to ER homeostasis, we aimed to employ a genomewide approach to identify S. cerevisiae mutants that improperly secrete the ER lumenal chaperone, Kar2p (Figure 1A). Previous attempts to isolate such mutants used a traditional genetic selection approach whereby the secreted enzyme invertase was appended with an HDEL ER retention signal, yielding two mutants, erd1 and erd2 (for ER retention defective), which showed secreted invertase activity (Pelham et al. 1988; Semenza et al. 1990). We designed a colony immunoblotting assay to screen a genomic collection of haploid MATα deletion strains that contains individual disruptions in each nonessential gene (Winzeler et al. 1999). In preliminary screens we identified a number of strains that secreted more Kar2p than wild-type cells; secondary screening with fluorescent secondary antibodies allowed quantification of Kar2p secretion in each of these strains, yielding a Kar2p secretion index that represents the fold change of Kar2p secretion over wild-type cells (Table S1). To further validate our collection of Kar2p retention mutants, we also screened a second collection of deletion strains, composed of the MATa set of haploid deletants. There was a high degree of overlap between the two collections, with only a few strains unique to either collection (Table S1). We tested the gene disruptions for this nonoverlapping set of mutants and identified three cases in which there were anomalies in the insertion of the deletion cassette. In the MATαilm1Δ background (which gave a Kar2p secretion phenotype) the KanMX marker was correctly integrated whereas in the corresponding MATa strain, the locus was wild type, consistent with the absence of Kar2p secretion in this strain (data not shown). In two cases, pmt1Δ and pro1Δ, the strain that gave a Kar2p secretion phenotype contained the correct gene deletion whereas in the opposite mating type there appeared to be both a wild-type locus and a KanMX-disrupted locus, suggesting that a chromosomal duplication masked the effect of the gene deletion. In the other strains that gave a Kar2p secretion phenotype in a mating-type-dependent manner, both strains contained the correct genetic disruption, suggesting either mating-type specific effects on the endoplasmic reticulum or the presence of spontaneous suppressor mutations. This latter effect has been previously reported as a particular problem for the get mutants (Schuldiner et al. 2008). These discrepancies serve to highlight the utility of performing genomewide screens in multiple collections to identify false negatives and generate an independent data set to lend confidence in the original screen.
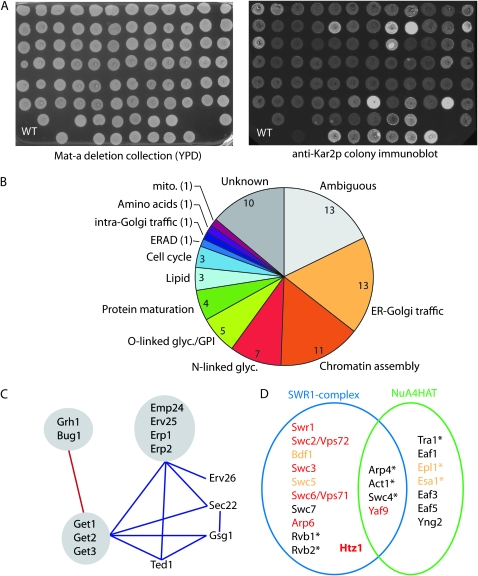
Figure 1.—
Overview of the Kar2p retention screen and categorization of mutants. (A) The haploid deletion collection was grown on YPD (left) and secreted Kar2p was quantified using fluorescent secondary antibodies (right). Colonies secreting twofold more Kar2p than WT cells were selected for further characterization. (B) Functional distribution of Kar2p secretors and the number of genes isolated in each category. (C) Genetic interactions among Kar2p secretors with annotated functions in ER-Golgi traffic. Components that have been isolated as physical complexes are shown in gray. Aggravating interactions are shown in blue, alleviating interactions in red. (D) Subunit architecture of the SWR1 and NuA4 complexes. Kar2p secretors are shown in red, components that showed a less significant Kar2p secretion phenotype are shown in orange, essential genes are annotated with an asterisk. Adapted from Kobor et al. (2004).
We selected a Kar2p secretion index of 2 as a cutoff for further characterization: we consider this a conservative threshold since strains with a secretion index of 1.5 also showed consistently higher Kar2p secretion than wild-type cells, albeit with more variability from experiment to experiment. Our final analysis yielded 73 strains that secreted at least twofold more Kar2p than wild-type cells (Table 1). To similarly investigate genes that are lethal if perturbed in a haploid cell, we also screened a strain collection that contains ∼800 essential genes under the control of a tetracycline-regulated promoter. We tested Kar2p secretion following an 8-hr period of gene repression by doxycycline and identified another 14 strains that secrete Kar2p (Table 2 and Table S2). Among these were several strains that secreted Kar2p even in the absence of doxycycline, likely because altered expression from the Tet-responsive promoter perturbs cellular function. These combined screens yielded a number of expected components with known functions in Kar2p retention, including the KDEL receptor, Erd2p, as well as many mutants of unknown or poorly defined function that may represent novel components in ER homeostasis.
TABLE 2.
Kar2p retention mutants identified from a Tet-repressible collection of essential genes
| Gene | ORF | Functional categorya | Kar2p secretion indexb |
|---|---|---|---|
| Erd2 | YBL040C | ER-Golgi traffic | 8.0 |
| Ost2 | YOR103C | Protein maturation | 5.6 |
| Iqg1 | YPL242C | Ambiguous | 5.0 |
| Pgi1 | YBR196C | Gluconeogenesis | 4.4 |
| Gpi1 | YGR216C | O-linked glyc./GPI | 4.2 |
| Alg11 | YNL048W | Protein maturation | 4.2 |
| Srp72 | YPL210C | Protein maturation | 4.0 |
| Gpi17 | YDR434W | O-linked glyc./GPI | 3.8 |
| Rft1 | YBL020W | Protein maturation | 3.6 |
| Sec39 | YLR440C | ER-Golgi traffic | 3.4 |
| Mot1 | YPL082C | Transcription | 2.6 |
| Sec17 | YBL050W | ER-Golgi traffic | 2.4 |
| Gpi19 | YDR437W | O-linked glyc./GPI | 2.4 |
| Abd1 | YBR236C | mRNA processing | 2.2 |
Functional category assigned from SGD annotations.
Kar2p secretion index was determined by quantitative immunoblotting following an 8-hr period of treatment with doxycycline and represents a fold increase over wild type.
We considered that cell wall defects might cause cell lysis during the colony immunoblotting procedure, yielding a Kar2p secretion signal that was in fact derived from intracellular pools of Kar2p. We tested this by monitoring Kar2p secretion in the presence of osmotic support (1 m sorbitol) during the growth and colony overlay phases of the experiment (data not shown). We tested the majority of MATa Kar2p retention mutants under these conditions and only one strain, she10Δ, showed a rescue of Kar2p secretion on sorbitol medium, suggesting that Kar2p secretion in most mutants did not result from cell lysis. We also aimed to determine whether the Kar2p secretion phenotype applied more broadly to other HDEL-containing ER residents. To this end we probed a subset of mutants for secretion of another ER chaperone, Pdi1p. In a qualitative manner, we saw similar trends in the degree of Pdi1p secretion; however, the relatively low affinity and specificity of the α-Pdi1p antibodies precluded more quantitative analysis (data not shown).
We focused our efforts on further characterizing the nonessential Kar2p retention mutants. This set of mutants was enriched in components with defined functions in ER-Golgi traffic and various aspects of secretory protein biogenesis (Figure 1B). Within the functional category of ER-Golgi transport, we identified multiple examples of shared phenotypes among distinct members of the same protein complex, including the GET complex (Get1p, Get2p, and Get3p), the p24 family (Emp24p, Erv25p, Erp1p, and Erp2p), and the Bug1p/Grh1p complex. Furthermore, this subset of mutants showed extensive genetic interactions with each other as defined by systematic genetic analysis (Schuldiner et al. 2005), suggesting that the gene products function in shared pathways (Figure 1C). One unexpected functional category that was overrepresented in our set of Kar2p retention mutants was chromatin assembly, including components of the SWR1-complex that deposit the variant histone, Htz1p, itself also identified (Figure 1D). Moreover, additional members of the SWR1 complex and two essential subunits of the NuA4 histone acetylation machinery that modifies Htz1p also secreted Kar2p when mutated, albeit to a lesser extent than our twofold cutoff (Table S1). Some of these chromatin assembly mutants also showed a vacuolar protein sorting (VPS) phenotype, suggesting pleiotropic defects in secretory pathway function that may stem from disregulation of a specific subset of genes.
UPR-dependence of the Kar2p secretion phenotype:
Secretion of Kar2p can result from activation of the unfolded protein response (UPR), which dramatically upregulates ER chaperones, among other targets. Mutations that cause the accumulation of misfolded proteins and subsequent activation of the UPR may induce secretion of Kar2p simply as a result of Kar2p overexpression (Belden and Barlowe 2001). Conversely, mutations that result in defective ER retention may in turn lead to an activated UPR; inappropriate leakage of chaperones would deplete the essential folding factors within the ER such that protein folding is impaired and the UPR induced. Thus, simply monitoring UPR activation in these strains is unlikely to resolve whether Kar2p secretion causes or results from ER stress. To overcome this conundrum and determine which mutants secrete Kar2p largely because of an active UPR, we introduced a deletion allele of HAC1, the transcription factor that mediates the UPR, into each of the Kar2p retention mutants. For viable double mutants, we quantified Kar2p secretion relative to a hac1Δ single mutant. Approximately 40% of Kar2p retention mutants exhibited significant Hac1p-independent secretion of Kar2p, designated as a Kar2p secretion signal in the double mutant at least twofold greater than that of a hac1Δ strain (Table 1). We propose that these mutants correspond to proteins with roles in the retention and/or retrieval of Kar2p rather than in the maturation of secretory proteins. It is somewhat surprising that these mutants that secrete a key ER chaperone are still viable absent the ability to upregulate the synthesis of additional ER residents via the UPR. These strains may be able to survive with lowered steady-state levels of Kar2p, or may be able to upregulate ER chaperones via a Hac1p-independent pathway.
Conversely, a number of mutants no longer secreted Kar2p in the absence of functional Hac1p, suggesting a contribution of the UPR to Kar2p secretion in the original single mutant strain. Indeed, several of these mutants showed a constitutive UPR as measured by a β-galactosidase reporter assay (Table S1). Finally, a subset of mutants exhibited a severe synthetic growth defect when combined with the hac1Δ allele (Table 1), suggesting a requirement for the UPR to maintain viability in the single mutants. Under conditions of impaired protein folding, the UPR can become essential to cope with the increased burden on the ER (Spear and Ng 2003), and several of the Kar2p retention mutants previously identified as synthetic sick/lethal in combination with hac1Δ also possess a constitutive UPR (Schuldiner et al. 2005). We demonstrated that the UPR was activated in each of the strains that showed a synthetic lethal interaction with hac1Δ (Table S1), suggestive of roles in protein folding and/or degradation within the ER. Some of these mutants (eos1Δ, lhs1Δ, alg3Δ, mnn11Δ, and ubx2Δ) have known defects in protein glycosylation or folding, whereas several mutants (pho88Δ, csf1Δ, mrpl16Δ, and ylr065cΔ) correspond to novel candidates for similar roles. Furthermore, another recent genomewide analysis of mutations that induce the UPR identified a large number of these strains as having consititutive UPR, consistent with a role in protein folding and/or biogenesis (Jonikas et al. 2009). Additional characterization of secretory pathway function provided more direct support for roles for three proteins (Csf1p, Mrpl16p, and Pho88p) in the maturation of secretory proteins (Figure 2) and for the final protein, Ylr065cp, in protein quality control (Figure 3).
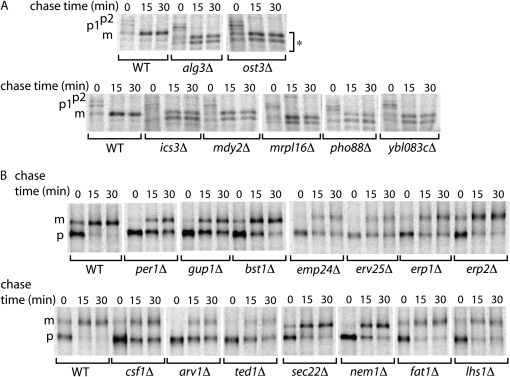
Figure 2.—
Maturation of CPY and Gas1p is delayed in some Kar2p secretion mutants. (A) CPY maturation from ER (p1) to Golgi (p2) and vacuolar (p3) forms was monitored by pulse-chase analysis. Several mutants accumulated underglycosylated forms of CPY (marked with *). (B) Maturation of the GPI-anchored protein, Gas1p, from ER-localized precursor (p) to Golgi-modified mature (m) forms was similarly monitored. A number of mutants showed a delay in the appearance of mature Gas1p. Only those strains exhibiting defects in protein maturation are shown.
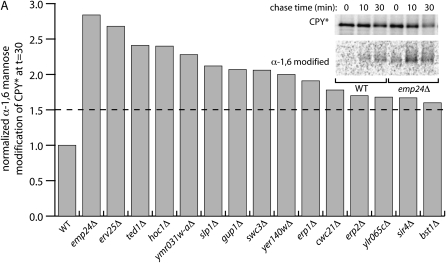
Figure 3.—
Quality control of a misfolded protein in Kar2p retention mutants. (A) Kar2p retention mutants expressing HA-CPY* were subjected to pulse-chase analysis (inset) and the degree of acquisition of α-1,6-mannose was quantified by sequential immunoprecipitation. Only those strains exhibiting defects in protein quality control are shown.
A subset of Kar2p secretors exhibit defects in post-translational modification of secretory proteins:
We tested secretory pathway function in each of the Kar2p retention mutants by monitoring the in vivo maturation of the cell wall GPI-anchored protein, Gas1p and the vacuolar hydrolase, CPY. Both of these model proteins are modified upon delivery to the Golgi, yielding an easily monitored marker for forward transport through the secretory pathway during a pulse-chase experiment. We identified a subset of mutants that showed glycosylation defects in CPY, resulting in the appearance of underglycosylated forms of the mature protein (Figure 2A). Alg3p and Ost3p are known components of the N-linked glycosylation machinery (Lehle et al. 2006), and a dubious ORF, YBL083C, partially overlaps with ALG3. The remaining mutants, ics3Δ, pho88Δ, mdy2Δ, and mrpl16Δ, have not previously been described as defective in processing of secretory proteins. Since Mrpl16p is localized to mitochondria and has a structural role in the mitochondrial ribosome, the effect on glycosylation of secretory proteins is likely to be indirect, perhaps by influencing the synthesis of lipid-linked oligosaccharide precursors. Conversely, Pho88p is localized to the ER, putting it in a prime position to participate directly in N-linked glycosylation. Furthermore, the synthetic lethality of pho88Δ and mrpl16Δ strains with hac1Δ further supports a role in protein folding through post-translational modification of secretory cargoes. The precise functions of these putative glycosylation components remain to be fully characterized.
We also identified a number of mutants with a delay in maturation of the cell wall GPI-anchored protein, Gas1p (Figure 2B). Some of the more severely affected mutants correspond to previously described components of the GPI-anchor attachment and remodeling machinery (Bst1p, Gup1p, and Per1p; Orlean and Menon 2007), or as putative cargo receptors for Gas1p (Emp24p; Muniz et al. 2000). Furthermore, related members of the p24 family of proteins were similarly affected; Erv25p, Erp1p, and Erp2p form a physical complex with Emp24p and play roles in both anterograde and retrograde traffic between the ER and Golgi (Belden and Barlowe 1996; Marzioch et al. 1999; Aguilera-Romero et al. 2008). The remaining mutants correspond to proteins with poorly described molecular functions in vivo: Ted1p, which may regulate the p24 proteins (Haass et al. 2007); Arv1p, involved in intracellular distribution of lipids (Kajiwara et al. 2008); and Csf1p, an uncharacterized protein with a single predicted transmembrane domain. Finally, several mutants showed less severe delays in the maturation of Gas1p: Lhs1p is an ER chaperone (Craven et al. 1996); Sec22p functions in protein localization in the early secretory pathway by mediating vesicle fusion (Bennett 1995); Fat1p and Nem1p play distinct roles in lipid biosynthesis, functioning in fatty acid transport and regulation, respectively (Zou et al. 2002; Santos-Rosa et al. 2005). Proteins that function in various aspects of lipid biosynthesis and trafficking likely impact Gas1p biogenesis through alterations in the attachment of the GPI anchor or the ability of Gas1p to enter into an appropriate lipid environment prior to forward transport. This accumulation of immature GPI-anchored proteins in the ER could overwhelm the protein folding and degradation machinery, causing induction of the UPR and subsequent upregulation of Kar2p. Similarly, failure to appropriately glycosylate CPY could result in UPR activation and Kar2p secretion. Indeed, many of these glycosylation and GPI-anchor mutants were synthetic lethal when combined with a hac1Δ null allele, suggesting they depend upon the UPR to maintain a functional secretory pathway (Table 1).
ER quality control is defective in a subset of Kar2p secretors:
An important function of the endoplasmic reticulum is to regulate protein secretion such that misfolded or unassembled proteins are not deployed within the cell (Ellgaard and Helenius 2003). We reasoned that Kar2p secretion from cells may result from a breakdown in this quality control checkpoint, either because this pathway also functions in the retention of ER residents or alternatively, because Kar2p may be bound to misfolded proteins that are improperly packaged into COPII vesicles and thereby secreted. We examined ER quality control in our panel of mutants by monitoring the glycosylation state of a model misfolded protein, CPY*, which is normally targeted for ERAD and thus does not acquire Golgi-specific modifications. We introduced an HA-tagged form of CPY* into each of the Kar2p retention mutants and measured the acquisition of α-1,6-mannose during a pulse-chase experiment, indicative of Golgi delivery of the aberrant protein (Figure 3). In wild-type cells, very little CPY* was immunoprecipitated with antibodies against the α-1,6-mannose moiety; however, in a small number of mutants we detected significant Golgi modification of this misfolded protein.
Among the mutants with impaired ER retention were the p24 family members, emp24Δ, erv25Δ, erp1Δ, and erp2Δ and the putative p24 regulator, ted1Δ. Similarly affected were two components of the GPI-anchor remodeling machinery, bst1Δ and gup1Δ. Mutation of either EMP24 (also known as BST2) or BST1 has been previously described as causing defects in ER retention of another misfolded secretory protein, the S11 form of invertase, consistent with broad defects in ER retention in these related sets of mutants (Elrod-Erickson and Kaiser 1996). Several other strains that showed more moderate defects in the ER retention of CPY* correspond to particularly interesting candidates in ER quality control. Yer140wp is a predicted membrane protein of unknown localization that physically interacts with Slp1p, another poorly characterized membrane protein (Collins et al. 2007). Ylr065cp is yet another predicted membrane protein that, when mutated, is synthetic lethal with hac1Δ, indicative of a role in protein folding and quality control. We propose that these proteins represent novel players that contribute to the fidelity of protein sorting in the ER.
Kar2p secretion can result from Golgi-ER retrieval defects:
Release of Kar2p to the cell surface could result from impaired retention of the ER population, or by altered retrieval of escaped Kar2p from the Golgi via the HDEL receptor, Erd2p (Townsley et al. 1994). We aimed to distinguish between defects in retention and retrieval by examining the localization of Erd2p, which we fused to GFP as integrated insertion into the ERD2 locus. This integration supports viability, suggesting that it functionally complements erd2 disruption. In wild-type cells grown in rich medium, Erd2p localizes largely to the ER (Schuldiner et al. 2005), with a minor population in punctate foci that likely correspond to the cis-Golgi (Figure 4A). In the majority of Kar2p retention mutants, Erd2p was similarly localized; however, there were several mutants that accumulated Erd2p-GFP in a single larger spot that may represent a distended Golgi structure, suggestive of a failure to recycle proteins back to the ER. These mutants include the ER-Golgi SNARE mutant, sec22Δ; SNARE proteins mediate the fusion of vesicles with the correct target compartment and Sec22p functions both in anterograde ER-Golgi and retrograde Golgi-ER traffic (Burri et al. 2003; Dilcher et al. 2003). Perturbation of Sec22p function likely directly impedes the retrieval of Erd2p-GFP, resulting in its steady-state Golgi localization. Erd2p-GFP was similarly mislocalized in mutants that correspond to all three members of the GET complex, Get1p, Get2p, and Get3p, which function in the ER integration of tail-anchored proteins (Schuldiner et al. 2008), including SNAREs, and thereby indirectly affect Golgi-ER retrieval. The get mutants have been previously described as defective in Erd2p localization (Schuldiner et al. 2005), among other pleiotropic phenotypes that are consistent with broad defects in many aspects of cellular function (Schuldiner et al. 2008). Five additional mutants, mdy2Δ, mga2Δ, rce1Δ, htz1Δ, and yor164cΔ, exhibited a similar aberrant distribution of Erd2p-GFP, albeit less striking than the get1Δ, get2Δ, get3Δ, and sec22Δ strains. Finally, the ilm1Δ strain showed a distinct Erd2-GFP localization pattern, with increased fluorescence in a distended structure that appeared to be continuous with the perinuclear ER and may represent a distended karmella-like domain. The known genetic interactions among the group of mutants that showed defective Erd2p-GFP localization are consistent with overlapping roles in Golgi-ER retrieval; many components show synthetic sick or aggravating interactions with the get mutants, suggesting distinct functional roles that are more deleterious when combined than when present as single mutations (Figure 4B). Conversely, genetic interactions among the get mutants are neutral or alleviating, consistent with shared roles in a single pathway (Boone et al. 2007).
Figure 4.—
Golgi-ER retrieval is perturbed in some Kar2p retention mutants. (A) Subcellular localization of Erd2p-GFP was examined in each of the Kar2p retention mutants. Unlike the ER localization seen in WT cells, a number of mutants accumulated Erd2p-GFP in punctate foci. In ilm1Δ cells, Erd2p-GFP accumulated in distended ER structures (arrowheads). (B) These Erd2p localization mutants show a variety of interactions, including phenotypic enhancement or synthetic lethality (PE/SL; blue), phenotypic suppression (PS; red), yeast 2-hybrid (Y2-H; green), and physical association (gray). (C) Subcellular localization of the tail-anchored protein, GFP-Frt1p, in WT cells and Kar2p retention mutants.
Five of the mutants that show defective Erd2p localization correspond to proteins that have been isolated as two separate physical complexes: Get1p, Get2p, and Get3p form an ER-localized protein complex (Schuldiner et al. 2005), and Mdy2p copurifies with Yor164cp (Fleischer et al. 2006; Krogan et al. 2006). High throughput yeast two-hybrid analysis identified an interaction between Get3p and Yor164cp (Ito et al. 2001), suggesting a link between these two complexes that may explain the shared Erd2p defect. Get3p has been proposed to function as a cytosolic receptor that binds the hydrophobic domains of newly synthesized tail-anchored proteins that subsequently docks with the ER localized receptors, Get1p and Get2p. The physical association of Yor164cp with Mdy2p, which in turn interacts with ribosomes (Fleischer et al. 2006), is suggestive of an intermediate role for the Yor164cp/Mdy2p complex linking the GET machinery directly to the ribosome. We examined the localization of a tail-anchored protein, GFP-Frt1p, in the yor164cΔ and mdy2Δ strains to determine if membrane integration of this class of proteins is perturbed. In wild-type cells, GFP-Frt1p localizes to punctate foci within the endoplasmic reticulum; in get mutant strains, we observed a more diffuse, cytosolic localization, with significant accumulation in a single large puncta, similar to that observed for other tail-anchored proteins in these mutants (Schuldiner et al. 2008). Similarly, in yor164cΔ and mdy2Δ cells, GFP-Frt1p, displayed diffuse cytosolic staining with a single focus (Figure 4C). Other Erd2p retrieval mutants did not show the same phenotype and closely resembled wild-type cells (data not shown), suggesting a specific role for the Yor164cp and Mdy2p proteins in this process. The precise function that Yor164cp and Mdy2p perform in protein insertion and how they interface with both the ribosome and Get3p remain to be explored. However, we propose that the Kar2p secretion phenotype of the mdy2Δ and yor164cΔ mutants stems largely from defective insertion of SNARE proteins that, like in the getΔ mutants, results in the impaired retrograde traffic of Golgi-localized Erd2p, thereby causing inefficient retrieval and secretion of Kar2p. Similar findings identifying Yor164cp and Mdy2p, newly renamed Get4p and Get5p, respectively, as components of the tail-anchored insertion pathway have recently been published (Jonikas et al. 2009).
Evidence for modulation of Kar2p secretion by the histone variant Htz1p:
One surprising set of mutants that was markedly enriched in our collection of Kar2p retention mutants corresponds to the SWR1 complex and the variant histone, Htz1p, that it acts on. Many of these mutants have pleiotropic defects associated with various deficiencies in the secretory pathway, including vacuolar protein sorting defects (Bonangelino et al. 2002), membrane remodeling defects (Wright et al. 2003), and sensitivity to exogenous lipids (Lockshon et al. 2007). Having added an additional defect, ER retrieval, to the list of htz1Δ phenotypes, we aimed to further dissect the nature of this mislocalization. One function of Htz1p is to antagonize the silencing activities of Sir2p and HMR at telomeres and the mating loci (Meneghini et al. 2003). We reasoned that if Erd2p mislocalization and Kar2p secretion resulted from inappropriate silencing in these regions, then normal function could be restored by deletion of either SIR2 or HMR in the htz1Δ background. However, Kar2p secretion was equivalent in the htz1Δ, htz1Δsir2Δ, and htz1ΔhmrΔ mutants (Figure 5A), suggesting that the specific regulatory function of Htz1p is likely to result from binding to individual promoters of candidate genes rather than a broader anti-silencing function.
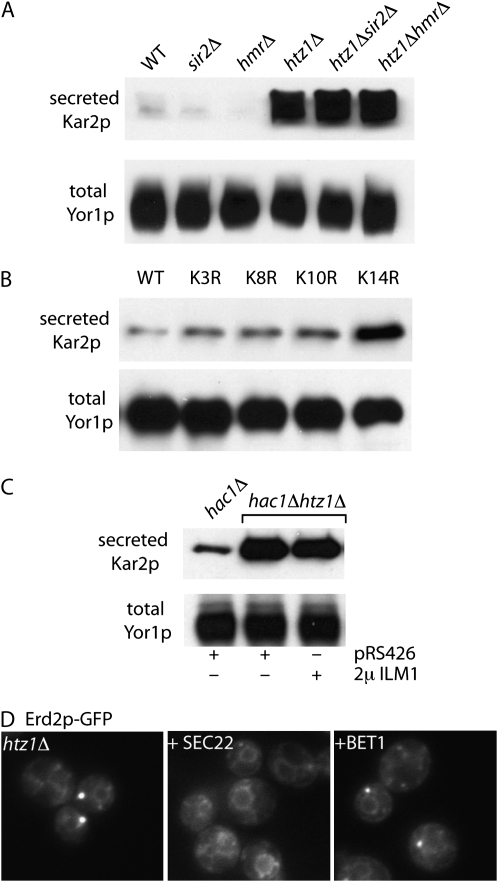
Figure 5.—
Htz1p contributes to Kar2p secretion via Golgi-ER retrieval. Secretion of Kar2p was detected in the indicated strains by immunoblotting of the extracellular medium (top) and total cellular protein was monitored by immunoblotting of the plasma membrane protein, Yor1p (bottom). (A) Kar2p secretion in htz1Δ cells was equivalent regardless of sir2Δ and hmrΔ deletion. (B) Secretion of Kar2p correlates specifically with the K14R acetylation-deficient mutant of Htz1p. (C) Kar2p secretion in an htz1Δ mutant was not rescued by overexpression of ILM1. Cells expressing the empty vector, pRS426, serve as a negative control. (D) The punctate distribution of Erd2p-GFP in htz1Δ cells (left) was restored to an ER localization by overexpression of SEC22 (middle), but not BET1 (right).
The shared Kar2p secretion phenotype of the SWR1 and NuA4 complexes (Figure 1D) led us to determine whether acetylation of Htz1p is required for normal ER retention and homeostasis (Babiarz et al. 2006; Keogh et al. 2006). We examined Kar2p secretion in strains where Htz1p was replaced with mutant forms that are unable to be acetylated on each of four key lysine residues, K3, K8, K10, and K14 (Babiarz et al. 2006). Compared with an isogenic wild-type strain, only the K14R variant of Htz1p showed significant Kar2p secretion (Figure 5B), suggesting a requirement for acetylation of K14 on Htz1p to maintain ER homeostasis or organization. To determine whether the Kar2p secretion phenotype of the htz1Δ strain stems from downregulation of any of the 87 known Kar2p retention mutants, we examined existing microarray data sets for genes that coincided with our collection of mutants (Meneghini et al. 2003). Only one gene that was downregulated in the htz1Δ and htz1Δsir2Δ mutants also showed a Kar2p retention defect when mutated: ILM1. We tested the ability of ILM1 to rescue the Kar2 secretion and Erd2p-GFP localization phenotypes when overexpressed in the htz1Δ strain, but were unable to detect any rescue (data not shown). We were concerned that overexpression of Ilm1p, an ER membrane protein, would induce the UPR and thereby upregulate Kar2p expression, masking any rescue of the phenotype. We repeated the ILM1 suppression experiment in a hac1Δ background, such that the UPR would be inactivated and Kar2p secretion would stem directly from the htz1Δ defect. Similar to wild-type cells, extracellular Kar2p was not abundant in hac1Δ cells, whereas hac1Δhtz1Δ double mutants showed significant Kar2p secretion, which was not rescued by overexpression of ILM1, suggesting that downregulation of ILM1 is not the primary cause of Kar2p secretion in htz1Δ cells (Figure 5C). Precisely how many genes contribute to the Kar2p secretion phenotype of the htz1Δ mutant remains to be determined, and Ilm1p disregulation may still play a part in this process. The pleiotropic phenotypes of the htz1Δ and SWR1-complex mutants suggest a relatively broad impact on the secretory pathway, consistent with a complex pattern of disregulation that would perturb a variety of processes.
Such broad defects in secretory pathway function in htz1Δ mutants might indicate a role in tail-anchored protein insertion, where multiple steps of membrane fusion would be impaired due to altered abundance of the SNARE fusion machinery. Using a variety of tail-anchored proteins, we did not observe any defect in intracellular distribution in htz1Δ mutant cells (data not shown). However, overexpression of the SNARE, Sec22p, which functions both in anterograde and retrograde traffic between the ER and Golgi, caused a redistribution of Erd2p-GFP to the ER (Figure 4D) and diminished the Kar2p secretion phenotype in htz1Δ cells (data not shown). Similar overexpression of additional anterograde SNAREs, Bet1p, Bos1p, and Sed5p, had no effect (Figure 4D and data not shown), consistent with a retrograde trafficking defect in the htz1Δ mutant. We speculate that in the absence of normal Htz1p function, retrograde vesicles are rendered less fusion competent and that upregulation of the SNARE machinery alleviates this defect. How Htz1p influences membrane trafficking events remains to be determined, but the pleiotropic phenotypes of htz1Δ cells, including aberrant formation of proliferated ER domains known as karmellae (Wright et al. 2003), a cold-sensitive growth phenotype and sensitivity to the presence of the fatty acid, oleate (Lockshon et al. 2007), are consistent with altered lipid metabolism that could lead to broad defects in secretory pathway function.
CONCLUSIONS
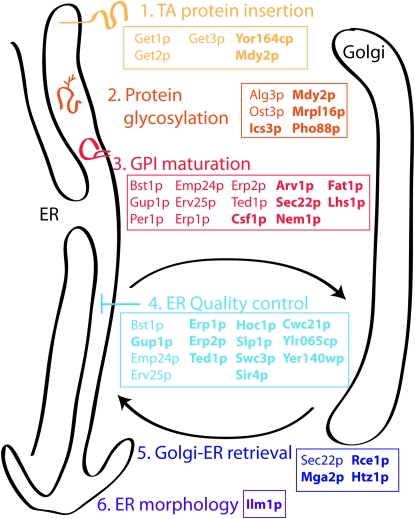
To define the cellular machinery that contributes to ER homeostasis, we employed a genomewide approach to identify mutants that are defective in the intracellular retention of the abundant lumenal chaperone, Kar2p. We identified 73 nonessential genes and 14 essential genes as candidate mediators of ER retention. This set of mutants encompassed known components of protein folding, post-translation modification, and protein trafficking pathways and identified novel candidates of protein glycosylation, GPI biogenesis, and ER quality control (Figure 6). Our characterization of the mislocalization of Erd2p-GFP led to the identification of a novel pair of regulators of tail-anchored protein insertion. Like the get mutants, yor164cΔ and mdy2Δ accumulate tail-anchored proteins in a large punctate spot that likely stems from aggregation of the hydrophobic domains when membrane insertion fails. Since Mdy2p associates with the ribosome, we propose that the Mdy2p/Yor164cp complex binds tail-anchored proteins as they come off the ribosome, working in conjunction with Get3p, which binds to the hydrophobic membrane domains to deliver them to the ER-localized Get1p/Get2p complex. Although the order of binding events and the precise role of each component remains to be fully elucidated, our findings expand our understanding of the machinery that facilitates biogenesis of this important class of membrane proteins.
Figure 6.—
Kar2p retention mutants encompass multiple pathways that contribute to ER homeostasis. Our genomewide screen identified components involved in various aspects of ER function, including protein biogenesis, protein glycosylation, GPI biogenesis, and ER quality control. Proteins annotated in boldface type represent novel functional designations for those components.
From the perspective of maintaining ER function, one of the more interesting subsets of mutants we identified were those with defective ER quality control. The effective retention of misfolded proteins plays an important role in maintaining accurate cellular function; however, an overzealous quality control checkpoint can have detrimental consequences. For example, cystic fibrosis can be caused by the stringent ER retention and degradation of mutant forms of CFTR, a membrane protein that can partially function if delivered to the cell surface (Welch 2004). The mutants that we identified as defective in the ER retention of CPY* represent candidates for broad regulators of ER quality control. In particular, several poorly characterized membrane proteins, Slp1p, Yer140wp, and Ylr065cp, have clear human orthologs and warrant further characterization with respect to additional misfolded proteins. Together, the suite of mutants that we have defined here represents a strong starting point from which we hope to gain further insight into the interrelated processes of secretory protein biogenesis, ER export, and protein quality control.
Acknowledgments
We thank Randy Schekman, Jasper Rine, Hiten Madhani, Liza Pon, Traude Beilharz, and Roy Buchanan for contributing strains, plasmids, and antibodies. Many thanks to Michael Wolfe for his early efforts in the screening process and to David Fidock for providing access to the microscopy facility. This work was supported in part by the National Institutes of Health grants GM-078186 (to E.A.M.) and GM-078855 (to M.D.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101105/DC1.
References
- Aguilera-Romero, A., J. Kaminska, A. Spang, H. Riezman and M. Muniz, 2008. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. J. Cell Biol. 180 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli, T., and R. Sitia, 2008. Protein quality control in the early secretory pathway. EMBO J. 27 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz, J. E., J. E. Halley and J. Rine, 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., 2003. Molecular recognition of cargo by the COPII complex: a most accommodating coat. Cell 114 395–397. [DOI] [PubMed] [Google Scholar]
- Beh, C. T., and M. D. Rose, 1995. Two redundant systems maintain levels of resident proteins within the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92 9820–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden, W. J., and C. Barlowe, 1996. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 271 26939–26946. [DOI] [PubMed] [Google Scholar]
- Belden, W. J., and C. Barlowe, 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell 12 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. K., 1995. SNAREs and the specificity of transport vesicle targeting. Curr. Opin. Cell Biol. 7 581–586. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., E. M. Chavez and J. S. Bonifacino, 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone, C., H. Bussey and B. J. Andrews, 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. 8 437–449. [DOI] [PubMed] [Google Scholar]
- Breitkreutz, B. J., C. Stark and M. Tyers, 2003. Osprey: a network visualization system. Genome Biol. 4 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri, L., O. Varlamov, C. A. Doege, K. Hofmann, T. Beilharz et al., 2003. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100 9873–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, S. R., K. J. Hill and A. A. Cooper, 2001. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 276 23296–23303. [DOI] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806–810. [DOI] [PubMed] [Google Scholar]
- Craven, R. A., M. Egerton and C. J. Stirling, 1996. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 15 2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Dilcher, M., B. Veith, S. Chidambaram, E. Hartmann, H. D. Schmitt et al., 2003. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22 3664–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and A. Helenius, 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 4 181–191. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson, M. J., and C. A. Kaiser, 1996. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell 7 1043–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer, T. C., C. M. Weaver, K. J. McAfee, J. L. Jennings and A. J. Link, 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle et al., 2003. Global analysis of protein expression in yeast. Nature 425 737–741. [DOI] [PubMed] [Google Scholar]
- Gurkan, C., S. M. Stagg, P. Lapointe and W. E. Balch, 2006. The COPII cage: unifying principles of vesicle coat assembly. Nat. Rev. Mol. Cell. Biol. 7 727–738. [DOI] [PubMed] [Google Scholar]
- Haass, F. A., M. Jonikas, P. Walter, J. S. Weissman, Y. N. Jan et al., 2007. Identification of yeast proteins necessary for cell-surface function of a potassium channel. Proc. Natl. Acad. Sci. USA 104 18079–18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas, M. C., S. R. Collins, V. Denic, E. Oh, E. M. Quan et al., 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323 1693–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara, K., R. Watanabe, H. Pichler, K. Ihara, S. Murakami et al., 2008. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell 19 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan et al., 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid, M. M., and A. A. Cooper, 2007. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol. Biol. Cell 18 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J., 2000. Transport between ER and Golgi. Curr. Opin. Cell Biol. 12 445–449. [DOI] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo et al., 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440 637–643. [DOI] [PubMed] [Google Scholar]
- Lee, M. C., E. A. Miller, J. Goldberg, L. Orci and R. Schekman, 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20 87–123. [DOI] [PubMed] [Google Scholar]
- Lehle, L., S. Strahl and W. Tanner, 2006. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 45 6802–6818. [DOI] [PubMed] [Google Scholar]
- Lockshon, D., L. E. Surface, E. O. Kerr, M. Kaeberlein and B. K. Kennedy, 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch, M., D. C. Henthorn, J. M. Herrmann, R. Wilson, D. Y. Thomas et al., 1999. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell 10 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Mezzacasa, A., and A. Helenius, 2002. The transitional ER defines a boundary for quality control in the secretion of tsO45 VSV glycoprotein. Traffic 3 833–849. [DOI] [PubMed] [Google Scholar]
- Muniz, M., C. Nuoffer, H. P. Hauri and H. Riezman, 2000. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D. T., E. D. Spear and P. Walter, 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean, P., and A. K. Menon, 2007. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48 993–1011. [DOI] [PubMed] [Google Scholar]
- Otte, S., and C. Barlowe, 2004. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell. Biol. 6 1189–1194. [DOI] [PubMed] [Google Scholar]
- Pagant, S., L. Kung, M. Dorrington, M. C. Lee and E. A. Miller, 2007. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol. Biol. Cell 18 3398–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H. R., K. G. Hardwick and M. J. Lewis, 1988. Sorting of soluble ER proteins in yeast. EMBO J. 7 1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, D., 2002. Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. J. Clin. Invest. 109 443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., J. Leung, N. Grimsey, S. Peak-Chew and S. Siniossoglou, 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati et al., 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123 507–519. [DOI] [PubMed] [Google Scholar]
- Schuldiner, M., J. Metz, V. Schmid, V. Denic, M. Rakwalska et al., 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134 634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, J. C., K. G. Hardwick, N. Dean and H. R. Pelham, 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61 1349–1357. [DOI] [PubMed] [Google Scholar]
- Singh, I., R. Pass, S. O. Togay, J. W. Rodgers and J. L. Hartman, IV, 2009. Stringent mating-type-regulated auxotrophy increases the accuracy of systematic genetic interaction screens with Saccharomyces cerevisiae mutant arrays. Genetics 181 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear, E. D., and D. T. Ng, 2003. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell 14 2756–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley, F. M., G. Frigerio and H. R. Pelham, 1994. Retrieval of HDEL proteins is required for growth of yeast cells. J. Cell Biol. 127 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman et al., 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101 249–258. [DOI] [PubMed] [Google Scholar]
- Vashist, S., W. Kim, W. J. Belden, E. D. Spear, C. Barlowe et al., 2001. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 155 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, W. J., 2004. Role of quality control pathways in human diseases involving protein misfolding. Semin. Cell Dev. Biol. 15 31–38. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Wright, R., M. L. Parrish, E. Cadera, L. Larson, C. K. Matson et al., 2003. Parallel analysis of tagged deletion mutants efficiently identifies genes involved in endoplasmic reticulum biogenesis. Yeast 20 881–892. [DOI] [PubMed] [Google Scholar]
- Zou, Z., C. C. DiRusso, V. Ctrnacta and P. N. Black, 2002. Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J. Biol. Chem. 277 31062–31071. [DOI] [PubMed] [Google Scholar]