Abstract
Insects use an amazing variety of genetic systems to control sexual development. A Y-linked male determining gene (M) controls sex in the Australian sheep blowfly Lucilia cuprina, an important pest insect. In this study, we isolated the L. cuprina transformer (Lctra) and transformer2 (Lctra2) genes, which are potential targets of M. The LCTRA and LCTRA2 proteins are significantly more similar to homologs from tephritid insects than Drosophila. The Lctra transcript is alternatively spliced such that only females make a full-length protein and the presence of six TRA/TRA2 binding sites in the female first intron suggest that Lctra splicing is autoregulated as in tephritids. LCTRA is essential for female development as RNAi knockdown of Lctra mRNA leads to the development of male genitalia in XX adults. Analysis of Lctra expression during development shows that early and midstage male and female embryos express the female form of Lctra and males express only the male form by the first instar larval stage. Our results suggest that an autoregulatory loop sustains female development and that expression of M inhibits Lctra autoregulation, switching its splicing to the male form. The conservation of tra function and regulation in a Calliphorid insect shows that this sex determination system is not confined to Tephritidae. Isolation of these genes is an important step toward the development of a strain of L. cuprina suitable for a genetic control program.
INSECTS have developed a great variety of genetic systems to determine sex (Marin and Baker 1998; Schütt and Nöthiger 2000; Saccone et al. 2002; Shearman 2002; Sanchez 2008). One of them consists of a Y-linked male determining factor whose activity represses female development and promotes the male phenotype. This system controls sex determination in the Mediterranean fruitfly Ceratitis capitata, the Olive fruitfly Bactrocera oleae, and the house fly Musca domestica. In the latter, a Y-linked dominant male factor M, which can be autosomal in some rare strains, represses F, the key gene for female sex determination, leading to male development. In the absence of M, F is activated, resulting in female development (Dübendorfer et al. 2002). The zygotic activation of F requires maternal activity of the F gene and is highly dose sensitive. Indeed, zygotes that are heterozygous for F develop as normal females if they derive from a mother with two functional F alleles, while those derived from heterozygous mothers cannot sustain female development. Thus, F appears to be autoregulated (Dübendorfer and Hediger 1998). The Australian sheep blowfly Lucilia cuprina, is an economically important pest insect belonging to the Caliptratae subsection of dipterans and thus closely related to the house fly M. domestica (Beck et al. 1985; Heath and Bishop 2006). Sex in L. cuprina is determined by a male determining region that is located near the Y chromosome centromere (Bedo and Foster 1985). However, the nature of the male determining factor as well as the subordinate genes that compose the sex determination cascade in this species are unknown.
The insect sex determination system that has been best characterized is that of the fruitfly Drosophila melanogaster. Until recently, it was thought that the ratio of X chromosomes to sets of autosomes constitutes the primary signal for sex determination in this insect (Cline 1993; Penalva and Sanchez 2003). However, recent evidence points to the number of X chromosomes rather than the X:A ratio as the primary signal (Erickson and Quintero 2007). According to this view, the male or female dose of X chromosomes is defined by the collective concentrations of four X-linked signal element (XSE) proteins in the zygote, which function to activate the Sex-lethal gene (SXL-F) in females, thereby promoting female development through a short cascade of downstream genes. SXL-F regulates the splicing of tra pre-mRNA such that only females produce an RNA that codes for a full-length and functional TRA protein (Sosnowski et al. 1989). TRA forms a complex with TRA2, a cofactor that is constitutively expressed in both sexes and promotes the female-specific splicing of doublesex pre-mRNA (dsx), the last component of the regulatory hierarchy. Interestingly, a feedback loop has been described to maintain SXL-F activity in Drosophila, where the SXL-F protein is capable of activating the splicing of its own pre-mRNA as well as that of tra, and reciprocally, female TRA from either maternal or zygotic expression stimulates Sxl-positive autoregulation (Siera and Cline 2008). In the absence of functional SXL protein, male-specific splicing of tra occurs by default, and hence, male-specific splicing of dsx, resulting in the development of the male phenotype. The male and female DSX proteins, DSXM and DSXF, are transcription factors that promote sexual development by activating the transcription of sex-specific differentiation genes.
Wilkins (1995) proposed that the sex determination gene hierarchy evolved from the bottom up. Consistent with this model, orthologs of Drosophila Sxl have been found in several dipterans including Megaselia scalaris (Sievert et al. 1997), C. capitata (Saccone et al. 1998), B. oleae (Lagos et al. 2005), M. domestica (Meise et al. 1998), and the Calliphoridae species Chrysomya rufifacies (Muller-Holtkamp 1995) and L. cuprina (P. Atkinson, personal communication). However, Sxl isn't sex-specifically spliced in these species and does not appear to have a role in sex determination. In contrast, at the bottom of the sex determination hierarchy, dsx is sex-specifically spliced in Apis mellifera (Cho et al. 2007), Bombyx mori (Suzuki et al. 2001), Anopheles gambiae (Scali et al. 2005), M. scalaris (Kuhn et al. 2000), M. domestica (Hediger et al. 2004) Anastrepha obliqua (Ruiz et al. 2005), C. capitata (Saccone et al. 2002), B. tryoni (Shearman and Frommer 1998), and B. oleae (Lagos et al. 2005).
Outside of the genus Drosophila, the transformer gene has been isolated from the tephritids C. capitata (Cctra) (Pane et al. 2002), B. oleae (Botra) (Lagos et al. 2007), and from several species from the genus Anastrepha (e.g., the West Indian fruit fly A. obliqua) (Ruiz et al. 2007). The genomic organization and sex-specific splicing of tra is similar in all of these tephritid species. As in Drosophila, only females produce an RNA that codes for a full-length TRA protein. Further, TRA is essential for female development in C. capitata and B. oleae as was shown by RNAi experiments (Pane et al. 2002; Lagos et al. 2007). Interestingly, the tephritid TRA genes contain several putative TRA/TRA2 binding sites within the male-specific exons and their flanking introns. These findings suggested an autoregulatory mechanism for the maintenance of female-specific expression of tra in these species. Moreover, a recent study shows that C. capitata tra2 is also required for maintaining the positive feedback regulation of Cctra during development and is therefore necessary for establishing female sex determination in female embryos (Salvemini et al. 2009). A proposed model for tra autoregulation in tephritids suggests that the binding of the TRA/TRA2 complex to male-specific exon sequences in tra mRNA causes a blockage of the male-specific splice acceptor sites to the general splicing machinery preventing the incorporation of the male exons into the mature tra mRNA (Pane et al. 2002). However, since overexpression of C. capitata tra in XY Drosophila leads to the female-specific splicing pattern of dsx (Pane et al. 2005), it is likely that medfly TRA has also retained the splicing enhancer function described for Drosophila TRA.
In this article we have isolated and characterized the transformer and transformer2 genes of L. cuprina with a long-term aim of understanding the genetic mechanism controlling sex determination in this important pest species and its evolution from that of other Diptera. We have found that Lctra shares several common features with tephritid tra genes, such as the presence of six TRA/TRA2 binding sites in its pre-mRNA and a unique N-terminal domain, which is absent in the tra homologs of all the Drosphila species. The function of Lctra in selecting and maintaining the female pathway of development is conserved, showing that the tephritid sex determination system is present in a broader group of insects including the Calliphoridae family. The isolation of these genes will be useful for the development of modified strains of L. cuprina that can be employed in a genetic control program.
MATERIALS AND METHODS
Rearing of L.
cuprina strains: L. cuprina adults were maintained in the laboratory at a constant temperature of 21° under a 12/12 hr light/dark cycle. The flies were fed water and a protein-rich cookie and given lamb liver every 4 days for egg laying. The larvae were grown in commercial jelly meat pet food at 27° until the wandering third instar larval stage. The pet food was then removed and the pupae incubated at 27° until eclosion. The HS14 transgenic line of L. cuprina carries the Lchsp83-ZsGreen marker on the X chromosome and is maintained in a stable manner under the same conditions.
PCR, RACE, and recombinant DNA:
To isolate the L. cuprina sex determination genes our general strategy was to perform two rounds of PCR using nested primers with cDNA templates. The degenerate primers were designed against conserved amino acid blocks and were designated F1 and R1 for the first round and F2 and R2 for the second round of PCR. To make the cDNA template, total RNA was extracted with TRIZOL reagent (Invitrogen) following the manufacturer's instructions. The RNA was Turbo-DNAse treated (Ambion), phenol/chloroform extracted, ethanol precipitated, and resuspended in nuclease-free water to be used directly for RT–PCR. Adult male and female RNA was further purified by affinity chromatography with oligo (dT) cellulose (Sigma). First strand cDNA synthesis was performed using an oligo (dT) primer and Expand Reverse Transcriptase (Roche). Cycling conditions for both PCR rounds were denaturation 95° for 2 min, then 30 cycles (denaturation 95° for 25 sec, annealing at 48° for 30 sec, and extension at 68° for 2 min), and finally extension at 68° for 5 min. Subcloning and sequencing of the candidate fragments were carried out by standard procedures.
The degenerate primers were:
Tra-F1 5′ TTY CAA MGW GAT GAT ATW GTD GTD AAT CC 3′.
Tra-F2 5′ GAA AAA RTT CCH TAT TTY RTT GAT GAA RTT MGW GAA 3′.
Tra-R1 5′ GG WAC DGG AAC DGG AAT WGT AAT WAT TTG DGG 3′.
Tra-R2 5′ GG TTG DGG DGG YAA ACC ATA DGG DGG 3′.
Tra2-F1 5′ TGT ATW GGT GTD TTY GGT TTR AAT ACH AAT AC 3′.
Tra2-F2 5′ TAT GGT CCH ATW GAA CGU ATW CAA GTD GTD 3′.
Tra2-R1 5′ TA HAC ACC DGG DGT DGG DGT ATG TGC ACG TTG 3′.
Tra2-R2 5′ TA AKC HAC ACG WAT ACG ACG ACC ATC HAC 3′.
To obtain full-length cDNA sequences, we extended the cDNA fragment on both sides by 5′ and 3′ RACE using the Smart RACE Kit and Advantage 2 taq DNA polymerase (Clontech). Two rounds of PCR were performed on the 5′ and 3′ RACE libraries with specific primers directed to the library's adaptors and gene-specific primers. The cycling conditions were denaturation 95° for 2 min, then 30 cycles (denaturation 95° for 25 sec, annealing at 65° for 30 sec, and extension at 72° for 2 min), and finally extension at 72° for 5 min. The gene-specific primers used were:
Tra-F1 5′-ACC TAT CGT CAT CAT CGT CGT CGT CAA CTG C-3′.
Tra-R1 5′-GTG GAG AAC GAG TTC GTG AAC GTG TTC TAG-3′.
Tra-F2 5′-CGG CGA AGA CGT TCA ACC AGT AGA GAT CGT-3′.
Tra-R2 5′-GAA CGA CGG CTT CTG TAG TCT CTT CTT ACG C-3′.
Luc-tra2-RV1 5′-CTG GTC GTC CCA TAT AAA CAC CAG GCG TTG-3′.
Luc-tra2-FW1 5′-CTC AAA CCG GAC GTT CTC GGG GTT TTT GTC-3′.
Luc-tra2-FW2 5′-GCA GCC TGT GAT AAT TGC TGT GGC ATG GAA-3′.
To analyze the expression pattern of Lctra and Lctra2 over development, cDNA templates were prepared from total RNA isolated from various developmental stages. Thermal cycling conditions were those used for RACE. Gene-specific primers were designed to different exons so that amplification products were significantly smaller than from any contaminating genomic DNA. Further, negative control templates were prepared by omitting reverse transcriptase from the first strand cDNA synthesis reaction. The gene-specific primers used were:
Luc-Tra-FW 5′-ATG GAC TCC ATT ACA ACA GGA TTG GCA GCA-3′.
Luc-Tra-RV 5′-CTA ATG TTG TGG GGG TAA ACC ACC ATA AGA CGC-3′.
Tra2-FW 5′-ATG AGT CCA CGT TCA CAT AGT CGT TCT GTT ACA CCA-3′.
Tra2-RV 5′-TTA ATG GTA TCG ATA ACG ATA ACG ACG TGG TGA-3′.
qRT–PCR analysis was performed with adult male or female total RNA template as described previously (Li et al. 2008). The primers used were:
M2 for 5′-CAACGCAGATTTGCTAAATATTTCGAATG-3′.
M1 for 5′-TAAGCTACTTTTAAAGCTAAATATTCGAATGG-3′.
Mrev 5′-TATCACGGGCATCTAGGGTTGTTTG-3′.
α-tub for 5′-GTGATTTGGCCAAGGTACAACGTG-3′.
α-tubrev 5′-CGACGTACCAGTGGACGAAAGC-3′.
RNAi:
A cDNA fragment of the female Lctra gene was amplified from cDNA template with primers that introduced a T7 promoter sequence at each of the product ends. The resulting 920-bp fragment, comprising part of exon 1, exons 2 and 3, and part of exon 4, was used to produce dsRNA fragments by in vitro transcription performed with the Megascript kit (Ambion). The dsRNA was ethanol precipitated and resuspended in injection buffer (0.1 mm sodium phosphate (pH 6.8), 5 mm KCl) to a final concentration of 1 μg/μl. Embryos of a cross between males of the HS14 line and wild-type females were collected within 30 min of egg laying, microinjected, and allowed to develop at 21° until the stage of first instar larvae. The larvae were then observed under the fluorescent microscope and separated into fluorescent green larvae (XX individuals) and nonfluorescent larvae (XY individuals) and grown separately in pet food at 27°. After eclosion the adult flies were observed under the microscope for sexually dimorphic traits.
Sequence analysis:
Protein multiple sequence alignment was performed using CLUSTAL-W 1.83 software and analysis of the alignments was performed using BOX SHADE. Phylogenetic analysis was carried out using a basic neighbor-joining algorithm in the Geneious software package and FigTree v1.1.1 was used to draw the tree. The putative TRA-TRA2 binding sites were identified in Lctra gene sequence using MACVECTOR software for Macintosh. The accession numbers for the genes reported in this study are:
RESULTS
Isolation of Lctra:
We initially attempted two different strategies to isolate the L. cuprina homolog of tra. One approach exploited the very close linkage of the highly conserved gene l(3)73Ah with the tra gene in Drosophila species, C. capitata, and B. oleae. Indeed, this was the strategy that was used to isolate Cctra and Botra (Pane et al. 2002; Lagos et al. 2007). Although we were able to isolate the L. cuprina homolog of l(3)73Ah and flanking sequences, the tra gene does not appear to be closely linked with this gene (data not shown). The second approach involved designing degenerate PCR primers on the basis of the few amino acid motifs that were conserved in the tra homologs of several Drosophila species and C. capitata tra (Pane et al. 2002). PCR reactions were performed with cDNA templates prepared from adult female poly(A)+ RNA using several different primer combinations. However, none of the PCR reactions produced a fragment that had any sequence similarity to Cctra or Dmtra. We then modified the design of degenerate primers on the basis of conserved amino acid blocks in the TRA proteins of the two tephritid species but not Drosophila. The rationale was that since both these species and L. cuprina use a Y-linked male determining gene to determine sex, Lctra might be more similar to Cctra and Botra than to Dmtra. With new primer combinations, a 416-bp amplification product was obtained from female cDNA templates. Sequencing of the subcloned DNA fragment confirmed that we had isolated Lctra. To obtain full-length cDNA sequences, 5′ and 3′ RACE–PCR were performed with adult male and female RNA templates. From the assembled sequences, one female transcript of 1577 nt and two male transcripts of 1888 nt (male 1) and 1734 nt (male 2) were identified. The female transcript comprises a long open reading frame encoding a predicted protein of 377 amino acids. The LcTRA protein contains a high proportion of serine and arginine amino acids, characteristic of the SR family of splicing regulators. The male 1 and 2 transcripts encode short proteins of 88 and 76 amino acids, respectively, which are presumably nonfunctional as they lack the SR motifs involved in splicing regulation. Male 2 appeared to be a minor transcript as it was only detected in ∼1 in 10 cloned PCR products. qRT–PCR analysis with primers that are specific for the M1 or M2 transcript confirmed that there is 8.4 times more of the M1 than the M2 transcript in adult males. Our results show that as in Drosophila and some tephritid insects, Lctra is sex-specifically spliced and the female TRA protein is likely to have a splicing regulator function.
Characterization of the Lctra gene:
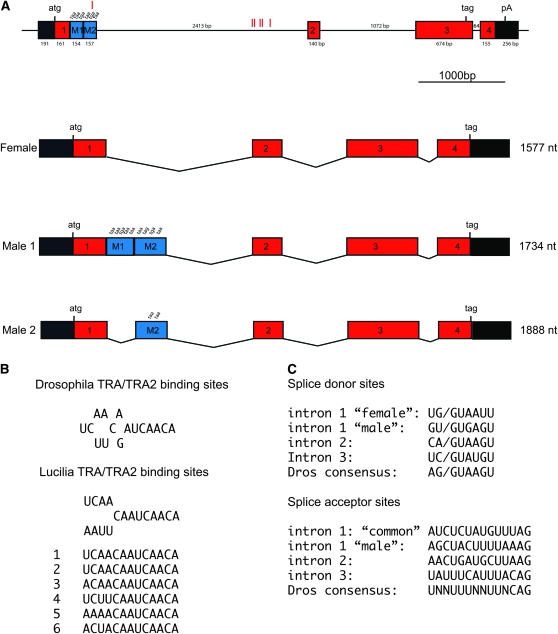
The genomic organization of Lctra was revealed by PCR amplification of genomic DNA using exon-specific primers. An alignment of genomic and cDNA sequences showed that Lctra consists of six exons and three introns that comprise 5427 bp of genomic DNA (Figure 1). The exons designated as one, two, three, and four are included in the mature transcripts of both sexes, while the exons M1 and M2 are male specific. The splicing patterns of the female and the major male 1 transcript are identical except that the splice donor sites in the first intron are different. In females the first donor site is used in splicing of exon 1 to exon 2. The major male 1 transcript arises from the use of a downstream splice donor site that is joined to the same splice acceptor as used in females. This results in the incorporation of the M1 and M2 sequences, which contain multiple in-frame translation stop codons (Figure 1). An additional splicing event excises the M1 sequence and gives rise to the minor male 2 transcript. This splicing event uses the same splice donor site as is used in female splicing of intron 1.
Figure 1.—
Schematic drawing of the genomic organization and the structure of the sex-specific splice variants of Lctra. (A) The top diagram represents the genomic DNA comprising the Lctra locus (to scale). The position of the exons is shown as square boxes, with exons 1, 2, 3, and 4 in red representing common exons to both female and male mRNAs. Exons M1 and M2 in blue represent male specific exons. Introns are represented by solid lines and the 5′- and 3′-untranslated regions are represented by black boxes. Exon and intron sizes are indicated and the translational start and stop sites are marked, but for clarity not all stop sites are shown. The position of putative TRA/TRA2 binding sites within the M2 exon and the first intron is represented by red vertical lines. The splicing patterns of the male and females transcripts are shown below the gene organization diagram (introns are not to scale). (B) Sequence of the six TRA/TRA2 binding sites found in the Lctra genomic DNA sequence and comparison with the D. melanogaster and L. cuprina consensus. (C) Splice donor and acceptor sites for all Lctra introns. The intron 1 “female” donor site is used to make both the female and male 2 transcripts whereas the intron 1 “male” donor site is used only in males. The intron 1 “male” acceptor site is used only to produce the minor male 2 transcript. Both sexes use the intron 1 “common” acceptor site to produce the major transcripts.
The position of the three introns in the female pre-mRNA is well conserved between Lctra and Cctra but has no correlation with the pattern shown for the tra gene in Drosophila. The first and third introns occur at identical positions in Lctra, Cctra, Botra, and Aotra (Figure 2). The second intron in Lctra is located near the position of the second intron in Cctra, Botra, and Aotra. However, in the region of the exon 2/exon 3 junction the LCTRA protein does not align well with the tephritid TRA proteins, making it difficult to precisely compare the relative locations of intron 2. Thus in general, the organization of the Lctra gene is very similar to the tephritid tra genes (supporting information, Figure S1). However, the splicing patterns are somewhat simpler in L. cuprina as the female form and major male form of Lctra transcripts differ only in the choice of splice donor site for intron 1.
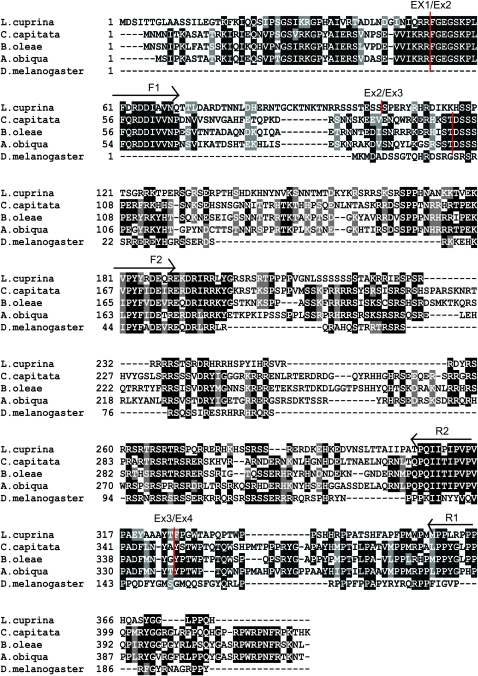
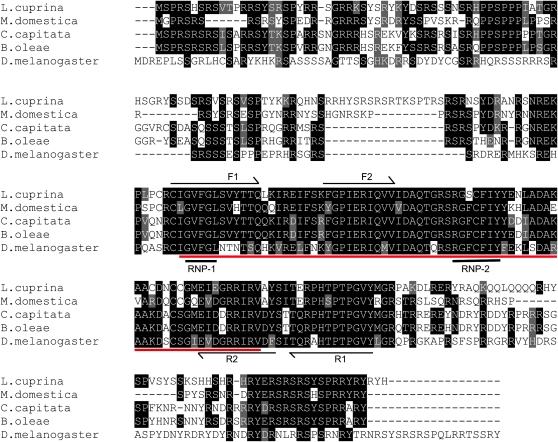
Figure 2.—
Multiple sequence alignment of TRA proteins from L. cuprina, C. capitata, B. oleae, A. obliqua, and D. melanogaster. Identical amino acids are shaded in black while similar amino acids are shaded in gray. Vertical red lines indicate the corresponding locations of the exon/intron boundaries in the L. cuprina and tephritid tra genes. Arrows indicate the conserved motifs that were the basis of the degenerate primers that were used to amplify Lctra cDNA sequence.
A ClustalW multiple sequence alignment was performed using the amino acid sequences for L. cuprina TRA, C. capitata TRA (GenBank AAM88673), B. oleae TRA (GenBank CAG29243), A. obliqua TRA (GenBank ABW04165), and D. melanogaster TRA (GenBank P11596). The alignment shows that LcTRA is more similar to TRA from the tephritid species than to Drosophila TRA (Figure 2). LcTRA is 30–33% identical and 13–14% similar to the tephritid TRA proteins but only 17% identical and 8% similar to Drosophila TRA. Most strikingly, the LcTRA amino terminal domain (aa 18–70) is very similar to the amino terminal domains of the tephritid TRA proteins but has no homology to Drosophila TRA. Indeed, Drosophila TRA, which is considerably shorter than LcTRA, doesn't appear to contain a region corresponding to the LcTRA amino terminal domain. LcTRA also contains a 16-amino-acid arginine-rich motif (PYYRDEQREKDRIRRL) and an 11-amino-acid proline-rich motif (PQIIPIPVPVP), which are well conserved in tephritid and Drosophila TRA proteins. A phylogenetic analysis was performed using the TRA protein sequences from the species mentioned above. As anticipated the results showed that the three tephritid TRA proteins form a cluster and are more closely related to LcTRA than DmTRA (Figure S2).
Developmental expression of Lctra:
To determine when the sex-specific splicing patterns are established during Lucilia development, RT–PCR was performed with primers that amplify across the first intron of Lctra, generating products of different size for the female and male transcripts (Figure 3A). For this experiment, individuals from a cross of line HS14 transgenic males with wild-type virgin females were used. Line HS14 contains a single X-linked insertion of the constitutively expressed Lchsp83-ZsGreen marker gene (M. C. Concha, E. J. Belikoff and M. J. Scott, unpublished results). Fluorescence from the ZsGreen marker is detected from midembryogenesis onward in XX individuals (9 hr at 21°), while XY individuals do not show fluorescence due to the absence of the ZsGreen gene. Total RNA was isolated from males and females at different stages of development and from early embryos (4 hr at 21°), which were mixed sex. As shown in Figure 3B, only the female form of Lctra was detected in early embryos of mixed sex as well as in midstage embryos from both sexes. The main male splice form of Lctra was not detectable until the first instar larvae stage, in which males made only the male form of Lctra RNA. From this stage onward differential sex-specific forms of tra RNA are found in males and females of L. cuprina. To determine if there was a significant maternal contribution of female Lctra transcript, RNA was isolated from unfertilized eggs laid by virgin females and from precellular embryos (<1 hr at 21°). Only the female form of Lctra was detected in both samples (Figure 3C). These results demonstrate that embryos of both sexes contain maternal RNA pools coding for the female TRA protein and that in XY individuals the male form of Lctra appears sometime between midembryogenesis and the beginning of the first instar larvae stage. This finding raises the question of how the male pathway is then selected in the XY embryo and suggests that the function of the Y-linked male determining factor is essential in early male sex determination.
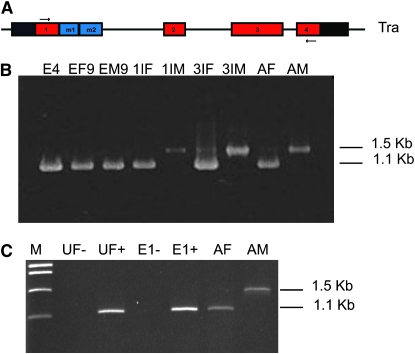
Figure 3.—
Analysis of the expression of Lctra over development by RT–PCR. (A) Position of primers in exons 1 and 4 of Lctra, designed to amplify products of different sizes for female and male transcripts. (B and C) RT–PCR amplification of Lctra on total RNA obtained from different developmental stages of L. cuprina. Male transcripts are 1.5 kb whereas female transcripts are 1.1 kb in size. Stages in B are: E4 early embryos of mixed sexes at 4 hr of development, EF9 midstage female embryos at 9 hr, EM9 midstage male embryos at 9 hr, 1IF female first instar larvae, 1IM male first instar larvae, 3IF female third instar larvae, 3IM male third instar larvae, AF female adults, and AM male adults. In panel C, RNA was isolated from unfertilized eggs (UF) or fertilized precellular embryos at 30–60 min of development (E1) and cDNA prepared either with (+) or without (−) reverse transcriptase.
The Lctra gene is essential for female development:
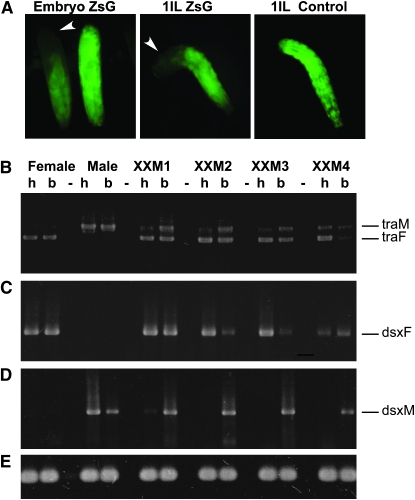
In tephritid species and in Drosophila, tra is an essential gene for female development. Injection of tra dsRNA into early C. capitata and B. oleae embryos led to the development of masculinized XX individuals (Pane et al. 2002; Lagos et al. 2007). In these species TRA is proposed to have an autoregulatory function, which could explain why a transient reduction in RNA levels so effectively blocked female development. Therefore, we next investigated whether the function of tra was conserved in L. cuprina. To test this we compromised Lctra gene expression during early development using the RNA interference technique. A 950-bp Lctra dsRNA was injected into the posterior end of preblastoderm embryos obtained from a cross of line HS14 transgenic males with wild-type virgin females. In this way, XX first instar larvae that developed from injected embryos were identified by fluorescence and readily separated from nonfluorescent XY siblings. In a control experiment, 116 fluorescent larvae were selected from uninjected embryos. One hundred six of them developed into adults and all were female. Of the 89 XX adults that developed from injected embryos, 68 showed evidence of sex reversal (Figure 4 and Table 1). Seventy-two percent developed with external male genitalia but with female interocular width. Two percent had both male genitalia and male interocular width. Twenty-six percent appeared to be phenotypically normal females. Of the 64 XY adults that developed from injected embryos, 100% showed a normal male phenotype. It is of note that Pane et al. (2002) observed that injections of Cctra dsRNA into the posterior end of embryos led to the development of some adult XX individuals with male genitalia but heads with a female bristle pattern. As a control of the RNAi technique, dsRNA directed against ZsGreen was injected into the posterior end of preblastoderm embryos of a cross between line 56 males and wild-type virgin females (Figure 5A). Line 56 contains an autosomal single insertion of the Lchsp83-ZsGreen marker. Fluorescence from the ZsGreen marker is observed in both males and females from midembryogenesis onward. Figure 5A shows loss of fluorescence in the posterior end but not in the anterior end of injected late embryos and first instar larvae. The knockdown of ZsGreen is only transient as injected individuals show complete fluorescence by the stage of third instar larvae (data not shown). The situation observed for ZsGreen RNAi correlates with that observed for tra RNAi, where the effect of gene knockdown is mostly observed in the posterior end but not in the anterior end of the animal. This effect can be attributed to the failure of the dsRNA solution to diffuse to the opposite end of the long Lucilia embryo, causing a knockdown of gene expression only in the rear. The control RNAi experiment also suggests that the strong transformation of XX individuals into males is caused by a transient knockdown of tra during early development.
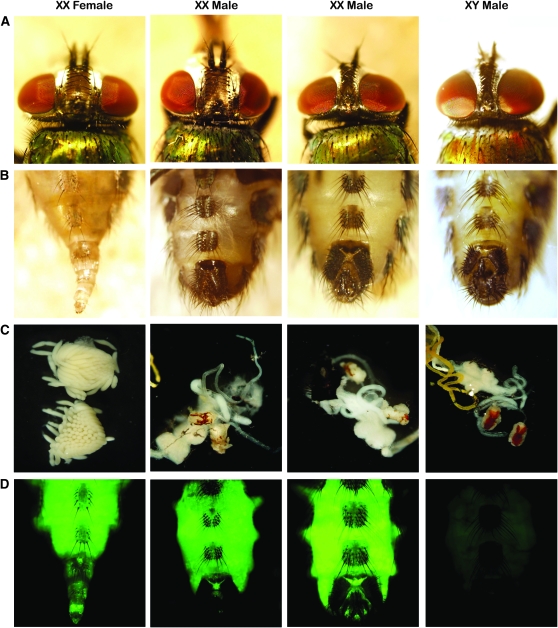
Figure 4.—
Injection of Lctra dsRNA into the posterior end of preblastoderm embryos causes female-to-male sex reversal. Phenotypically wild-type females (XX female, A and B) can be recognized from males by a wider interocular distance in the head and by the presence of an ovipositor in the genitalia, whereas males (XY male, A and B) have a pigmented copulatory apparatus with characteristic clasps. Internally, adult females present two ovaries with a large number of eggs (XX female, C) and males present red pigmented testes (XY male, C). Only XX individuals carry the X-linked ZsGreen marker gene. Most frequently, transformed XX males show male genitalia and gonads but conserve the characteristic female head (XX male 1, A, B, C, and D) while a small percentage of the injected XX individuals develop as completely transformed males (XX male 4, A, B, C, and D).
TABLE 1.
Injection of Lctra dsRNA into embryos blocks female development
| Genotype | Percentage (N) | Interocular width | Genitalia | Gonads |
|---|---|---|---|---|
| XX | 72 (64) | F | M | Testes |
| XX | 2 (2) | M | M | Testes |
| XX | 26 (23) | F | F | Ovaries |
| XY | 100 (65) | M | M | Testes |
Figure 5.—
Analysis of the splicing patterns of Lctra and Lcdsx in XX transformed males and wild-type individuals. (A) Control for the RNAi technique; embryos from a cross between Lchsp83-ZsGreen males and wild-type females were injected with dsRNA for ZsGreen. Fluorescence is lost in the posterior end of injected embryos and larvae but not in the anterior end. (B) RT–PCR amplification with Lctra-specific primers on total RNA isolated from heads (h) and bodies (thorax plus abdomen) (b) of transformed XX males presenting male genitalia and female head (XXM1, XXM2, and XXM3), a completely transformed XX male (XXM4) and of a wild-type female and male. The primers used amplify different size products for male (traM) and female transcripts (traF). (C and D) RT–PCR amplification with Lcdsx female and male specific primers, using the same total RNA samples as in B. The sex-specific amplification products are labeled dsxF for female and dsxM for male, respectively. (E) RT–PCR control using α-tubulin-specific primers. The RNAi knockdown of Lctra in XX males sets Lctra splicing in the male mode in the posterior end of injected individuals, which in turn changes the splicing pattern of Lcdsx from the female to the male form.
Some of the XX flies with external male genitalia were dissected and found to have testes of normal morphology (Figure 4). Since the L. cuprina Y chromosome appears to be devoid of genes that encode fertility factors (Bedo and Foster 1985), we wondered whether any of the transformed XX flies were fertile. Out of 10 crosses between single XX transformed males and wild-type virgin females, 1 was fertile. As anticipated, all of the offspring from this cross developed into females. These results show that tra is essential for female development in L. cuprina and that the Y chromosome is not essential for fertility. Since a transient reduction in female Lctra RNA in Lucilia embryos causes a strong sex reversal in adult flies, it is likely that the role of Lctra in selecting the sexual fate occurs early in development.
Regulation of Lctra activity:
A feature of the tephritid tra genes is the presence of multiple putative TRA/TRA2 binding sites within the male-specific exons and flanking introns (Ruiz et al. 2007) (Figure S1). A bioinformatics search of the Lctra gene sequence identified 6 putative TRA/TRA2 sites that were a perfect match to the consensus (T/A)(C/A)(A/T)(A/T)CAATCAACA (Figure 1). Five of the sites were clustered over a 233-bp region that is 2063 bp downstream of the female splice donor site but 341 bp upstream of the intron 1 splice acceptor site. The sixth TRA/TRA2 site is located within the M2 male-specific exon sequence, 70 nt upstream of the male-specific splice donor site. That only one TRA/TRA2 site is in a male exon is in contrast to the Cctra, Botra, and Aotra genes where most of the TRA/TRA2 sites are in the male-specific exons (Figure S1). The presence of these multiple TRA/TRA2 sites in Lctra unspliced transcripts suggests a potential for autoregulation of Lctra RNA splicing.
To investigate how tra activity is regulated in L. cuprina we tested for the presence of sex-specific transcripts of tra and its downstream target gene dsx in transformed XX males. Transcripts from the Lcdsx gene are sex-specifically spliced in a similar fashion as the M. domestica dsx RNAs (M. C. Concha and M. J. Scott, unpublished results). Total RNA was isolated from dissected heads and remaining bodies (thorax plus abdomen) of transformed XX individuals (XXM1–4). Primers were designed that amplified different size PCR products for the male and female forms of the Lctra and Lcdsx RNAs. The results of the RT–PCR analysis are shown in Figure 5, B–D. Both male and female forms of Lctra were detected in RNA from bodies but a majority of the female form was detected in head RNA. XXM1, XXM2, and XXM3 presented female interocular width and male external genitalia, while XXM4 was a completely transformed XX male. In this individual, head RNA seems to contain a slightly more abundant fraction of male tra than in the other tested flies. In the case of the dsx gene, both male and female forms of Lcdsx were detected in bodies but only the female form was detected in heads of all tested individuals. In XXM4, female Lcdsx is present in scarce amounts in both head and body RNA. These results show that knockdown of female Lctra expression in early development causes the activation of the male mode of splicing of Lctra and a change from the female to the male mode of splicing in Lcdsx.
Isolation and characterization of the L.
cuprina tra2 homolog: To obtain a more complete understanding of sex determination in L. cuprina we isolated the tra2 homolog. Using a similar strategy to that used to isolate Lctra, an amplification product of the expected size was obtained from cDNA templates and sequence comparisons confirmed that we had isolated the L. cuprina homolog of tra2, which was designated Lctra2. To obtain full-length cDNA sequences 5′ and 3′ RACE was performed with adult male and female RNA templates. The sequences were assembled into a unique 1499-nt transcript.
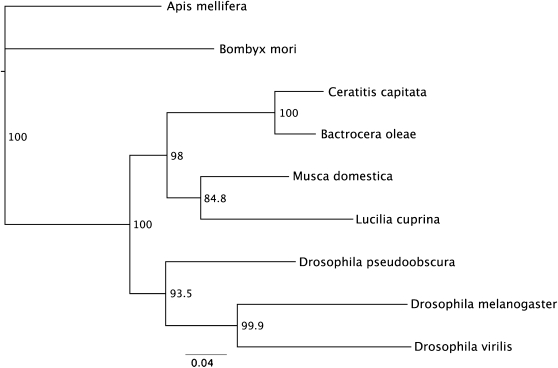
In D. melanogaster the tra2 gene encodes a protein with an RNA-recognition motif (RRM), flanked by two arginine-rich/serine-rich regions (RS domains), which mediate protein–protein interactions. Three tra2 transcripts arise due to alternative splicing and transcription start sites (Amrein et al. 1990; Mattox et al. 1990). In the medfly C. capitata and in the housefly M. domestica only a single tra2 transcript is detected (Burghardt et al. 2005). As in the latter insects, only a single Lctra2 transcript was detected in the RACE experiments and in RT–PCR performed on RNA isolated from different developmental stages (data not shown). A multiple sequence alignment was performed using CLUSTALW with amino sequences of the TRA2 proteins from L. cuprina, C. capitata (GenBank EU437408), B. oleae (GenBank AJ547623), M. domestica (GenBank AAW34233), and D. melanogaster (GenBank AAA62771). LcTRA2 is very similar to TRA2 from other Diptera (Figure 6). As expected, the strongest conservation is in the RRM domain and in the linker motif that immediately follows the RRM domain. The linker motif is highly conserved among TRA2 homologs and is considered a signature motif of TRA2 proteins (Dauwalder et al. 1996). With the notable exception of Drosophila TRA2, there are also regions of high sequence similarity in the arginine- and serine-rich amino and carboxyl terminal domains. To investigate the phylogenetic relationships of LCTRA2 to other insect TRA2 proteins a phylogenetic analysis was performed using the TRA2 sequences from the species mentioned above and additionally D. virilis (GenBank AAB58114), D. pseudoobscura (GenBank XP_001360605), A. mellifera (GenBank:XP_001121070), and B. mori (GenBank AAX47001). The results obtained using a neighbor-joining algorithm are shown in Figure 7. LCTRA2 clusters with M. domestica TRA2 as expected since both species belong to Calyptratae, a subsection of Schizophora in the insect order Diptera. L. cuprina and M. domestica TRA2 proteins are more closely related to the homologs from tephritid species than Drosophila species, which is consistent with previous analysis (Gomulski et al. 2008). Both tephritid and Drosophila species belong to the Acalptratae subsection of Schizophora. However, it has been hypothesized that Calliphorids are more closely related to Tephritids than Drosophilids (Crampton 1944).
Figure 6.—
Multiple sequence alignment of TRA2 proteins in L. cuprina, M. domestica, C. capitata, B. oleae, and D. melanogaster. Identical amino acids are shaded in black while similar amino acids are shaded in gray. Arrows indicate the conserved motifs that were the basis of the degenerate primers that were used to obtain Lctra2 DNA sequence. The RNA recognition motif (RRM) is underlined in red. RNP-1 and RNP-2 are the highly conserved ribonucleoprotein identifier sequences found in RRM motifs. The TRA2 proteins of all these insects are highly conserved in the RRM as well as in the flanking RS domains.
Figure 7.—
Neighbor-joining tree of insect TRA2 amino acid sequences. The numbers represent bootstrap support values from 1000 replicates. The scale represents the mean character distance.
DISCUSSION
In the present study we have isolated and characterized the transformer gene from the pest insect species L. cuprina. As in other Diptera, Lctra is alternatively spliced such that only the female transcript codes for a full-length protein. Interestingly, LcTRA presents little sequence similarity with the TRA proteins of the Drosophila species and shows a high degree of homology with TRA of Tephritidae. Indeed, as in tephritid insects, Lctra is essential for female development and its activity is autoregulated, suggesting that in L. cuprina tra is likely the top switch of the female development cascade. Since L. cuprina is a member of the Calliphoridae family of insects, this regulatory system in which tra acts as the female master switch, is not confined to Tephritidae but appears to have a much wider distribution among Diptera. We have also isolated the transformer2 gene and found that it is not alternatively spliced, giving rise to a single transcript in males and females, as is the case in M. domestica tra2 and C. capitata tra2, and to the contrary of D. melanogaster, which presents several forms of tra2 in somatic and germline cells. Overall, both Lctra and Lctra2 resemble more closely its homologs in the tephritid species of insects than in D. melanogaster.
Regulation of the Lctra gene:
The organization of the Lctra gene is similar to that of the C. capitata, B. oleae, and A. obliqua tra genes but is simpler. In the tephritid species, the tra gene contains three or more male exons and the male splicing patterns are complex, particularly in A. obliqua. In L. cuprina, the difference between the female and major male splicing patterns is simply that females chose the first donor site in intron 1 and males chose a downstream site. Both sexes splice to the same acceptor site. The presence of six putative TRA/TRA2 binding sites in the regulated first intron suggests that Lctra RNA splicing is autoregulated. This hypothesis is supported by the results from the RNAi experiments as injection of Lctra dsRNA lead to XX individuals switching from the female to male form of Lctra splicing.
Regulation of Lctra RNA splicing could be achieved by the binding of TRA/TRA2 to the female primary transcript to either block the use of the male splice donor site or activate the use of the female donor site. As the female splice donor site (UG|GUAAUU) is a better match to the Drosophila consensus (AG|GUAAGU) (Weir and Rice 2004) than the male donor site (GU|GUGAGU) and most of the TRA/TRA2 binding sites are closer to the male donor site than the female site, it is more likely that LcTRA could regulate its own splicing by inhibiting the use of the male splice donor site. This explanation is consistent with the study on C. capitata tra presented by Pane et al. (2002), who proposed that TRA negatively regulates the male mode of splicing in females. The fact that the amino terminal domain of TRA is well conserved in L. cuprina and the tephritid species but absent in Drosophila TRA is consistent with the suggestion that this domain is involved in TRA autoregulation.
Ruiz et al. (2007) identified putative RBP1 and TRA2-ISS binding sites in the regulated first female intron of the C. capitata, B. oleae, and A. obliqua tra genes. They proposed that RBP1 and TRA2 somehow combine with the TRA/TRA2 complex to repress the male-splicing pattern. We searched the Lctra gene sequence but found only one putative RBP1 type A site (TCAACTTTTA) and one TRA2-ISS site (CAAGA) in the female first intron. Although there were several matches to the RBP1 type B site (ATCYNNA), there were no more than expected for a short AT-rich sequence in the 2728-bp female first intron that is 71% AT. Thus it would seem unlikely that this model could explain how splicing of the Lctra gene is regulated.
Clearly a high priority is to develop a splicing assay system to determine if sequence elements other than the identified putative TRA/TRA2 sites are required for sex-specific splicing. It would also be of interest to determine if the spatial arrangement of the TRA/TRA2 sites relative to the major male splice donor site is important for regulated splicing. For example, if TRA/TRA2 bound to the male exon interacted with TRA/TRA2 bound to sites in the intron this could lead to looping out of the intervening sequence. As a consequence the male donor site would be inaccessible to the splicing apparatus or the female donor site might be brought into close spatial proximity with the acceptor site.
Lctra function and expression:
The RNAi knockdown of female Lctra mRNA in early embryos caused the selection of the male splicing mode of Lctra and Lcdsx, particularly in the bodies of adults derived from embryos injected in the posterior end. The female splice variants of these genes were also found in the heads and to a lesser extent in the bodies of adult transformed XX males. The presence of the female forms in bodies is most likely because this included thoracic tissue derived from cells from the anterior half of the injected embryos. Female Lctra and Lcdsx transcripts in heads could be because the relatively rapid development and long length of L. cuprina embryos (Gregor et al. 2005) would have limited diffusion of the dsRNA to the anterior end of the embryo, furthest from the site of injection. Consequently, it would be anticipated that LcTRA protein levels would be low at the posterior end and high at the anterior end of injected embryos, as is observed in the ZsGreen control experiments. These observations suggest that there may be a threshold of LcTRA protein level to be attained to activate the female splicing of Lctra and Lcdsx.
The availability of a transgenic line expressing a strong X-linked ZsGreen marker gene allowed the identification of females and males from midembryogenesis onward. Using this system we found that both sexes contain female Lctra mRNA during embryonic stages and that the male form of Lctra is expressed in males from the first instar larvae stage onward. Moreover, the presence of female Lctra mRNA in unfertilized eggs and in very early precellular embryos of mixed sex showed that there is maternal inheritance of female Lctra RNA in embryos of both sexes. These results raise the question of how the female pathway of development is initiated in XX individuals and how it is prevented in XY individuals. Our results suggest that female development is established during embryogenesis by a maternal pool of female Lctra mRNA that is translated into protein in zygotes to initiate an autoregulatory loop, thus promoting its own splicing and activity. This system allows LcTRA to accumulate in female embryos over a certain threshold required for its maintenance and for the activation of the female RNA splicing mode of its target gene dsx, thereby promoting female differentiation. It should be noted that it remains to be shown that maternal Lctra RNA is translated into protein in developing embryos. In males, this autoregulation would be blocked by the activity of M, thereby selecting the male pathway of development by default. Pane et al. (2002) proposed that in C. capitata males, M blocks Cctra autoregulation by directly inhibiting splicing of Cctra transcripts or by acting on the CcTRA protein. Alternatively, M could transiently inhibit Cctra transcription or translation of Cctra transcripts. Similar models could explain how M blocks Lctra autoregulation in male L. cuprina embryos. However, it would seem unlikely that M acts to inhibit the translation of maternal Lctra RNA, thereby preventing the initiation of the autoregulatory loop. Only the female form of Lctra RNA was detected in males at midembryogenesis, well after the onset of general zygotic transcription. If M acts solely to block translation of maternal Lctra transcripts, it would be anticipated that some male Lctra transcripts would be detectable by the midembryo stage. Thus, while it is possible that M could inhibit translation of Lctra RNA, it is more likely that M regulates Lctra at the splicing or, temporarily, at the transcriptional level thus preventing the accumulation of female LcTRA protein over a threshold required for the maintenance of the autoregulatory loop.
The genetic control of sex determination in L. cuprina presents many similarities to the system used by C. capitata. Both species of flies use a dominant Y-linked male determining gene to control sexual development and in both the tra gene functions as a master switch that controls the female fate by establishing an autoregulatory loop (Pane et al. 2002). Moreover, both species present maternal inheritance of female tra transcripts in embryos of both sexes further supporting its conserved role in the initiation of tra autoregulation. The use of alternative splice donor sites and exon skipping in Lctra and Cctra is in contrast to the Drosophila tra 3′ alternative splicing mechanism. Furthermore, in both species of insects RNAi knockdown of tra results in fertile XX males, indicating that in contrast to Drosophila, the Y chromosome is not essential for fertility in these flies. Sex determination in L. cuprina also resembles that of its closer relative M. domestica. In the latter, female sex determination is controlled by the F gene, whose activity is maintained by an autoregulatory loop and is strongly dependent on maternally inherited F product (Dübendorfer and Hediger 1998). Although the isolation of the M. domestica tra gene has not yet been published it has been proposed that F corresponds to tra (Burghardt et al. 2005). Moreover, RNAi mediated knockdown of tra2 in Musca embryos results in transformed XX males that are fertile. Consequently, this sex determination system where tra acts as a master switch may be more widely spread to include other groups such as the Muscidae and Calliphoridae.
Interestingly, while transient RNAi knockdown of Lctra resulted in a high proportion of XX individuals developing male genitalia, only 1 in 10 tested was fertile. A possible explanation for this low fertility rate is that although the Lucilia Y chromosome does not contain essential fertility genes, it may contain genes involved in male fitness and performance in mating. Alternatively, since the heads of transformed XX individuals contained mostly the female forms of Lctra and Lcdsx transcripts, the sexual courtship behavior may have been less than optimal. It is well documented that the dsx and fruitless (fru) genes are involved in sexual courtship behavior in Drosophila and that the control of this behavior resides in specific areas of the brain (Shirangi et al. 2006). Splicing of fru transcripts is also regulated by TRA/TRA2 in Drosophila (Heinrichs et al. 1998) and C. capitata (Salvemini et al. 2009). It would be of interest to study courtship behavior and male fitness in a transgenic line of Lucilia expressing a stable inducible tra dsRNA construct.
Evolution of Lctra and sex determining systems:
From a detailed comparative morphological study of male terminalia, Crampton (1944) found that within the Diptera order, the Calyptratae group, to which L. cuprina belongs, shared many features with some members of the Acalyptratae, including species from the superfamily Tephritoidea. That Calyptratae may be more similar to Tephritidae (Acalyptratae) than to Drosophilidae (Acalyptratae) has been supported by studies of the white (Gomulski et al. 2001), glucose-6-phosphate dehydrogenase (Soto-Adames et al. 1994), alcohol dehydrogenase (Brogna et al. 2001), and tra2 genes (Gomulski et al. 2008). Our results strongly support this evolutionary hypothesis.
It will be of interest to determine whether tra acts as a master switch of sex determination in other important Calyptratae species such as M. domestica, tsetse flies, and the screwworm fly Cochliomyia hominivorax. The latter species have been the subject of major control efforts using the sterile insect technique (Krafsur 1998). Since these species are more closely related to L. cuprina than tephritid species, we would predict tra function and regulation would be conserved in at least some other Calyptratae.
Genetic control of L. cuprina:
L. cuprina is an economically important insect species that constitutes a major pest to the sheep industries in Australia and New Zealand (Beck et al. 1985; Heath and Bishop 2006). This insect has long been considered to be a good target for a genetic control program (Scott et al. 2004). Indeed, considerable effort was made to develop a “field female killing system” that was shown to be effective in reducing a L. cuprina island population in a large field trial (Davidson 1989; Foster et al. 1991). The isolation of the Lctra gene will facilitate the development of genetically modified strains that would have certain advantages for genetic control programs. For example, induction of expression of Lctra double-stranded RNA could lead to the development of a male population of flies, which could be sterilized by radiation before field release. We have recently isolated L. cuprina heat inducible gene promoters for this purpose. Alternatively, the regulated first intron of Lctra could be used to control the sex-specific expression of a tetracycline-repressible lethal gene, as recently developed for C. capitata (Fu et al. 2007).
Acknowledgments
We thank Anja Schiemann for performing qRT–PCR analysis, Fang Li for RT–PCR of RNA from unfertilized eggs, Brandi-lee Carey for maintenance of Lucilia cuprina cultures, Esther Belikoff for embryo collection and making beveled quartz glass needles for micro-injection, Simon Hills for performing a phylogenetic analysis of insect TRA/TRA2 protein sequences, and Helen Fitzsimons and Maria Imschenetzky for comments on the manuscript. This research has been supported by contract EC456 from Australian Wool Innovation.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.100982/DC1.
References
- Amrein, H., T. Maniatis and R. Nöthiger, 1990. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. EMBO J. 9 3619–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., B. Moir and T. Meppem, 1985. The cost of parasites to the Australian sheep industry. Q. Rev. Rural Econ. 7 336–343. [Google Scholar]
- Bedo, D. G., and G. G. Foster, 1985. Cytogenetic mapping of the male-determining region of Lucilia cuprina (Diptera: Calliphoridae). Chromosoma 92 344–350. [Google Scholar]
- Brogna, S., P. V. Benos, G. Gasperi and C. Savakis, 2001. The Drosophila alcohol dehydrogenase gene may have evolved independently of the functionally homologous medfly, olive fly, and flesh fly genes. Mol. Biol. Evol. 18 322–329. [DOI] [PubMed] [Google Scholar]
- Burghardt, G., M. Hediger, C. Siegenthaler, M. Moser, A. Dübendorfer et al., 2005. The transformer2 gene in Musca domestica is required for selecting and maintaining the female pathway of development. Dev. Genes Evol. 215 165–176. [DOI] [PubMed] [Google Scholar]
- Cho, S., Z. Y. Huang and J. Zhang, 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T. W., 1993. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 9 385–390. [DOI] [PubMed] [Google Scholar]
- Crampton, G. C., 1944. A comparative morphological study of the terminalia of male calyptrate cyclorrhaphous diptera and their acalyptrate relatives. Bull. Brooklyn Entomol. Soc. 34 1–34. [Google Scholar]
- Dauwalder, B., F. Amaya-Manzanares and W. Mattox, 1996. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc. Natl. Acad. Sci. USA 93 9004–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, S., 1989. Sheep blowfly control by genetic sabotage. Rural Res. 145 19–24. [Google Scholar]
- Dübendorfer, A., and M. Hediger, 1998. The female-determining gene F of the housefly, Musca domestica, acts maternally to regulate its own zygotic activity. Genetics 150 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dübendorfer, A., M. Hediger, G. Burghardt and D. Bopp, 2002. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int. J. Dev. Biol. 46 75–79. [PubMed] [Google Scholar]
- Erickson, J. W., and J. J. Quintero, 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5 e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, G. G., G. L. Weller and G. M. Clarke, 1991. Male crossing over and genetic sexing systems in the Australian sheep blowfly Lucilia cuprina. Heredity 67 365–371. [DOI] [PubMed] [Google Scholar]
- Fu, G., K. C. Condon, M. J. Epton, P. Gong, L. Jin et al., 2007. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol. 25 353–357. [DOI] [PubMed] [Google Scholar]
- Gomulski, L. M., R. J. Pitts, S. Costa, G. Saccone, C. Torti et al., 2001. Genomic organization and characterization of the white locus of the Mediterranean fruitfly, Ceratitis capitata. Genetics 157 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulski, L. M., G. Dimopoulos, Z. Xi, M. B. Soares, M. F. Bonaldo et al., 2008. Gene discovery in an invasive tephritid model pest species, the Mediterranean fruit fly, Ceratitis capitata. BMC Genomics 9 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor, T., W. Bialek, R. R. van Steveninck, D. W. Tank and E. F. Wieschaus, 2005. Diffusion and scaling during early embryonic pattern formation. Proc. Natl. Acad. Sci. USA 102 18403–18407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, A. C., and D. M. Bishop, 2006. Flystrike in New Zealand: an overview based on a 16-year study, following the introduction and dispersal of the Australian sheep blowfly, Lucilia cuprina Wiedemann (Diptera: Calliphoridae). Vet. Parasitol. 137 333–344. [DOI] [PubMed] [Google Scholar]
- Hediger, M., G. Burghardt, C. Siegenthaler, N. Buser, D. Hilfiker-Kleiner et al., 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 214 29–42. [DOI] [PubMed] [Google Scholar]
- Heinrichs, V., L. C. Ryner and B. S. Baker, 1998. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 18 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur, E. S., 1998. Sterile insect technique for suppressing and eradicating insect population: 55 years and counting. J. Agric. Entomol. 15 303–317. [Google Scholar]
- Kuhn, S., V. Sievert and W. Traut, 2000. The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome 43 1011–1020. [DOI] [PubMed] [Google Scholar]
- Lagos, D., M. F. Ruiz, L. Sanchez and K. Komitopoulou, 2005. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348 111–121. [DOI] [PubMed] [Google Scholar]
- Lagos, D., M. Koukidou, C. Savakis and K. Komitopoulou, 2007. The transformer gene in Bactrocera oleae: the genetic switch that determines its sex fate. Insect Mol. Biol. 16 221–230. [DOI] [PubMed] [Google Scholar]
- Li, F., A. H. Schiemann and M. J. Scott, 2008. Incorporation of the noncoding roX RNAs alters the chromatin-binding specificity of the Drosophila MSL1/MSL2 complex. Mol. Cell. Biol. 28 1252–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, I., and B. S. Baker, 1998. The evolutionary dynamics of sex determination. Science 281 1990–1994. [DOI] [PubMed] [Google Scholar]
- Mattox, W., M. J. Palmer and B. S. Baker, 1990. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 4 789–805. [DOI] [PubMed] [Google Scholar]
- Meise, M., D. Hilfiker-Kleiner, A. Dübendorfer, C. Brunner, R. Nöthiger et al., 1998. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development 125 1487–1494. [DOI] [PubMed] [Google Scholar]
- Muller-Holtkamp, F., 1995. The Sex-lethal gene homologue in Chrysomya rufifacies is highly conserved in sequence and exon-intron organization. J. Mol. Evol. 41 467–477. [DOI] [PubMed] [Google Scholar]
- Pane, A., M. Salvemini, P. Delli Bovi, C. Polito and G. Saccone, 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129 3715–3725. [DOI] [PubMed] [Google Scholar]
- Pane, A., A. De Simone, G. Saccone and C. Polito, 2005. Evolutionary conservation of Ceratitis capitata transformer gene function. Genetics 171 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva, L. O., and L. Sanchez, 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 67 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, M. F., R. N. Stefani, R. O. Mascarenhas, A. L. Perondini, D. Selivon et al., 2005. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae). Genetics 171 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, M. F., A. Milano, M. Salvemini, J. M. Eirin-Lopez, A. L. Perondini et al., 2007. The gene transformer of anastrepha fruit flies (Diptera, tephritidae) and its evolution in insects. PLoS ONE 2 e1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, G., I. Peluso, D. Artiaco, E. Giordano, D. Bopp et al., 1998. The Ceratitis capitata homologue of the Drosophila sex-determining gene sex-lethal is structurally conserved, but not sex-specifically regulated. Development 125 1495–1500. [DOI] [PubMed] [Google Scholar]
- Saccone, G., A. Pane and L. C. Polito, 2002. Sex determination in flies, fruitflies and butterflies. Genetica 116 15–23. [DOI] [PubMed] [Google Scholar]
- Salvemini, M., M. Robertson, B. Aronson, P. Atkinson, L. C. Polito et al., 2009. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53 109–120. [DOI] [PubMed] [Google Scholar]
- Sanchez, L., 2008. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52 837–856. [DOI] [PubMed] [Google Scholar]
- Scali, C., F. Catteruccia, Q. Li and A. Crisanti, 2005. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J. Exp. Biol. 208 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütt, C., and R. Nöthiger, 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127 667–677. [DOI] [PubMed] [Google Scholar]
- Scott, M. J., J. C. Heinrich and X. Li, 2004. Progress towards the development of a transgenic strain of the Australian sheep blowfly (Lucilia cuprina) suitable for a male-only sterile release program. Insect Biochem. Mol. Biol. 34 185–192. [DOI] [PubMed] [Google Scholar]
- Shearman, D. C., 2002. The evolution of sex determination systems in dipteran insects other than Drosophila. Genetica 116 25–43. [DOI] [PubMed] [Google Scholar]
- Shearman, D. C., and M. Frommer, 1998. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7 355–366. [DOI] [PubMed] [Google Scholar]
- Shirangi, T. R., B. J. Taylor and M. McKeown, 2006. A double-switch system regulates male courtship behavior in male and female Drosophila melanogaster. Nat. Genet. 38 1435–1439. [DOI] [PubMed] [Google Scholar]
- Siera, S. G., and T. W. Cline, 2008. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics 180 1963–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert, V., S. Kuhn and W. Traut, 1997. Expression of the sex determining cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome 40 211–214. [DOI] [PubMed] [Google Scholar]
- Sosnowski, B. A., J. M. Belote and M. McKeown, 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58 449–459. [DOI] [PubMed] [Google Scholar]
- Soto-Adames, F. N., H. M. Robertson and S. H. Berlocher, 1994. Phylogenetic utility of partial DNA sequences of G6pdh at different taxonomic levels in Hexapoda with emphasis on Diptera. Ann. Entomol. Soc. Am. 87 723–736. [Google Scholar]
- Suzuki, M. G., F. Ohbayashi, K. Mita and T. Shimada, 2001. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 31 1201–1211. [DOI] [PubMed] [Google Scholar]
- Weir, M., and M. Rice, 2004. Ordered partitioning reveals extended splice-site consensus information. Genome Res. 14 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, A. S., 1995. Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17 71–77. [DOI] [PubMed] [Google Scholar]